SUMMARY
Glioblastoma harbors a dynamic subpopulation of glioblastoma stem-like cells (GSCs) that can propagate tumors in vivo and is resistant to standard chemoradiation. Identification of the cell-intrinsic mechanisms governing this clinically important cell state may lead to the discovery of novel therapeutic strategies for this challenging malignancy. Here, we demonstrate that the mitotic E3 ubiquitin ligase CDC20-Anaphase-Promoting Complex (CDC20-APC) drives invasiveness and self-renewal in patient tumor-derived GSCs. Moreover, CDC20 knockdown inhibited and CDC20 overexpression increased the ability of human GSCs to generate brain tumors in an orthotopic xenograft model in vivo. CDC20-APC control of GSC invasion and self-renewal operates through pluripotency-related transcription factor SOX2. Our results identify a CDC20-APC/SOX2 signaling axis that controls key biological properties of GSCs, with implications for CDC20-APC-targeted strategies in the treatment of glioblastoma.
INTRODUCTION
Glioblastoma, the most common malignant primary brain tumor in adults remains a challenging disease with a poor prognosis (Wen and Kesari, 2008). Increasing appreciation of the cancer cell heterogeneity within glioblastomas has focused attention on a subpopulation of cells called tumor-initiating cells or glioblastoma stem-like cells (GSCs) (Singh et al., 2004). GSCs contribute to overall tumor growth as well as tumor recurrence following chemoradiation and exhibit elevated invasive potential compared to their non-stem cell counterparts (Bao et al., 2006; Chen et al., 2012; Cheng et al., 2011). GSCs also retain the genetic features of parental tumors, suggesting they are a faithful model system for human glioblastoma (Lee et al., 2006; Pollard et al., 2009).
The Anaphase-Promoting Complex (APC) E3 ubiquitin ligase functions with co-activator CDC20 to drive mitosis (Peters, 2006). CDC20-APC has been viewed as a potential strategic target in several human cancers (Wang et al., 2015). CDC20 mRNA is elevated in glioblastoma compared to low-grade gliomas, and CDC20 immunoreactivity in gliomas correlates with pathological grade, but little is known about the biological roles of CDC20-APC in glioblastoma (Bie et al., 2011; Marucci et al., 2008). Recent studies have revealed unexpected non-mitotic roles for CDC20-APC in the developing mammalian brain, indicating CDC20-APC executes functions beyond the cell cycle (Kim et al., 2009; Puram et al., 2011; Yang et al., 2009). These observations have important ramifications not only for brain development but also raise the possibility that CDC20-APC may function in the aberrant developmental state of GSCs.
Here we report CDC20-APC is required for GSC invasiveness and self-renewal in a manner distinct from its role in cell cycle control. We identify pluripotency-related transcription factor SOX2 as a CDC20-interacting protein and show CDC20-APC operates through SOX2 to regulate human GSC invasion and self-renewal. Finally, we demonstrate CDC20-APC is essential for GSC tumorigenicity in orthotopic xenografts and that CDC20 expression has prognostic value in a subset of glioblastoma patients. These results highlight a critical role for CDC20-APC in the maintenance of human GSC function and suggest that targeting this pathway in glioblastoma may disrupt the GSC state.
RESULTS
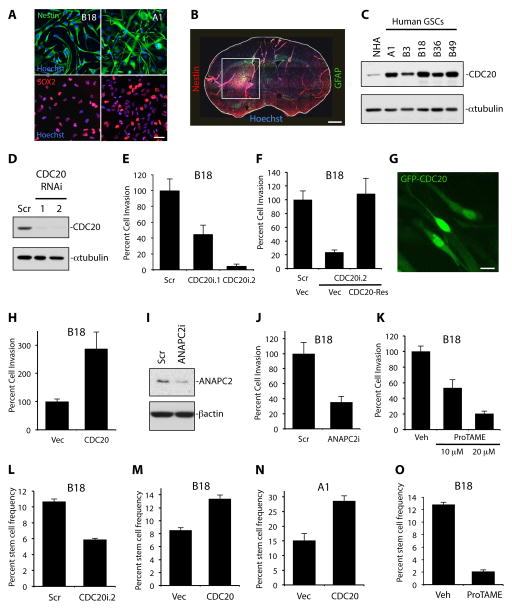
We have generated low-passage patient-derived glioblastoma stem-like cell lines (GSCs) (Table S1), which express neural stem cell markers (Figure 1A, S1A–C), exhibit self-renewal in vitro (Figure S1D), and form infiltrative brain tumors in immunocompromised mice (Figure 1B, S1E) (Pollard et al., 2009). We examined CDC20 expression by immunoblotting in multiple GSC lines and found increased protein levels in GSCs compared to primary human astrocytes (Figure 1C). To test the role of CDC20 in GSCs, we used RNA interference (RNAi) lentiviruses to target human CDC20 (CDC20i.1 and CDC20i.2), which resulted in efficient CDC20 knockdown (Figure 1D). We focused first on invasiveness, a defining clinical feature of gliomas. GSCs transduced with CDC20 RNAi were subjected to an in vitro Matrigel invasion assay, which quantitatively assesses invasion through an extracellular matrix-coated filter (Figure 1E). CDC20 knockdown using by two distinct RNAi viruses inhibited GSC invasiveness by 55% and 95%, respectively (Figure 1E).
Figure 1. CDC20-APC controls glioblastoma stem-like cell invasion and self-renewal.
(A) GSC lines were subjected to immunofluorescence with indicated antibodies and Hoechst nuclear stain. Bar = 50 μm.
(B) B18 GSCs were injected into the right putamen of NOD-SCID mice, and animals sacrificed after 3 months. Sectioned brains were subjected to immunohistochemistry with indicated antibodies. Nuclei were stained with DAPI. White box highlights tumor. Bar = 100 μm.
(C) Lysates from GSC lines and normal human astrocytes (NHA) were processed for immunoblotting using indicated antibodies.
(D) B18 GSCs were transduced with CDC20 RNAi (CDC20i.1 and CDC20i.2) or control scrambled (Scr) lentivirus. 7 days later, cell lysates were subjected to immunoblotting using indicated antibodies. Similar results were seen with control viruses SHC002 and LacZ RNAi (data not shown).
(E) GSCs treated as in (D) were subjected to the in vitro Matrigel transwell assay 6 days after infection. Data represent mean+SEM. CDC20 knockdown inhibited GSC invasiveness compared to control (ANOVA, P = 0.001 and P < 0.0001 for CDC20i.1 and CDC20i.2, respectively, n = 4).
(F) GSCs transduced with the indicated lentiviruses were subjected to the in vitro Matrigel transwell assay as in (E). Data represent mean+SEM. CDC20 RNAi decreased GSC invasiveness compared to control (ANOVA; P < 0.003, n = 3). Expression of CDC20-Res rescued the CDC20 RNAi-triggered invasion phenotype (ANOVA; P = 0.003). Vec = control vector virus.
(G) B18 GSCs transduced with GFP-CDC20-Res-expressing lentivirus were subjected to live fluorescence microscopy. Bar = 10 μm.
(H) GSCs transduced with GFP-CDC20-Res-expressing or control vector lentiviruses (Vec) were assessed for invasion 5 days later. Data represent mean+SEM. CDC20 overexpression increased GSC invasion compared to control (unpaired t-test, P = 0.01, n = 5).
(I) B18 GSCs were transduced with ANAPC2 RNAi (ANAPC2i) or control scrambled (Scr) lentivirus. 7 days later, cell lysates were subjected to immunoblotting using indicated antibodies. Similar results were seen using control virus SHC002 (data not shown).
(J) GSCs treated as in (I) were subjected to the in vitro Matrigel transwell assay 6 days later. Data represent mean+SEM. ANAPC2 knockdown inhibited GSC invasiveness compared to control (unpaired t-test, P = 0.001, n = 4).
(K) B18 GSCs were subjected to the in vitro Matrigel transwell assay in the presence of ProTAME or DMSO (Veh). Data represent mean+SEM. ProTAME inhibited invasion in a dose-dependent manner (ANOVA, P < 0.003, n = 3).
(L) GSCs infected with CDC20i.2 or control (Scr) virus were subjected to the extreme limiting dilution assay. 7 days later, the number of wells with spheres was counted and analyzed. Data represent mean+SEM. CDC20 RNAi decreased the percentage of self-renewing GSCs compared to control (unpaired t-test, P = 0.0002, n = 3).
(M) B18 GSCs infected with CDC20-expressing or control virus were treated as in (L). Data represent mean+SEM. CDC20 overexpression increased the percentage of self-renewing GSCs compared to control (unpaired t-test, P = 0.0005, n = 3).
(N) A1 GSCs treated as in (M) were subjected to the extreme limiting dilution assay. Data represent mean+SEM. CDC20 overexpression increased the percentage of self-renewing GSCs compared to control infection (unpaired t-test, P = 0.009, n = 3).
(O) GSCs were subjected to the extreme limiting dilution assay with 10 μM ProTAME or DMSO (Veh) and analyzed as in (L). Data represent mean+SEM. ProTAME decreased the percentage of self-renewing GSCs compared to vehicle (unpaired t-test, P < 0.0001, n = 3).
See also Table S1 and Figure S1–3.
To demonstrate the specificity of the CDC20 RNAi phenotype, we performed a rescue experiment using rat Cdc20 (herein CDC20-Res), which shares 94.8% amino acid identity with human CDC20 but harbors 4 base mismatches within the sequence targeted by CDC20i.2, rendering it insensitive to CDC20i.2 (Figure S2A). The inhibition of GSC invasiveness by CDC20 knockdown was reversed by co-expression of CDC20-Res, demonstrating the specificity of the CDC20 RNAi phenotype (Figure 1F). To test the generalizability of CDC20’s role in GSC invasion, we subjected two additional patient tumor-derived GSC lines to CDC20 knockdown and similarly found that CDC20 RNAi decreased invasiveness (Figure S2B,C). CDC20 overexpression also increased the invasive capacity of three human GSC lines (Figure 1G,H, S2D,E). Thus, through both loss-of-function and gain-of-function approaches, CDC20 is necessary and sufficient for GSC invasion in vitro.
We next determined if CDC20 operates with the APC to control GSC invasiveness. We knocked down Anaphase-Promoting Complex 2 (ANAPC2), the essential catalytic subunit of the APC, and found that ANAPC2 RNAi inhibited GSC invasiveness in three human GSC lines (Figure 1I, J, S2B,C). We also tested if the interaction between CDC20 and the APC is essential for GSC invasiveness by using a pharmacological inhibitor of the APC, ProTAME, which interferes with the binding of the CDC20 IR tail with the APC (Figure 1K, Figure S2F) (Zeng et al., 2010). We confirmed exposure to ProTAME disrupts the interaction between CDC20 and APC subunit CDC27 in GSCs (Figure S2F). ProTAME treatment inhibited invasiveness in three human GSC lines, suggesting CDC20 acts with the APC to control GSC invasion (Figure 1K, Figure S2G,H).
We next examined the role of CDC20 in GSC self-renewal, a property which often parallels tumorigenic potential (Suva et al., 2014). We performed the extreme limiting dilution assay to measure the frequency of self-renewing cells and found that CDC20 knockdown decreased the percentage of self-renewing GSCs by 45%. (Figure 1L) (Singh et al., 2004). In complementary experiments, CDC20 overexpression increased the frequency of self-renewing cells by 56% and 89% in two GSC lines, respectively (Figure 1M,N). Exposure to APC inhibitor ProTAME also inhibited GSC self-renewal (Figure 1O). Together, these experiments indicate CDC20 operates with the APC to promote GSC invasion and self-renewal.
We next asked if cell cycle perturbations triggered by CDC20-APC manipulations might be responsible for the observed effects on GSC invasion and self-renewal. Examination of cell cycle profile revealed little to no change in the distribution of cell cycle phases in CDC20 knockdown GSCs compared to that of control infected cells (Figure S3A). Additionally, the degree of CDC20 knockdown achieved in these experiments did not significantly alter cellular proliferation by the MTS assay, although ANAPC2 knockdown modestly decreased proliferation (Figure S3B). These data are consistent with the previously reported observation that CDC20 knockdown does not significantly alter mitotic transition until CDC20 levels drop below a critical threshold (Wolthuis et al., 2008). In other experiments, CDC20 overexpression had little to no effect on the cell cycle distribution or proliferation of GSCs (Figure S3C–F). These results support the hypothesis that CDC20 control of GSC invasiveness and self-renewal can be separated from CDC20 regulation of the cell cycle.
We next asked if CDC20-APC control of GSC function might be a consequence of decreased cellular survival. Importantly, we found that the degree of CDC20 and ANAPC2 knockdown achieved did not significantly alter cell survival in GSCs (Figure S3G). Additionally, CDC20 RNAi did not significantly increase caspase-3 activity in GSCs, and ANAPC2 RNAi caused a mild increase in caspase-3 activity in only one of two GSC lines (Figure S3H,I). In other experiments, short-term treatment with APC inhibitor ProTAME revealed minimal to no cell death in two GSC lines (Figure S3J and data not shown). These results suggest alterations in cell survival were not significantly contributing to the invasion and self-renewal phenotypes observed with CDC20-APC manipulations.
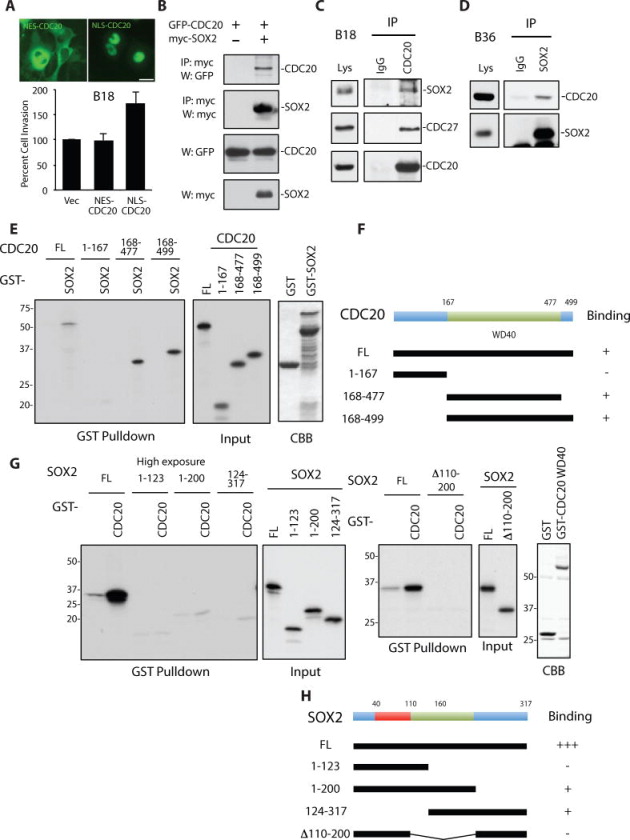
To understand the mechanism of CDC20-APC in GSC invasiveness, we turned to the question of where in the cell CDC20 operates to mediate invasiveness. Previous reports demonstrated that specific subcellular pools of CDC20 dictate distinct biological responses in neural development (Kim et al., 2009; Puram et al., 2011). CDC20 localizes to both cytoplasmic and nuclear compartments in GSCs (Figure 1G) (Kallio et al., 1998). To localize CDC20 to distinct subcellular locations, we generated viruses that express mutant CDC20 fusion proteins carrying either a nuclear localization sequence (GFP-NLS-CDC20) or nuclear export sequence (GFP-NES-CDC20), the latter localizing CDC20 to the cytoplasm (Figure 2A). Expression of nuclear CDC20 enhanced GSC invasiveness, whereas expression of cytoplasmic CDC20 did not significantly alter invasive capacity, suggesting CDC20-APC stimulates a nuclear program to drive invasion (Figure 2A).
Figure 2. CDC20-APC interacts with SOX2 through the WD40 repeat domain of CDC20.
(A) Top: B18 GSCs transduced with lentiviruses expressing indicated GFP-tagged mutant CDC20 proteins were subjected to live fluorescence microscopy. Bar = 25 μm. Bottom: GSCs treated as above were assessed for invasion 6 days later. Data represent mean+SEM. Expression of nuclear localized CDC20 (NLS-CDC20) but not cytoplasmic CDC20 (NES-CDC20) increased GSC invasiveness compared to control (ANOVA, P = 0.005, n = 5).
(B) Lysates of 293 cells transfected with GFP-CDC20 together with the myc-SOX2 expression plasmid or control vector were immunoprecipitated using myc antibody and immunoblotted with the indicated antibodies.
(C) GSC line B18 lysates were immunoprecipitated with the CDC20 or control IgG antibody and immunoblotted with the indicated antibodies.
(D) GSC line B36 lysates were immunoprecipitated with the SOX2 or control IgG antibody and immunoblotted with the indicated antibodies.
(E) In vitro translated, 35S-methionine-labeled CDC20 mutant proteins were used in GST pull-down assays using recombinant GST-SOX2 and GST proteins (left panel). The middle panel (Input) confirms comparable levels of CDC20 mutants. CDC20 proteins were visualized by fluorography. Similar amounts of GST and GST-SOX2 were used for pull-downs (Coomassie brilliant blue staining (CBB), right panel).
(F) Schematic depicting CDC20 domain structure (top) and summary of in vitro binding experiments (bottom).
(G) In vitro translated, 35S-methionine-labeled SOX2 mutant proteins were used in GST pull-down assays using recombinant GST-CDC20 WD40 repeat domain (aa 168–477) (referred to as GST-CDC20 in this panel) and GST proteins. SOX2 proteins were visualized by fluorography. The first panel shows a low exposure and the second panel a high exposure of a representative experiment. Input panels confirm SOX2 mutants were produced at comparable levels. Similar amounts of GST and GST-CDC20 were used for pull-downs (CBB, far right panel).
(H) Schematic depicting SOX2 domain structure (top) and summary of in vitro binding experiments (bottom).
To elucidate the signal transduction pathway downstream of CDC20-APC, we considered nuclear proteins implicated in GSC invasiveness and self-renewal. The stem cell regulatory gene SOX2 has received recent attention in the glioblastoma field due to its critical roles in glioblastoma self-renewal, invasion, and tumor propagation (Alonso et al., 2011; Gangemi et al., 2009). We first tested if a physical interaction exists between CDC20 and SOX2. Remarkably, epitope-tagged CDC20 and SOX2 were found in a complex in transfected 293 cells (Figure 2B). Moreover, we found CDC20 endogenously interacts with SOX2 in two distinct GSC lines (Figure 2C,D). APC subunit CDC27 was also found in an endogenous complex with SOX2, suggesting CDC20-APC interacts with SOX2 (Figure 2C and data not shown). To determine if CDC20 binds directly to SOX2, we performed GST-pull down assays using recombinant GST-SOX2 fusion proteins and in vitro translated CDC20, which revealed a robust direct interaction (Figure 2E). Deletion mapping indicated the WD40 repeat domain of CDC20 interacts directly with SOX2 in vitro (Figure 2E,F). Reciprocal GST pull-down assays using GST-fusion proteins carrying the WD40 repeat domain of CDC20 (GST-CDC20(WD40)) and in vitro translated deletion mutants of SOX2 revealed CDC20(WD40) binds to SOX2 aa1–200 and aa124–317, suggesting SOX2 aa124–200 are required for CDC20 binding (Figure 2G,H). Indeed, the SOX2 deletion mutant lacking aa110–200 failed to bind CDC20(WD40) (Figure 2G,H). These data indicate CDC20-APC endogenously interacts with SOX2 in GSCs likely via direct binding between SOX2 amino acids 124–200 and the WD40 repeat domain of CDC20, suggesting a mechanistic link between CDC20-APC and SOX2.
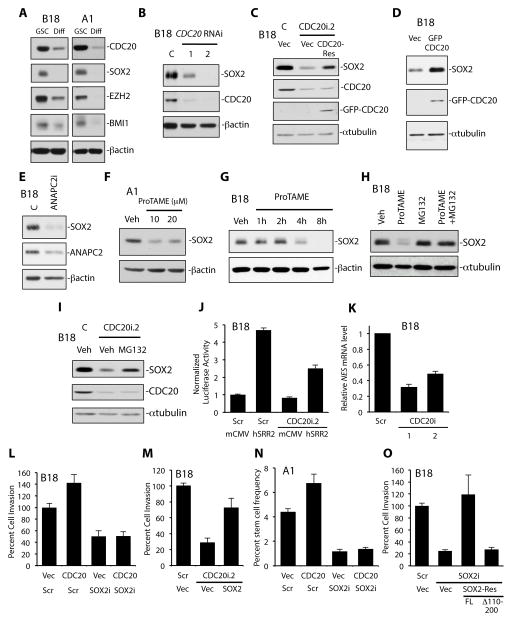
Differentiation of human GSCs in culture led to a dramatic decrease in CDC20 protein levels, suggesting that as with SOX2, CDC20 is enriched in the GSC state (Figure 3A). To test if CDC20-APC regulates SOX2 in GSCs, we subjected GSCs to CDC20 knockdown (Figure 3B, S4A). Intriguingly, CDC20 RNAi decreased SOX2 protein levels in GSCs, and co-expression of RNAi-resistant CDC20-Res with CDC20 RNAi reversed this decrease, suggesting CDC20 specifically promotes SOX2 protein expression (Figure 3B,C, S4A). Conversely, CDC20 overexpression in two GSC lines increased SOX2 protein (Figure 3D, Figure S4B). In other experiments, both ANAPC2 knockdown and APC inhibitor ProTAME decreased SOX2 protein in two GSC lines, suggesting collectively that CDC20 collaborates with the APC to maintain SOX2 levels (Figure 3E–G, Figure S4C).
Figure 3. CDC20-APC regulation of SOX2 protein and transcription controls GSC invasion and self-renewal.
(A) GSCs (B18, A1) were maintained in GSC or differentiating medium (Diff) (containing fetal bovine serum and no growth factors) for 14 days. Cell lysates were subjected to immunoblotting using indicated antibodies.
(B) B18 GSCs were transduced with CDC20 RNAi (CDC20i.1 and CDC20i.2) or control LacZ RNAi (C) lentivirus. 7 days later, cell lysates were subjected to immunoblotting using indicated antibodies.
(C) GSCs were transduced with the indicated lentiviruses. 7 days later, cell lysates were subjected to immunoblotting using indicated antibodies. Expression of CDC20-Res rescued the CDC20 RNAi-triggered decrease in SOX2 protein. C = SHC002 virus. Vec = control vector virus.
(D) GSCs transduced with CDC20-expressing lentivirus or control vector virus (Vec) were maintained in RHB-A media for 5 days. Cell lysates were subjected to immunoblotting using indicated antibodies.
(E) GSCs were transduced with ANAPC2 RNAi or control LacZ RNAi (C) lentivirus. 7 days later, cell lysates were subjected to immunoblotting using indicated antibodies.
(F) GSCs were treated with ProTAME or DMSO (Veh) for 12 hours. Cell lysates were subjected to immunoblotting using indicated antibodies.
(G) GSCs were treated with 20 μM of ProTAME or DMSO (Veh) as indicated. Cell lysates were subjected to immunoblotting using indicated antibodies.
(H) GSCs were treated with 20 μM of ProTAME, 5 μM of proteasome inhibitor MG132, or a combination of both for 8 hours. Cell lysates were subjected to immunoblotting using indicated antibodies. Veh = DMSO.
(I) GSCs were transduced with CDC20 RNAi (CDC20i.2) or control SHC002 (C) lentivirus for 7 days and treated with 10 μM of MG132 or DMSO (Veh) for 6 hours. Cell lysates were subjected to immunoblotting using indicated antibodies.
(J) GSCs stably infected with the SOX2 transcriptional reporter (hSRR2) or control reporter (mCMV) were transduced with CDC20 RNAi (CDC20i.2) or control scrambled (Scr) lentivirus. 7 days later, luciferase assays were performed. Luciferase values were normalized by total protein, and fold-change calculated by scaling to Scr + mCMV values (=1). Data represent mean+SEM. CDC20 RNAi decreased SOX2 reporter activity compared to control (ANOVA, P < 0.0001, n = 3).
(K) GSCs were infected with CDC20 RNAi (CDC20i.1 and CDC20i.2) or control scrambled (Scr) lentivirus. RNA was harvested 7 days later and reverse transcribed into cDNA. qPCR was performed on samples using specific primers for human NES. GAPDH and ACTB were used as reference genes. Data represent mean+SEM. CDC20 RNAi decreased NES mRNA in GSCs compared to control. (ANOVA, P < 0.0001, n = 3).
(L) GSCs infected with the CDC20-expressing or control vector (Vec) lentivirus together with the SOX2 RNAi (SOX2i) or control SHC002 RNAi (Scr) virus were subjected to the in vitro Matrigel transwell assay 7 days later. Data represent mean+SEM. Expression of CDC20 increased invasion compared to control infection (ANOVA, P = 0.007, n = 6). Expression of CDC20 plus SOX2 RNAi reduced invasion compared to infection with Scr plus either the CDC20-expressing or control vector virus. (ANOVA; P < 0.0001 and P = 0.001, respectively).
(M) GSCs infected with SOX2-expressing or control vector (Vec) lentivirus together with CDC20 RNAi or control SHC002 RNAi (Scr) were treated as in (L). Data represent mean+SEM. CDC20 knockdown decreased invasiveness compared to control (ANOVA, P < 0.0001, n = 4). Expression of SOX2 plus CDC20 RNAi increased invasion compared to infection with Vec plus CDC20 RNAi viruses (ANOVA; P = 0.011).
(N) GSCs infected as in (L) were subjected to the extreme limiting dilution assay as in Figure 1L. Data represent mean+SEM. Expression of CDC20 plus Scr increased self-renewal compared to control (ANOVA, P = 0.004, n = 3). Expression of CDC20 plus SOX2 RNAi reduced self-renewal compared to infection with Scr plus either the CDC20-expressing or control virus. (ANOVA; P < 0.0001 and P = 0.001, respectively).
(O) GSCs were transduced with the indicated lentiviruses and subjected to the in vitro Matrigel transwell assay as in (L). Data represent mean+SEM. SOX2 RNAi plus control vector virus (Vec) decreased GSC invasiveness compared to control (ANOVA; P < 0.001, n = 6). Expression of full-length SOX2-Res (FL) rescued the SOX2 RNAi-triggered defect in invasiveness (ANOVA; P < 0.0001) whereas SOX2-ResΔ110–200 did not (ANOVA; P = 0.9).
See also Figure S4.
We next turned to the question of how CDC20-APC regulates SOX2 protein levels and examined the effect of APC inhibitor ProTAME on SOX2 protein over time in GSCs (Figure 3G and data not shown). SOX2 protein levels began to decrease about 4 hours after ProTAME exposure (Figure 3G and data not shown), but SOX2 mRNA demonstrated little to no change after ProTAME treatment over a similar timeframe (Figure S4D,E). We therefore examined the possibility that CDC20-APC controls SOX2 protein stability. Consistent with this hypothesis, treatment with proteasome inhibitor MG132 reversed the decrease in SOX2 protein triggered by both ProTAME and CDC20 RNAi in two GSC lines (Figure 3H,I, Figure S4F). Similar results were seen using a different proteasome inhibitor, bortezomib, in the setting of ProTAME, suggesting CDC20-APC stabilizes SOX2 protein (data not shown).
To determine the biochemical consequences of CDC20-APC control of SOX2, we established a SOX2 transcriptional activity reporter using a lentiviral GFP T2A luciferase expression vector driven by the SOX2-responsive human SOX2 regulatory region 2 enhancer (hSRR2) (Figure 3J) (Sikorska et al., 2008). We confirmed GSCs infected with this SOX2 reporter virus exhibited a hSRR2-specific GFP and luciferase signal compared to control reporter-infected cells; COS-1 cells, which do not express SOX2, did not exhibit a hSRR2-dependent signal (Figure S4G). CDC20 knockdown in GSCs substantially decreased the hSRR2-driven luciferase signal compared to control RNAi, suggesting CDC20 promotes SOX2-mediated transcription (Figure 3J). Accordingly, CDC20 knockdown and APC inhibitor ProTAME decreased the mRNA levels of SOX2 target gene NES (Nestin) in GSCs (Figure 3K, Figure S4H) (Berezovsky et al., 2014). Together, these data indicate CDC20-APC positively regulates SOX2 transcriptional activity in GSCs.
To determine the biological consequences of CDC20 regulation of SOX2, we performed epistasis experiments using the Matrigel invasion assay. We first confirmed that SOX2 knockdown decreases GSC invasiveness in three GSC lines (Figure 3L, S4I,J) (Alonso et al., 2011). SOX2 RNAi did not significantly affect cellular survival or health by the propidium iodide exclusion, MTS, and caspase-3 activity assays, consistent with a prior report (Figure S4L,M and data not shown) (Gangemi et al., 2009). Whereas CDC20 overexpression increased GSC invasiveness, the combination of CDC20 overexpression and SOX2 RNAi decreased invasiveness to a level similar to that of SOX2 RNAi alone (Figure 3L). In a second GSC line, SOX2 RNAi also inhibited the ability of CDC20 overexpression to enhance invasiveness (Figure S4K). Conversely, SOX2 overexpression partially but significantly reversed the CDC20 RNAi-induced invasion phenotype (Figure 3M), together suggesting that SOX2 acts downstream of CDC20 to drive invasiveness. The increase in GSC self-renewal triggered by CDC20 overexpression was also inhibited by SOX2 knockdown in two GSC lines (Figure 3N and data not shown), indicating SOX2 functions downstream of CDC20 to control self-renewal. To test if the binding of CDC20 to SOX2 is critical for GSC invasion, structure-function experiments were performed in the setting of SOX2 RNAi (Figure 3O, S4N). Using a SOX2 cDNA carrying 7 base mismatches in the sequence targeted by SOX2 RNAi (SOX2-Res), we generated lentiviruses that express full-length SOX2-Res and mutant SOX2-ResΔ110–200, the latter of which does not bind CDC20 in vitro (Figure 2G,H). Whereas expression of full-length SOX2-Res rescued the SOX2 RNAi-triggered deficit in invasion, SOX2-ResΔ110–200 did not, suggesting the binding of SOX2 to CDC20 is important for GSC invasion (Figure 3O).
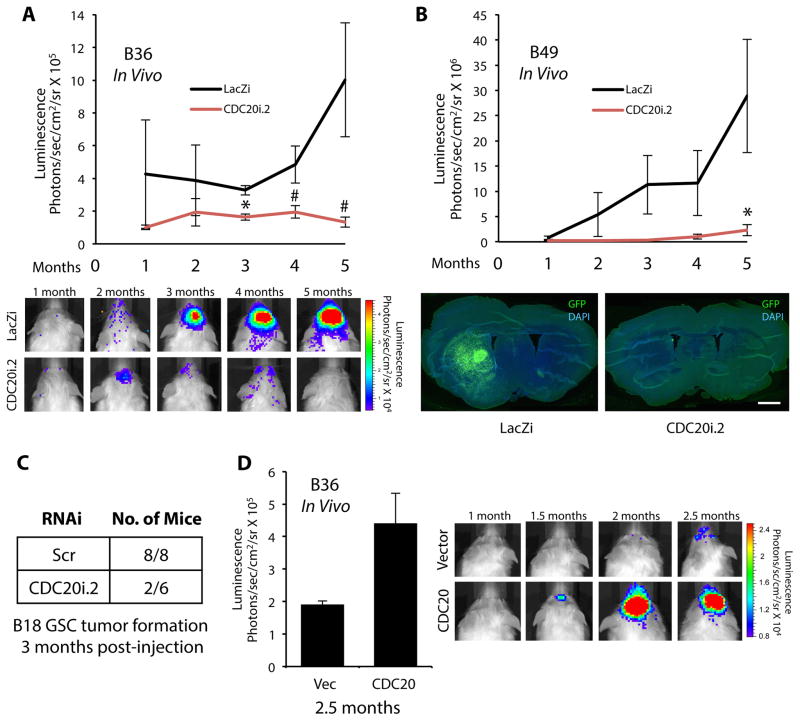
To examine the relevance of CDC20-APC in GSC tumorigenicity in vivo, we used two GSC lines stably expressing GFP T2A luciferase, enabling GFP immunofluorescence as well as bioluminescence imaging (BLI) in live animals to monitor tumor burden (Figure 4A,B). GSCs infected with CDC20 RNAi or control virus were injected into the brains of NOD-SCIDγ mice. BLI performed over several months revealed CDC20 knockdown inhibited brain tumor formation (Figure 4A,B). GFP immunofluorescence in brain sections of injected mice demonstrated infiltrative tumors corresponding to the BLI signal (Figure 4B and data not shown). In other experiments, we infected a third GSC line with CDC20 RNAi or control virus, injected these cells into the brains of NOD-SCID mice, and sacrificed mice 3 months later to assess tumorigenicity by immunofluorescence (Figure 4C, S5). Control-infected GSCs formed brain tumors in all animals (8/8), while CDC20 RNAi-infected GSCs formed tumors in only 2 of 6 animals, suggesting again that CDC20 is critical for the tumor-initiating potential of GSCs (Figure 4C). In complementary experiments, GSCs stably expressing luciferase were infected with CDC20-expressing or control virus, injected into NOD-SCIDγ mice, and assessed for brain tumor growth by BLI, which showed that CDC20 overexpression enhances tumor growth in vivo (Figure 4D). Together, these experiments indicate CDC20 drives the in vivo tumorigenicity of human GSCs.
Figure 4. CDC20 drives GSC tumorigenicity in vivo.
(A) B36 GSCs stably infected with CMV-driven GFP T2A luciferase lentivirus were transduced with CDC20 RNAi or control LacZ RNAi (LacZi) lentivirus and injected into the right putamen of NOD-SCIDγ mice. Injected mice were subjected to live bioluminescence imaging (BLI). Top: Data represent mean±SEM (n = 5 animals per condition). CDC20 knockdown decreased GSC tumorigenicity compared to control (unpaired t-test, *P < 0.01, #P < 0.05). Bottom: Representative animals subjected to BLI are shown.
(B) B49 GSCs stably infected with CMV-driven GFP T2A luciferase lentivirus were treated as in (A) and injected into the brains of NOD-SCIDγ mice. Injected mice were subjected to live BLI. Top: Data represent mean±SEM (n = 5 animals per condition). CDC20 knockdown decreased GSC tumorigenicity compared to control (unpaired t-test, *P < 0.05). Bottom: Injected mice were sacrificed at 5 months. Representative coronal brain sections subjected to GFP immunofluorescence to visualize tumor are shown. Nuclei were stained with DAPI. Bar = 100 μM
(C) B18 GSCs infected with CDC20i.2 or control SHC002 (Scr) virus were injected into the brains of NOD-SCID as in (A). 3 months after injection, animals were sacrificed, and brains were processed for immunohistochemistry using antibodies against NES and GFAP. Nuclei were stained with Hoechst 33342. The number of animals harboring a brain tumor in each treatment group is indicated. CDC20 knockdown decreased GSC tumorigencity compared to control (Fisher’s exact test, P = 0.015).
(D) B36 GSCs stably infected with CMV-driven GFP T2A luciferase lentivirus were transduced with GFP-CDC20-expressing or control vector lentivirus and injected into the brains of NOD-SCIDγ mice as in (A). Injected mice were subjected to live BLI. Left: Data presented are mean±SEM (n = 4 animals per condition). CDC20 overexpression increased tumor formation compared to control (unpaired t-test, P < 0.04). Right: Representative animals subjected to BLI are shown.
See also Figure S5.
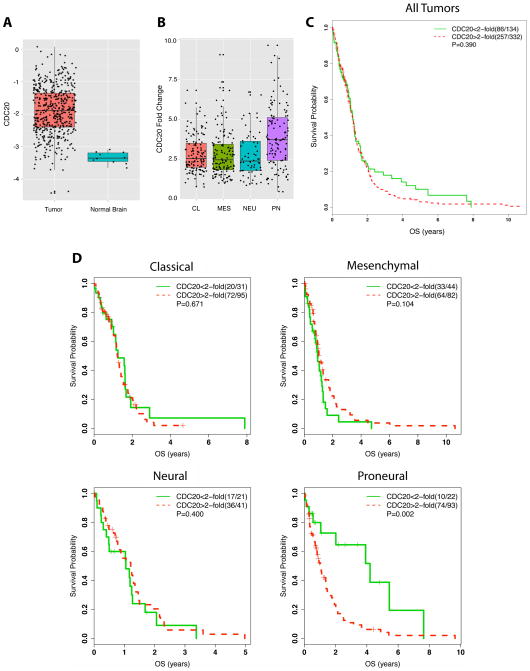
We interrogated The Cancer Genome Atlas (TCGA) to investigate if CDC20 expression correlates with clinical outcomes in glioblastoma patients. Consistent with prior reports, we found CDC20 mRNA is significantly elevated in glioblastomas compared to normal brain (Figure 5A) (Bie et al., 2011; Marucci et al., 2008). We then assessed CDC20 expression in the four TCGA-based molecular subtypes—Proneural, Mesenchymal, Classical, and Neural—and found the Proneural subtype demonstrated significantly higher CDC20 expression compared to the other subtypes (Figure 5B) (Verhaak et al., 2010). We stratified the glioblastoma patients with valid survival data into high (two-fold-change or greater compared to normal brain) and low CDC20 mRNA groups and observed that CDC20 expression in the entire population was not significantly associated with overall survival (OS) (Figure 4C). We then performed Kaplan-Meier survival analyses on patients with high or low CDC20 mRNA expression within each subtype (Figure 4D). Although CDC20 expression did not correlate with OS within the Mesenchymal, Classical, or Neural subtypes, patients with high CDC20-expressing Proneural tumors exhibited a substantially shorter OS (median 53.9 weeks) compared to that of patients with low CDC20-expressing tumors (median 219.6 weeks) (Figure 4D). We confirmed this association using a Cox proportional hazard model to identify an optimal cutoff for CDC20 expression in relation to OS, which also indicated a significant correlation between high CDC20 expression and shorter OS specifically in the Proneural subtype (Figure S6A, B).
Figure 5. High CDC20 expression is associated with decreased overall survival (OS) in Proneural subtype glioblastomas.
(A) Box plot (median and middle 50% of data represented in each box) for CDC20 mRNA expression in TCGA glioblastoma (n = 473) and normal brain tissue samples (n = 10). CDC20 expression is higher in glioblastoma samples compared to normal tissue (unpaired t-test, P = 9.62 X 10−14).
(B) Box plot for normalized CDC20 gene expression (compared to normal samples) demonstrates the highest level of CDC20 expression in the Proneural subtype compared to other subtypes (Holm’s adjustment for multiplicity, P <0.0001).
(C) Kaplan-Meier curves showing OS of 466 newly diagnosed glioblastoma patients from the TCGA based on CDC20 expression. High CDC20 represents two-fold or greater expression and low CDC20 less than two-fold expression compared to mean CDC20 expression in normal brain samples (log-rank test, P = 0.390).
(D) Kaplan-Meier curves showing OS of TCGA patients separated by molecular subtype based on CDC20 expression. Data were analyzed as in (C). High CDC20 expression was associated with decreased OS only in patients with Proneural tumors (log-rank test, P = 0.002).
See also Figure S6.
Somatic mutations in the isocitrate dehydrogenase 1 gene (IDH1) are found in a subset of Proneural patients with longer OS than patients with IDH1-wildtype tumors (Hartmann et al., 2010; Parsons et al., 2008; Yan et al., 2009). We asked if CDC20 expression might interact with IDH1 mutation status or represent an independent prognostic marker in Proneural glioblastomas (Figure S6C,D). When IDH1 MUT tumors were included, Proneural tumor patients with high CDC20 expression again had a poorer prognosis (Figure S6C). When IDH1 MUT tumors were excluded, the number of Proneral tumor patients with low CDC20 expression was small (6 patients with 4 censored), but the OS of patients with high and low CDC20 tumors was not appreciably different, suggesting an interaction between IDH1 mutation and CDC20 expression (Figure S6C). We then examined gene expression data for Proneural tumors only and found IDH1 MUT tumors exhibit significantly lower CDC20 expression compared to that of IDH1 WT tumors (Figure S6D). Together, these data indicate CDC20 expression is prognostic of OS in Proneural glioblastomas and, in a limited subset analysis, appears to interact with IDH1 mutation status.
DISCUSSION
In this study, we have demonstrated CDC20-APC operates through SOX2 to control human GSC invasion and self-renewal. Additionally, we have found CDC20 is critical for human GSC tumorigenicity in vivo. Interrogation of the TCGA revealed high CDC20 expression was associated with decreased overall survival in Proneural subtype glioblastomas.
CDC20-APC has been intensively studied in the cell cycle field and is viewed as a promising target in several human cancers (Wang et al., 2015). As proof of concept, conditional Cdc20 knockout in mouse models of skin cancer and fibrosarcoma caused mitotic arrest and apoptotic tumor regression (Manchado et al., 2010). Intriguingly, the essential role of CDC20 in GSC invasiveness and self-renewal appears to be separable from CDC20’s known role in cell cycle regulation; the CDC20 manipulations used herein did not obviously affect proliferation or cell cycle parameters, consistent with the previous finding that only a minimal level of CDC20 is needed for mitotic transition (Wolthuis et al., 2008). More recently, CDC20 knockdown was shown to sensitize cancer cells to chemotherapy and radiation therapy (Wan et al., 2014). Our results reinforce the rationale for the development of CDC20-APC inhibitors in glioblastoma not only to reduce tumor burden through cell cycle and cell death mechanisms but also to disrupt key functional properties of GSCs.
As with SOX2, CDC20 protein is enriched in human GSCs compared to glioblastoma cells differentiated in vitro. This finding, which remains to be validated in human tumor samples ex vivo, raises interesting questions about how CDC20 is regulated in the GSC state. Downstream of CDC20, regulation of SOX2 appears to occur at two—not necessarily mutually exclusive—levels: CDC20 binding to SOX2 and CDC20-APC control of SOX2 protein stability. SOX2 binding to CDC20 appears to be important for SOX2 control of GSC invasiveness (Figure 2H, 3O). It is possible that CDC20 binding enhances SOX2 function, perhaps through the CDC20-APC-dependent recruitment of transcriptional activators (Turnell et al., 2005). The exact mechanistic link between CDC20-APC and SOX2 protein stability remains an important open question. SOX2 regulation by the ubiquitin-proteasome system has only recently begun to be examined in the context of non-cancerous cells, such as embryonic stem (ES) cells. Whereas SOX2 acetylation and methylation increase SOX2 degradation, phosphorylation of murine SOX2 at Thr118 (Thr116 in human SOX2) by AKT stabilizes SOX2 protein, raising the possibility that CDC20-APC might affect SOX2 stability by altering SOX2 post-translational modifications (Baltus et al., 2009; Fang et al., 2014; Jeong et al., 2010). Alternatively, CDC20-APC may act indirectly on SOX2 by ubiquitinating and destroying a critical E3 ligase, which targets SOX2. Only two E3 ligases that target and degrade SOX2 have been reported so far. One is FZR1 (also CDH1), an alternative co-activator of the APC, which is responsible for G1 maintenance (Fukushima et al., 2013). However, we found little to no change in SOX2 protein levels in the setting of CDH1 RNAi in GSCs, and ANAPC2 RNAi and APC inhibitor ProTAME, which inhibit both CDH1-APC and CDC20-APC, decreased SOX2 protein, suggesting a dominant role for CDC20-APC in SOX2 protein regulation in GSCs (Figure 3E–G and data not shown). More recently, WWP2 was identified as a SOX2 ubiquitin ligase in ES cells (Fang et al., 2014). Whether WWP2 or other E3 ligases contribute to SOX2 stability in glioblastoma remains to be determined.
Our results have several intriguing implications for CDC20-APC’s role in the transcriptional networks governing glioblastoma molecular subtypes as well as non-cancerous stem/progenitor cells. Interestingly, we have found in the TCGA dataset that CDC20 expression is particularly elevated in the Proneural subtype. Since SOX2 is a known Proneural signature gene, the finding that CDC20-APC promotes SOX2-dependent transcription raises the intriguing hypothesis that CDC20-APC stimulates Proneural signature gene transcription (Verhaak et al., 2010). The CDC20-APC/SOX2 mechanism might therefore be particularly relevant for the biology underlying this molecular subtype. As a prognostic marker, CDC20 expression appears to interact with IDH1 mutation, suggesting a potential mechanistic link between IDH1 mutant status and low CDC20 expression. But the exact relationship between CDC20 expression and survival in the bulk tumor data of the TCGA and the CDC20-APC/SOX2 mechanism in GSCs requires further investigation. For instance, in contrast to bulk tumor, human GSCs cluster into predominantly two molecular subtypes—Proneural and Mesenchymal (Bhat et al., 2013). The molecular subtyping of the human GSC lines utilized in this study suggests the control of core GSC functions by the CDC20-APC/SOX2 signaling axis is generalizable and independent of GSC subtype (Figure S1A). Additionally, current mRNA and genome-based bulk tumor datasets may not reflect the SOX2 protein regulatory mechanisms reported herein, which will require interrogation of proteomic datasets. More speculatively, the CDC20-APC/SOX2 pathway may play a role in the transcriptional program in other cellular contexts, including the regulation of neural stem cells and potentially, the maintenance of pluripotency in embryonic or induced pluripotent stem cells (Lewitzky and Yamanaka, 2007; Pevny and Nicolis, 2010).
Although the mechanisms of SOX2’s critical role in self-renewal have been extensively investigated in the context of stem cell biology and cancer (He et al., 2009), the downstream mechanisms that specifically drive SOX2-dependent invasion in glioblastoma remain to be identified. SOX2 has been implicated in promoting the invasive potential of other cancers, raising the possibility that CDC20-APC control of SOX2 might regulate invasion in diverse cancers (Forghanifard et al., 2014; Girouard et al., 2012; Han et al., 2012; Lou et al., 2013; Xia et al., 2014). Future analyses of SOX2 transcriptional targets will be important to elucidate the precise mechanisms of SOX2-mediated invasiveness specifically in glioblastoma. Moreover, given the multitude of identified CDC20-APC substrates, it is likely that additional, SOX2-independent mechanisms contribute to CDC20-APC regulation of GSC invasiveness and self-renewal.
EXPERIMENTAL PROCEDURES
Cell Culture
The generation of adherent human GSC cultures has been described (Pollard et al., 2009). Briefly, tumor samples obtained directly from surgery were dissociated by mincing and incubation in Accutase (SIGMA) for 20–60 minutes at 37°C. Cell suspensions were passed through a 70 micron cell strainer (Falcon) and plated using Ndiff RHB-A media (Stem Cell, UK) with EGF and FGF-2 (Peprotech) (hereafter “GSC media”), each at 20 ng/ml, on polyornithine and laminin (SIGMA)-coated Primaria dishes/flasks (BD Bioscience). Media was replaced with half fresh GSC media every 2–3 days. Cells were routinely used between passages 5 and 20. Informed consent was obtained from patients for use of human tissue and cells, and all human tissue-related protocols used in this study were approved by the Institutional Review Board (Washington University). Primary human astrocytes (Lonza) were cultured in astrocyte growth media (Lonza). Human embryonic kidney 293 cells were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (FBS) and penicillin/streptomycin (Life Technologies). All cell lines were incubated at 37°C with 5% CO2. Lentiviral transduction was performed by adding virus with 4 μg/mL of polybrene for 4 hours to cells. For rescue or epistasis experiments, GSCs were transduced with RNAi or control lentivirus one day after plating and then transduced with CDC20-Res expression virus or control virus the following day. Cells were selected in 2 μg/mL of puromycin 1–2 days after infection. For self-renewal and in vivo tumorigenicity experiments, GSCs were utilized 4 days following indicated viral infections.
Cell Invasion Assay
The in vitro cell invasion assay was performed using Matrigel-coated invasion chambers (BD Bioscience) (Valster et al., 2005). In 24-well plates, 5 × 104 GSCs in GSC media was added to the upper chamber of a rehydrated, Matrigel-coated polycarbonate membrane filter. The bottom chamber of the well was pre-filled with RHB-A media containing 10% FBS as chemoattractant. After 24 hrs, non-invasive cells from the upper side of the filter were removed using a moist cotton swab. The invasive cells on the reverse side of the filter were then fixed and stained with DAPI nuclear dye, and images of the cells were captured in a blinded fashion in 3 different low-power fields (5X objective) per condition using a fluorescence microscope (Leica Microsystems, DMI4000 B). Quantitation of invasion was also performed in a blinded fashion using Image J software (NIH).
Extreme limiting dilution analysis
Cells were plated at five-fold dilutions (3000, 600, 120, 24, 5 or 1 cell/well) in Corning ultra-low attachment 96-well plates. 7–10 days later, the number of wells containing spheres was counted and used to calculate the frequency of self-renewing GSCs by online software (http://bioinf.wehi.edu.au/software/elda/) (Hu and Smyth, 2009; Singh et al., 2004).
Xenotransplantation
Animals were used in accordance with a protocol approved by the Animal Studies Committee of the Washington University School of Medicine per the recommendations of the Guide for the Care and Use of Laboratory Animals (NIH). 250,000 cells per animal (unless otherwise noted) were injected stereotactically into the right putamen of approximately 6-week-old male NOD-SCIDγ mice (for B36, B49) or female NOD-SCID mice (for B18) (Hope Center Animal Surgery Core, Washington University). The coordinates used were: 1 mm rostral to bregma, 2 mm lateral, and 2.5 mm deep.
Statistics
All images are representative of results from 3 independent experiments unless otherwise stated. Statistical analyses were performed with XLSTAT (Addinsoft), Excel (Microsoft), or R Version 3.1.1 software. The unpaired Student’s t-test was used for comparisons in experiments with only two groups. In experiments with more than two comparison groups, analysis of variance (ANOVA) was performed followed by Fisher’s least significant difference or the Bonferroni test for pairwise comparisons among three and greater than three groups, respectively.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant K08NS081105, American Cancer Society-Institutional Research Grant, Voices Against Brain Cancer, the Elsa U. Pardee Foundation, the Concern Foundation, the Duesenberg Research Fund, (to A.H.K.), National Institutes of Health Grant K01AG033724 (to H.Y.), and National Institutes of Health Grant P50 CA094056 (to S.A.). We thank members of the Kim and Yano laboratories for helpful discussions and critical reading of the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
D.D.M., A.D.G., T.M., and A.H.K. performed and analyzed experiments; H.Y., Y.P., T.B., E.A.T., S.A., and A.T. contributed to experiments; J.L. and I.C. performed bioinformatic analyses; A.H.K., E.C.L., M.R.C., K.M.R., J.L.D., G.J.Z., and R.G.D. provided clinical material; H.Y., G.P.D., and D.D.T. edited the manuscript; and A.H.K. conceived the research project, analyzed the data, and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alonso MM, Diez-Valle R, Manterola L, Rubio A, Liu D, Cortes-Santiago N, Urquiza L, Jauregi P, Lopez de Munain A, Sampron N, et al. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PloS one. 2011;6:e26740. doi: 10.1371/journal.pone.0026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltus GA, Kowalski MP, Zhai H, Tutter AV, Quinn D, Wall D, Kadam S. Acetylation of sox2 induces its nuclear export in embryonic stem cells. Stem Cells. 2009;27:2175–2184. doi: 10.1002/stem.168. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Berezovsky AD, Poisson LM, Cherba D, Webb CP, Transou AD, Lemke NW, Hong X, Hasselbach LA, Irtenkauf SM, Mikkelsen T, et al. Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic differentiation. Neoplasia. 2014;16:193–206. 206 e119–125. doi: 10.1016/j.neo.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bie L, Zhao G, Cheng P, Rondeau G, Porwollik S, Ju Y, Xia XQ, McClelland M. The accuracy of survival time prediction for patients with glioma is improved by measuring mitotic spindle checkpoint gene expression. PloS one. 2011;6:e25631. doi: 10.1371/journal.pone.0025631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Wu Q, Guryanova OA, Huang Z, Huang Q, Rich JN, Bao S. Elevated invasive potential of glioblastoma stem cells. Biochemical and biophysical research communications. 2011;406:643–648. doi: 10.1016/j.bbrc.2011.02.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Zhang L, Wei W, Jin X, Wang P, Tong Y, Li J, Du JX, Wong J. A Methylation-Phosphorylation Switch Determines Sox2 Stability and Function in ESC Maintenance or Differentiation. Molecular cell. 2014;55:537–551. doi: 10.1016/j.molcel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- Forghanifard MM, Ardalan Khales S, Javdani-Mallak A, Rad A, Farshchian M, Abbaszadegan MR. Stemness state regulators SALL4 and SOX2 are involved in progression and invasiveness of esophageal squamous cell carcinoma. Medical oncology. 2014;31:922. doi: 10.1007/s12032-014-0922-7. [DOI] [PubMed] [Google Scholar]
- Fukushima H, Ogura K, Wan L, Lu Y, Li V, Gao D, Liu P, Lau AW, Wu T, Kirschner MW, et al. SCF-mediated Cdh1 degradation defines a negative feedback system that coordinates cell-cycle progression. Cell reports. 2013;4:803–816. doi: 10.1016/j.celrep.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- Girouard SD, Laga AC, Mihm MC, Scolyer RA, Thompson JF, Zhan Q, Widlund HR, Lee CW, Murphy GF. SOX2 contributes to melanoma cell invasion. Laboratory investigation; a journal of technical methods and pathology. 2012;92:362–370. doi: 10.1038/labinvest.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Fang X, Lou X, Hua D, Ding W, Foltz G, Hood L, Yuan Y, Lin B. Silencing SOX2 induced mesenchymal-epithelial transition and its expression predicts liver and lymph node metastasis of CRC patients. PloS one. 2012;7:e41335. doi: 10.1371/journal.pone.0041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Hentschel B, Wick W, Capper D, Felsberg J, Simon M, Westphal M, Schackert G, Meyermann R, Pietsch T, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta neuropathologica. 2010;120:707–718. doi: 10.1007/s00401-010-0781-z. [DOI] [PubMed] [Google Scholar]
- He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annual review of cell and developmental biology. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. Journal of immunological methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Jeong CH, Cho YY, Kim MO, Kim SH, Cho EJ, Lee SY, Jeon YJ, Lee KY, Yao K, Keum YS, et al. Phosphorylation of Sox2 cooperates in reprogramming to pluripotent stem cells. Stem Cells. 2010;28:2141–2150. doi: 10.1002/stem.540. [DOI] [PubMed] [Google Scholar]
- Kallio M, Weinstein J, Daum JR, Burke DJ, Gorbsky GJ. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. The Journal of cell biology. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, Wong M, Rowitch D, Bonni A. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Lewitzky M, Yamanaka S. Reprogramming somatic cells towards pluripotency by defined factors. Current opinion in biotechnology. 2007;18:467–473. doi: 10.1016/j.copbio.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Lou X, Han X, Jin C, Tian W, Yu W, Ding D, Cheng L, Huang B, Jiang H, Lin B. SOX2 targets fibronectin 1 to promote cell migration and invasion in ovarian cancer: new molecular leads for therapeutic intervention. Omics: a journal of integrative biology. 2013;17:510–518. doi: 10.1089/omi.2013.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Guillamot M, de Carcer G, Eguren M, Trickey M, Garcia-Higuera I, Moreno S, Yamano H, Canamero M, Malumbres M. Targeting mitotic exit leads to tumor regression in vivo: Modulation by Cdk1, Mastl, and the PP2A/B55alpha,delta phosphatase. Cancer cell. 2010;18:641–654. doi: 10.1016/j.ccr.2010.10.028. [DOI] [PubMed] [Google Scholar]
- Marucci G, Morandi L, Magrini E, Farnedi A, Franceschi E, Miglio R, Calo D, Pession A, Foschini MP, Eusebi V. Gene expression profiling in glioblastoma and immunohistochemical evaluation of IGFBP-2 and CDC20. Virchows Archiv: an international journal of pathology. 2008;453:599–609. doi: 10.1007/s00428-008-0685-7. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nature reviews Molecular cell biology. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Pevny LH, Nicolis SK. Sox2 roles in neural stem cells. The international journal of biochemistry & cell biology. 2010;42:421–424. doi: 10.1016/j.biocel.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell stem cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Puram SV, Kim AH, Ikeuchi Y, Wilson-Grady JT, Merdes A, Gygi SP, Bonni A. A CaMKIIbeta signaling pathway at the centrosome regulates dendrite patterning in the brain. Nature neuroscience. 2011;14:973–983. doi: 10.1038/nn.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorska M, Sandhu JK, Deb-Rinker P, Jezierski A, Leblanc J, Charlebois C, Ribecco-Lutkiewicz M, Bani-Yaghoub M, Walker PR. Epigenetic modifications of SOX2 enhancers, SRR1 and SRR2, correlate with in vitro neural differentiation. Journal of neuroscience research. 2008;86:1680–1693. doi: 10.1002/jnr.21635. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnell AS, Stewart GS, Grand RJ, Rookes SM, Martin A, Yamano H, Elledge SJ, Gallimore PH. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature. 2005;438:690–695. doi: 10.1038/nature04151. [DOI] [PubMed] [Google Scholar]
- Valster A, Tran NL, Nakada M, Berens ME, Chan AY, Symons M. Cell migration and invasion assays. Methods. 2005;37:208–215. doi: 10.1016/j.ymeth.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Tan M, Yang J, Inuzuka H, Dai X, Wu T, Liu J, Shaik S, Chen G, Deng J, et al. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Developmental cell. 2014;29:377–391. doi: 10.1016/j.devcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang J, Wan L, Zhou X, Wang Z, Wei W. Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacology & therapeutics. 2015 doi: 10.1016/j.pharmthera.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen PY, Kesari S. Malignant gliomas in adults. The New England journal of medicine. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- Wolthuis R, Clay-Farrace L, van Zon W, Yekezare M, Koop L, Ogink J, Medema R, Pines J. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Molecular cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- Xia Y, Wu Y, Liu B, Wang P, Chen Y. Downregulation of miR-638 promotes invasion and proliferation by regulating SOX2 and induces EMT in NSCLC. FEBS letters. 2014;588:2238–2245. doi: 10.1016/j.febslet.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kim AH, Yamada T, Wu B, Bilimoria PM, Ikeuchi Y, de la Iglesia N, Shen J, Bonni A. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science. 2009;326:575–578. doi: 10.1126/science.1177087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC, Hathaway N, Dimova N, Cuny GD, King RW. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.