Abstract
The ~25 N-terminal “HAfp” residues of the HA2 subunit of the influenza virus hemagglutinin protein are critical for fusion between the viral and endosomal membranes at low pH. Earlier studies of HAfp in detergent support (1) N-helix/turn/C-helix structure at pH 5 with open interhelical geometry and N-helix/turn/C-coil structure at pH 7; or (2) N-helix/turn/C-helix at both pHs with closed interhelical geometry. These different structures led to very different models of HAfp membrane location and different models of catalysis of membrane fusion by HAfp. In this study, the interhelical geometry of membrane-associated HAfp is probed by solid-state NMR. The data are well-fitted to a population mixture of closed and semiclosed structures. The two structures have similar interhelical geometries and are planar with hydrophobic and hydrophilic faces. The different structures of HAfp in detergent vs membrane could be due to the differences in interaction with the curved micelle vs flat membrane with better geometric matching between the closed and semiclosed structures and the membrane. The higher fusogenicity of longer sequences and low pH is correlated with hydrophobic surface area and consequent increased membrane perturbation.
Influenza virus is enveloped by a membrane which contains the hemagglutinin (HA) protein composed of the HA1 and HA2 subunits.4 HA2 is a monotopic integral membrane protein, and HA1 is bound to the extraviral region of HA2. Infection of a host epithelial cell begins with HA1 binding to a cellular sialic acid receptor, and this binding triggers virion endocytosis. Endosomal pH is reduced to 5–6 via cell physiology, and deprotonation of HA2 acidic groups leads to refolding of HA2. The ~25 N-terminal “fusion peptide” (HAfp) residues of HA2 are highly conserved and important in fusion.5 The HAfp becomes exposed after HA2 refolding and binds to a membrane.6 Vesicle fusion is induced both by HAfp sequences as well as by larger HA2 constructs which include the HAfp, and there is greater fusion at acidic pH.7 There have been several HAfp structures in detergent-rich media at different pH's and effort to correlate pH-dependent structural differences with membrane fusion.1,2 However, there are large differences among the detergent structures so that structure/function correlation is unclear. The present work provides critical information about the HAfp structure in membrane. There are significant differences with the detergent structures, and the data support a role for HAfp hydrophobic surface area in fusion.
One structure/function model is based on the 20-residue HA3fp20 peptide (GLFGAIAGFIENGWEGMIDG) from the H3 viral subtype. The structures in detergent are N-helix/turn/C-helix at pH 5 and N-helix/turn/C-coil at pH 7.1 The pH 5 structure is “open” as evidenced by the oblique interhelical angle (Figure 1A). EPR data were interpreted to support insertion of the N-helix to the membrane center at pH 5 with shallower insertion at pH 7. Relative to pH 7, greater fusion at pH 5 was explained by C-coil to C-helix change with formation of an open structure with a hydrophobic interhelical pocket and deep N-helix insertion. The pocket and insertion result in membrane perturbation and fusion.8 A different fusion model was developed for the 23-residue HA1fp23 peptide (GLFGAIAGFIEGGWTGMIDGWYG) from the H1 viral subtype.2 Relative to HA3fp20, HA1fp23 contains G12N, E15T, and additional WYG C-terminal residues. Unlike HA3fp20 which shows pH-dependent structure and open structure at pH 5, HA1fp23 has a “closed” N-helix/turn/C-helix structure in detergent at both pH 4 and 7 with tightly packed antiparallel N- and C-terminal helices (Figure 1B). Formation of closed HA1fp23 vs open HA3fp20 structure was attributed to the respective presence vs absence of C-terminal WYG.9,10 The closed structure is amphipathic and would reasonably lie on the membrane surface and potentially induce membrane perturbation. HA1fp23 in detergent at pH 4 also has a ~0.2 fraction of open structure with fast closed/open exchange.11
Figure 1.
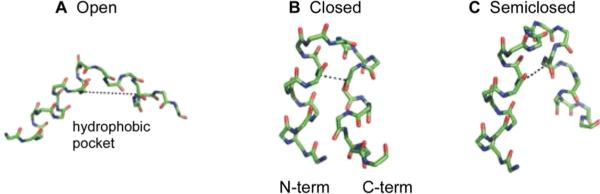
Backbone structural models of (A) open HA3fp20, (B) closed HA1fp23, and (C) semiclosed HA1fp23.1–3 C, N, and O atoms are respectively represented by green, blue, and red vertices. The dashed lines are between F9 N and G16 CO with distances ro = 11.5 Å, rc = 3.9 Å, and rs = 5.5 Å.
The different functional models are based on different structures in detergent and motivate the present work to understand the HAfp structure in membrane. HAfp induces fusion of membranes but not detergent micelles, so the membrane structures are more relevant for function. The present work builds on earlier solid-state NMR (SSNMR) studies of HA3fp20 in membranes showing N-helix/turn/C-helix structure at both pH 5 and 7, i.e., no C-coil structure at pH 7 as is found in detergent.12 The structure was observed in both fluid- and gel-phase membranes. At pH 5, the interhelical separation of HA3fp20 in membrane is much less than for a HA3fp20 open structure in detergent.3 The separation is consistent with a mixture of populations of closed structure and a somewhat different semiclosed structure, and therefore supports different HA3fp20 structures in membrane vs detergent (Figure 1C). Both the closed and semiclosed structures have a N-helix from residues 1–11 and C-helix from residues 14–22 and only differ in the residue 12/13 turn (Table S7).
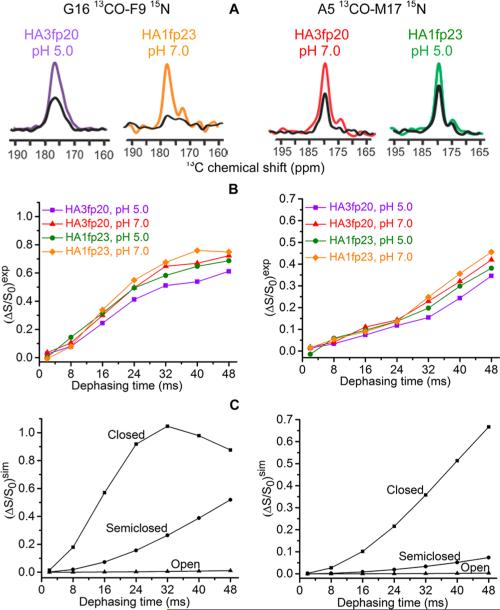
The present study focuses on HA3fp20 and HA1fp23 in membrane: (1) to understand structural dependence on viral subtype amino acid differences, sequence length, and pH; and (2) to correlate structural features with fusion. Earlier work only showed N-helix/turn/C-helix structure for HA3fp20 in membrane at low and neutral pH, so the present work focuses on interhelical separation via rotational-echo double-resonance (REDOR) SSNMR measurement of the dipolar couplings (d's) of samples with labeled (lab) G16 13CO-F9 15N or A5 13CO-M17 15N spin pairs. The d depends on the 13CO–15N distance (r) as d(Hz) = 3066/r(Å)3. A sample is at pH 5 (fusion pH in the endosome) or pH 7. A sample contains membrane-associated HA3fp20 or HA1fp23 with one labeling scheme (SI).13,14 S0 and S1 13C REDOR spectra are acquired as a function of dephasing time (τ) and the S0 and S1 13CO intensities are used to calculate dephasing ΔS/S0 = (S0 – S1)/S0 at each τ. For temperatures ≥0 °C, motion reduces ΔS/S0 and greatly complicates determination of r (Figure S7).15 Temperature of –30 °C is therefore used to attenuate motion.
Figure 2 displays experimental spectra and (ΔS/S0)exp vs τ buildups. The G16 and A5 13CO peak shifts are respectively 177 and 179 ppm and correlate with helical structure.2,12,16 The buildups reflect intra- rather than intermolecular spin pairs as evidenced by similar (ΔS/S0) for samples with either all labeled or a 1:1 labeled:unlabeled mixture of HA3fp20 (Figure S3). For each labeling scheme, the (ΔS/S0)exp buildups are comparable for HA3fp20 and HA1fp23 samples at both pH's which support similar structures in all samples with minimal dependence on subtype sequence, pH, or the C-terminal WYG residues. Similar structures in membrane contrast with different open vs closed structures for HA3fp20 vs HA1fp23 in detergent at low pH. Additional insight is obtained from comparison with (ΔS/S0)sim vs τ in the closed, semiclosed, and open structures. In contrast to detergent, the open structure is never dominant in membrane.
Figure 2.
(A) 13C-detect/15N-dephase REDOR S0 (colored) and S1 (black) spectra for membrane-associated HAfp with 40 ms dephasing time. (B) Experimental and (C) simulated (ΔS/S0) vs dephasing time.
The 13CO intensities include dominant lab and minor natural abundance (na) signals with (ΔS/S0)exp = [flab × (ΔS/S0)lab] +[fna × (ΔS/S0)na] and flab ≈ 0.75 and fna ≈ 0.25. The most quantitative structural information is obtained from analysis of the (ΔS/S0)lab, which is determined using the above equation and accurate estimates of (ΔS/S0)na. The (ΔS/S0)lab is always close to the corresponding (ΔS/S0)exp with typical (ΔS/S0)lab/(ΔS/S0)exp ≈ 1.15 (Tables S1 and S2). Each (ΔS/S0)na is an average over the ~25 different na sites, with the (ΔS/S0) of each site calculated using the na 13CO-lab 15N distance of the closed structure (SI).
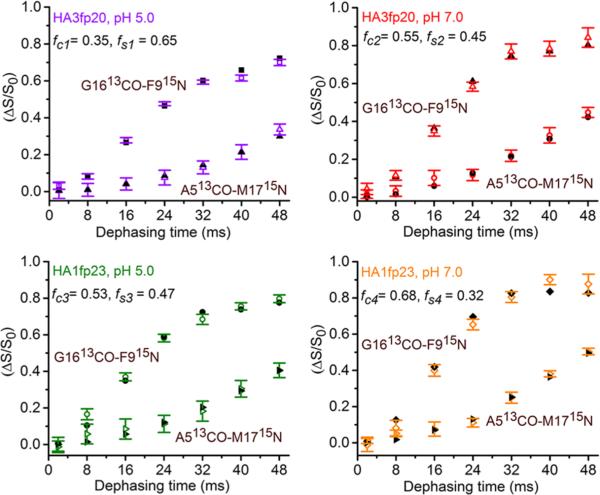
The (ΔS/S0)lab buildups do not quantitatively match the (ΔS/S0)sim buildups of the closed, semiclosed, or open structures. However, quantitative fitting is obtained for all buildups with a model for which a fraction (fc) of the peptides in each sample type (sequence + pH) have closed structure and the remaining fraction (fs) have semiclosed structure (Figure 3). In addition to the best-fit fractions shown in Figure 3, fitting includes best-fit rcG = 3.9 Å and rsG = 5.4 Å common to the four G16/F9 samples and best-fit rcA = 5.4 Å and rsA = 8.2 Å common to the four A5/M17 samples. These distances agree very well with the respective 3.9, 5.5, 5.4, and 8.2 Å values calculated from the closed HA1fp23 structure in detergent and the semiclosed HA3fp20 structure in membrane (Figure 1). The SI provides a full description of the fitting including best-fit parameter uncertainties and χ2. Fitting is always worse with inclusion of an open structure population.
Figure 3.
Plots of experimental (ΔS/S0)lab (colored) and best-fit (ΔS/S0)sim (black) from the closed/semiclosed model.
Significant differences between the structures in membrane vs detergent include: (1) presence vs absence of semiclosed structure; (2) absence vs presence of open structure; (3) mixture of closed and semiclosed structures for both HA3fp20 and HA1fp23 vs predominant open structure for HA3fp20 and closed structure for HA1fp23. The membrane and detergent samples are at thermodynamic equilibrium so the different structural populations reflect free energy differences between the two media. Some of these differences may be due to a locally flat membrane surface vs a locally curved detergent micelle surface (Figure 4). The closed and semiclosed structures are amphipathic with flat hydrophobic and hydro-philic surfaces on opposite faces that are geometrically matched to the surface of the amphipathic membrane. The presence of both closed and semiclosed structures in membrane may correlate to their similar hydrophobic surfaces and consequent similar protein/membrane interaction energies. There is less favorable matching with the curved micelle, particularly for the semiclosed structure which has more extended surfaces. The detergent micelle is also more plastic than the membrane with lower energy penalty for detergent relative to lipid relocation to shield the hydrophobic pocket of the open structure from water. For pH 7, there is good agreement between fc ≈ 0.7 for HA1fp23 in gel-phase membrane and fc ≈ 1 in bicelles with detergent:lipid ≈ 2:1 mole ratio.17
Figure 4.

Models of detergent micelle and membrane locations of closed structure HA1fp23. Dashed lines are the hydrocarbon core.
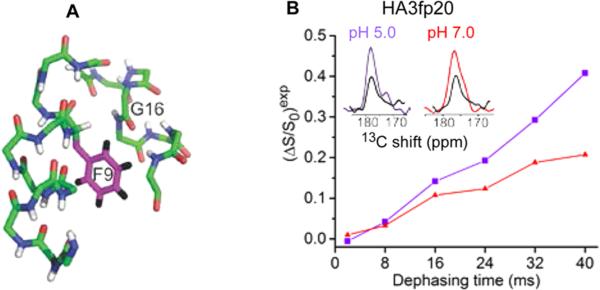
The similar closed and semiclosed populations in membrane reflect comparable free-energies of the two structures. Relative to HA3fp20, the larger fc's of HA1fp23 may be due to stabilization of the tight N-helix/C-helix packing via the longer C-helix containing the additional WYG residues.9,18 For either construct, larger fc's at pH 7 and larger fs's at pH 5 correlate with the protonation of E11 (pKa ≈ 5.9) adjacent to the turn.19 Stabilization of the closed structure by E11 –COO– and the semiclosed structure by –COOH also correlates with the most stable structures observed in MD simulations of HA3fp20 in implicit membrane.20 Computational energy minimization of the semiclosed structure resulted in retention of the semiclosed backbone and insertion of the F9 ring in the interhelical cavity (Figure 5A). This insertion is also observed in the MD structures with E11 –COOH. Insertion was probed by 13CO–2H REDOR of HA3fp20 with G16 13CO and F9 ring 2H labeling (Figure 5B). There was greater buildup at pH 5 than pH 7, which correlates with (1) calculated G16 CO-F9 ring center distance of ~5 Å in the semiclosed and ~8 Å in the closed structure; and (2) a larger fs in the pH 5 sample (Figure 3). The pH 5 buildup was well-fitted by a model with fc = 0.35 and fs = 0.65 and 13CO–2H dcD = 0 and best-fit dsD = 19(1) Hz (Figure S8). This corresponds to rsD ≈ 6 Å and supports location of the F9 ring in the interhelical cavity of the semiclosed structure. There is also hydrophobic F9/M17 interaction (Figure S9).
Figure 5.
(A) Model of insertion of the F9 ring in the semiclosed structure. (B) 13C-detect/2H-dephase REDOR of HA3fp20 samples with G16 13CO/F9 ring 2H labeling. The typical uncertainty is 0.02. S0 (colored) and S1 (black) spectra are for 40 ms dephasing time.
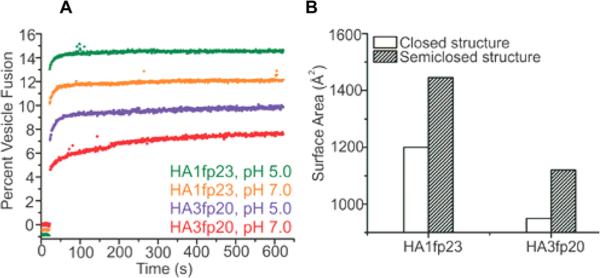
Structure–function correlation was probed with assays of HAfp-induced vesicle fusion under the four sample conditions used for SSNMR (Figure 6A). Significant fusion is observed for all conditions, and the fusion extents are ordered (HA1fp23, pH 5) > (HA1fp23, pH 7) > (HA3fp20, pH 5) > (HA3fp20, pH 7), which is consistent with earlier work.21 Relative to HA3fp20, the higher fusion of HA1fp23 supports a contribution from the C-terminal WYG residues. For either HA3fp20 or HA1fp23, there is higher fusion at pH 5 than pH 7 which correlates to larger fs and smaller fc at the lower pH and evidence higher fusion catalysis by the semiclosed structure. These data support a contribution to fusion catalysis from hydrophobic interaction between HAfp and the membrane (Figure 4). The mechanism is reduction in activation energy because the perturbed bilayer of the HAfp/membrane complex resembles the fusion transition state. The calculated HAfp hydrophobic surface area (Sa) is the quantity used to represent this hydrophobic interaction (Figure 6B). Sa(HA1fp23) > Sa(HA3fp20) because of the additional WYG residues, and Sa(semiclosed) > Sa(closed) because of the more open interhelical geometry of the semiclosed structure. The Sa of each sample is calculated using the experimentally derived fc and fs, and the ordering of these Sa's is the same as the fusion extents (Table S8). The Sa's and fusion extents for larger HA2 constructs also support the importance of protein hydrophobic surface area in fusion. One example is FHA2, the 185-residue extraviral domain of the HA2 subunit protein that includes HAfp.7 The calculated Sa(FHA2):Sa(HA1fp23) ≈ 5 and FHA2 is a much better fusion catalyst than HA1fp23.7
Figure 6.
(A) HAfp-induced vesicle fusion for 1:50 peptide:lipid mole ratio. (B) Calculated HAfp hydrophobic surface areas.
Although most fusion peptide structures are in detergents, a structure in membrane is very important because fusion is induced between membranes but not micelles. The present and previous studies support substantial structural differences in membrane vs detergent. HAfp and fusion peptides from other viruses with very different sequences are α helical monomers in detergent and form α monomers as well as antiparallel β sheet oligomers in membrane.15,22–24 The relative α and β populations are determined by membrane composition, e.g., inclusion of cholesterol often results in higher β population.22,25 For the present study without cholesterol, both HA3fp20 and HA1fp23 are mixtures of closed and semiclosed α structures which are different than the dominant open HA3fp20 and closed HA1fp23 structures in detergent. Similar membrane fusion by HA3fp20 and HA1fp23 correlates much better with their similar structures in membrane than with their very different structures in detergent.
Previous studies on fusion induced by the full-length HA protein support the importance of the HAfp in catalyzing the early hemifusion (membrane joining) step of fusion.26 Vesicle fusion resembles hemifusion, and HAfp-induced vesicle fusion is consistent with an important role for HAfp in hemifusion. The mixture of closed and semiclosed structures for HAfp in membrane is likely reflective of HAfp structure in full-length HA2 during virus/endosome fusion as evidenced by (1) the N-terminal 20- or 23-residue HAfp has autonomous folding in membrane, and the residue 34–175 C-terminal region has autonomous folding in aqueous solution; and (2) the HAfp and the residue 186–210 TM domain are the only HA regions which are deeply membrane-inserted after viral fusion.6,27 HA is minimally trimeric, but the three HAfp helices do not contact one another in HA2 subunit ectodomain trimers.28 HA2 probably contains α HAfp monomers at least during early hemifusion with the possibility of a second structural population of antiparallel β sheet oligomers.22,29 HAfp fusion activity may also relate to large ratios of hydrophobic to hydrophilic surface areas. For HA3fp20, the ratio is 2.8 for closed and 4.2 for semiclosed structure, and for HA1fp23, the ratios are 2.4 and 3.7. Large ratios for amphipathic peptides are correlated to stabilization of negative membrane curvature which is a feature of fusion intermediates.30,31 The semiclosed structures have the largest ratios so their greater fusogenicity may be due to curvature stabilization. The closed and semiclosed structures may also interconvert at ambient temperature with coupling to increased lipid motion and disorder which aid catalysis.17
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant AI047153. NMR technical assistance was provided by Dr. Daniel Holmes.
Footnotes
Supporting Information
Additional descriptions of experimental procedures and data analysis and results. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b04578.
The authors declare no competing financial interest.
REFERENCES
- 1.Han X, Bushweller JH, Cafiso DS, Tamm LK. Nat. Struct. Biol. 2001;8:715. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 2.Lorieau JL, Louis JM, Bax A. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11341. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh U, Xie L, Weliky DP. J. Biomol. NMR. 2013;55:139. doi: 10.1007/s10858-013-9709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White JM, Delos SE, Brecher M, Schornberg K. Crit. Rev. Biochem. Mol. Biol. 2008;43:189. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durell SR, Martin I, Ruysschaert JM, Shai Y, Blumenthal R. Mol. Membr. Biol. 1997;14:97. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- 6.Durrer P, Galli C, Hoenke S, Corti C, Gluck R, Vorherr T, Brunner J. J. Biol. Chem. 1996;271:13417. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- 7.Curtis-Fisk J, Preston C, Zheng ZX, Worden RM, Weliky DP. J. Am. Chem. Soc. 2007;129:11320. doi: 10.1021/ja073644g. [DOI] [PubMed] [Google Scholar]
- 8.Lai AL, Park H, White JM, Tamm LK. J. Biol. Chem. 2006;281:5760. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- 9.Lorieau JL, Louis JM, Bax A. Biopolymers. 2013;99:189. doi: 10.1002/bip.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du TP, Jiang L, Liu ML. J. Peptide Sci. 2014;20:292. doi: 10.1002/psc.2616. [DOI] [PubMed] [Google Scholar]
- 11.Lorieau JL, Louis JM, Schwieters CD, Bax A. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19994. doi: 10.1073/pnas.1213801109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, Weliky DP. J. Am. Chem. Soc. 2009;131:13228. doi: 10.1021/ja905198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gullion T, Schaefer J. J. Magn. Reson. 1989;81:196. [Google Scholar]
- 14.Cegelski L. Bioorg. Med. Chem. Lett. 2013;23:5767. doi: 10.1016/j.bmcl.2013.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodner ML, Gabrys CM, Parkanzky PD, Yang J, Duskin CA, Weliky DP. Magn. Reson. Chem. 2004;42:187. doi: 10.1002/mrc.1331. [DOI] [PubMed] [Google Scholar]
- 16.Zhang HY, Neal S, Wishart DS. J. Biomol. NMR. 2003;25:173. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 17.Lorieau JL, Louis JM, Bax A. J. Am. Chem. Soc. 2011;133:14184. doi: 10.1021/ja2045309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorieau JL, Louis JM, Bax A. J. Am. Chem. Soc. 2011;133:2824. doi: 10.1021/ja1099775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang DK, Cheng SF, Lin CH, Kantchev EAB, Wu CW. Biochim. Biophys. Acta. 2005;1712:37. doi: 10.1016/j.bbamem.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Panahi A, Feig M. J. Phys. Chem. B. 2010;114:1407. doi: 10.1021/jp907366g. [DOI] [PubMed] [Google Scholar]
- 21.Wharton SA, Martin SR, Ruigrok RWH, Skehel JJ, Wiley DC. J. Gen. Virol. 1988;69:1847. doi: 10.1099/0022-1317-69-8-1847. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Parkanzky PD, Bodner ML, Duskin CG, Weliky DP. J. Magn. Reson. 2002;159:101. doi: 10.1016/s1090-7807(02)00033-2. [DOI] [PubMed] [Google Scholar]
- 23.Yao HW, Hong M. J. Mol. Biol. 2013;425:563. doi: 10.1016/j.jmb.2012.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apellaniz B, Huarte N, Largo E, Nieva JL. Chem. Phys. Lipids. 2014;181:40. doi: 10.1016/j.chemphyslip.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiang W, Weliky DP. Biochemistry. 2009;48:289. doi: 10.1021/bi8015668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemble GW, Danieli T, White JM. Cell. 1994;76:383. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Skehel JJ, Wiley DC. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8967. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macosko JC, Kim CH, Shin YK. J. Mol. Biol. 1997;267:1139. doi: 10.1006/jmbi.1997.0931. [DOI] [PubMed] [Google Scholar]
- 29.Schmick SD, Weliky DP. Biochemistry. 2010;49:10623. doi: 10.1021/bi101389r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tytler EM, Segrest JP, Epand RM, Nie SQ, Epand RF, Mishra VK, Venkatachalapathi YV, Anantharamaiah GM. J. Biol. Chem. 1993;268:22112. [PubMed] [Google Scholar]
- 31.Smrt ST, Draney AW, Lorieau JL. J. Biol. Chem. 2015;290:228. doi: 10.1074/jbc.M114.611657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.