Summary
Ligands stimulate Notch receptors by inducing regulated intramembrane proteolysis (RIP) to produce a transcriptional effector. Notch activation requires unmasking of a metalloprotease cleavage site remote from the site of ligand binding, raising the question of how proteolytic sensitivity is achieved. Here, we show that application of physiologically relevant forces to the regulatory switch results in sensitivity to metalloprotease cleavage, and that bound ligands induce Notch signal transduction in cells only in the presence of applied mechanical force. Synthetic receptor-ligand systems that remove the native ligand-receptor interaction also activate Notch by inducing proteolysis of the regulatory switch. Together, these studies show that mechanical force exerted by signal-sending cells is required for ligand-induced Notch activation, and establish that force-induced proteolysis can act as a mechanism of cellular mechanotransduction.
Introduction
Notch signaling conveys information between cells using a mechanism that is conserved in organisms ranging from flies to humans. These signals influence a wide range of cell fate decisions both during development and in adult tissue homeostasis. In addition, a number of human diseases are associated with mutations of Notch pathway components that result in loss or gain of function.
Notch signaling occurs when a transmembrane ligand of the DSL family engages a transmembrane Notch receptor on a neighboring cell, inducing regulated intramembrane proteolysis (RIP) to produce a transcriptional effector (Kopan and Ilagan, 2009). During transport to the cell surface, Notch receptors are cleaved at site S1 by a furin-like protease, but are resistant to further proteolysis, because the activating cleavage site, called S2, is buried in an autoinhibited conformation within a negative regulatory region (NRR) consisting of three LNR modules and a juxtamembrane “heterodimerization domain” (HD) (Gordon et al., 2009; 2007). Ligand binding relieves autoinhibition by exposing S2 to ADAM metalloproteases (Brou et al., 2000; Mumm et al., 2000). Activating mutations of the Notch1 NRR that result in ligand-independent proteolysis are found frequently in human leukemias, highlighting the importance of tight control of metalloprotease access to the S2 site (Weng et al., 2004).
How ligand engagement relieves autoinhibition of Notch remains poorly understood. X-ray structures of the NRRs from Notch1 and Notch2 show that the S2 site near the C-terminal end of the HD is masked by the LNRs (Gordon et al., 2007; 2009), indicating that ligand binding must result in sufficient displacement of the LNRs to allow metalloprotease access to S2. Because the binding site for Notch ligands is centered on EGF repeats 11-12, more than 20 EGF modules away (Rebay et al., 1991), and because genetic and biochemical studies have established a requirement for endocytosis of ligand into signal sending cells (Musse et al., 2012), it has long been speculated that endocytic internalization of Notch-bound ligands delivers a pulling force that relieves autoinhibition by exposing S2 (Musse et al., 2012). It remains unknown, however, whether S2 proteolysis can be induced in the physiologic force regime or whether force is even required to activate ligand-bound receptors on cells.
In the work reported here, we develop a single-molecule assay to determine the force required for NRR proteolysis in vitro, and show using a cell-based magnetic tweezer assay we also developed that force is required for relief of Notch autoinhibition in cells. We also designed two synthetic ligand-receptor systems, which both show that signal-sending cells supply sufficient force to induce metalloprotease sensitivity in the NRR in the absence of native ligand-receptor interactions, indicating that ligand binding does not need to exert an allosteric effect on the sensitivity of the NRR in order for activating proteolysis to occur. These results show that mechanical force generated by signal-sending cells is sufficient to unfold the NRR and sensitize Notch to proteolytic activation.
Results
Physiologic forces sensitize the NRR to ADAM cleavage
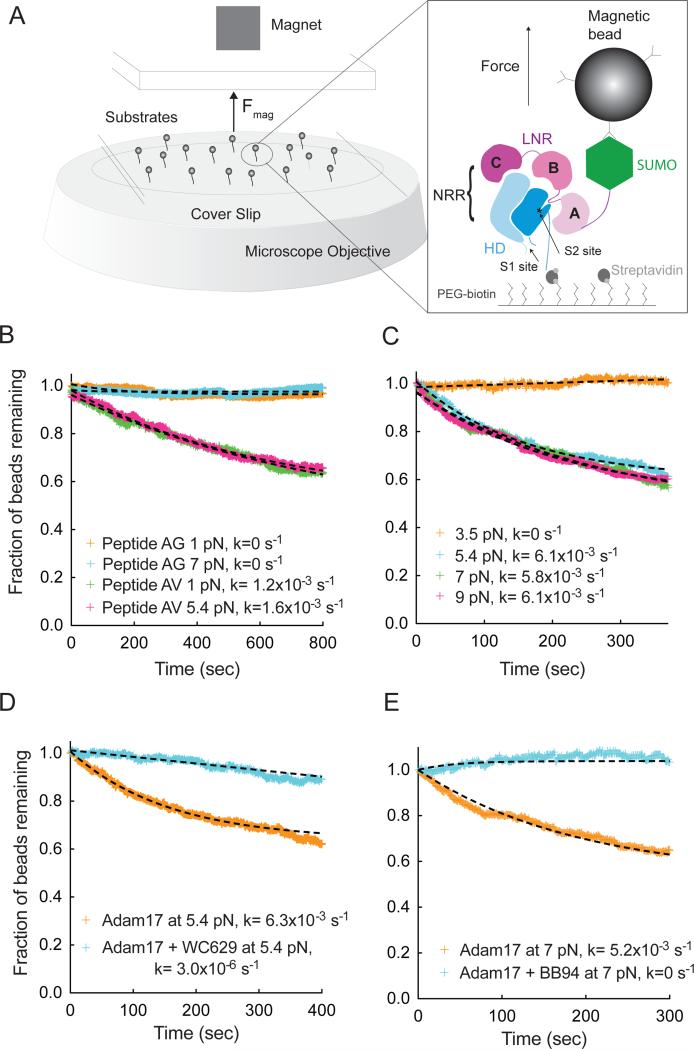
To test whether proteolytic cleavage of the activation switch occurs in a physiologic force regime, we developed a single-molecule, multiplexed magnetic tweezers assay to determine the proteolytic sensitivity of the isolated Notch1 NRR as a function of applied force (Figures 1 and S1). The Notch1 NRR, as well as control proteins intrinsically sensitive or resistant to Adam17 cleavage, was immobilized on the surface of a flow cell by streptavidin capture, tethered to magnetic beads coated with an anti-SUMO antibody, and subjected to ADAM17 delivered by syringe pump. Enzymatic cleavage of tethered molecules was determined as a function of applied magnetic force, monitoring bead loss by dark field microscopy.
Figure 1. Single molecule assay of Adam17-mediated proteolysis of the Notch1 NRR under force.
(A) Assay schematic and experimental design. Magnetic beads are tethered to proteins immobilized in a flowcell mounted on an inverted microscope. Force is applied to the beads by varying the distance between a magnet and the surface of the flowcell. Substrate proteolysis is monitored by determining the fraction of beads released over time. The expanded view in the right-hand panel illustrates the Notch1 NRR, captured on the flow cell with streptavidin and tethered to the magnetic bead using anti-SUMO antibodies. (B) Adam17-catalyzed proteolysis of biotinylated and SUMO tagged recombinant peptides, containing either the natural S2-cleavage site sequence (AV, green and pink), or a mutated sequence with a V1721G substitution (AG, orange and cyan) at the forces indicated. (C) Adam17-catalyzed proteolysis of the Notch1 NRR, monitored as a function of time at different levels of applied force. Traces shown represent averages of 2 or 3 replicates. (D, E) Effect of inhibitors on proteolysis of the Notch1 NRR in the single molecule cleavage assay. Traces shown represent a single experiment. (D) Effect of WC629, an anti-Notch1 inhibitory antibody that binds to the NRR, on the time course of Adam17-catalyzed NRR proteolysis. (E) Effect of BB94, an ADAM inhibitor, on the time course of Adam17-catalyzed NRR proteolysis. Additional control experiments are provided in Figure S1.
Proteolysis experiments using control peptides show that Adam17 cleaves a bead-tethered polypeptide that presents the native S2 processing site of Notch1 (“AV” peptide) when as little as 1 pN of force is applied. The kinetics of cleavage for the AV peptide are indistinguishable at 1 and 5.4 pN of applied force, already fully sensitive to Adam17 at 1 pN. In contrast, a control peptide with a mutated cleavage site (AG) is ADAM17 resistant up to 7 pN of force (Figure 1B).
When the intact Notch1 NRR is examined in this assay, it resists ADAM17 cleavage at a force of 3.5 pN, but undergoes proteolysis at forces equal to or greater than 5.4 pN, indicating that the transition from resistance to sensitivity occurs in a physiologically accessible regime between 3.5 and 5.4 pN of force (Figures 1C and S1D). Both an NRR conformation-specific inhibitory antibody WC629 and the metalloprotease inhibitor BB94 prevent proteolysis by Adam17, confirming that bead release results from metalloprotease cleavage at S2 (Figure 1D, E).
Force induces Notch activation in cells
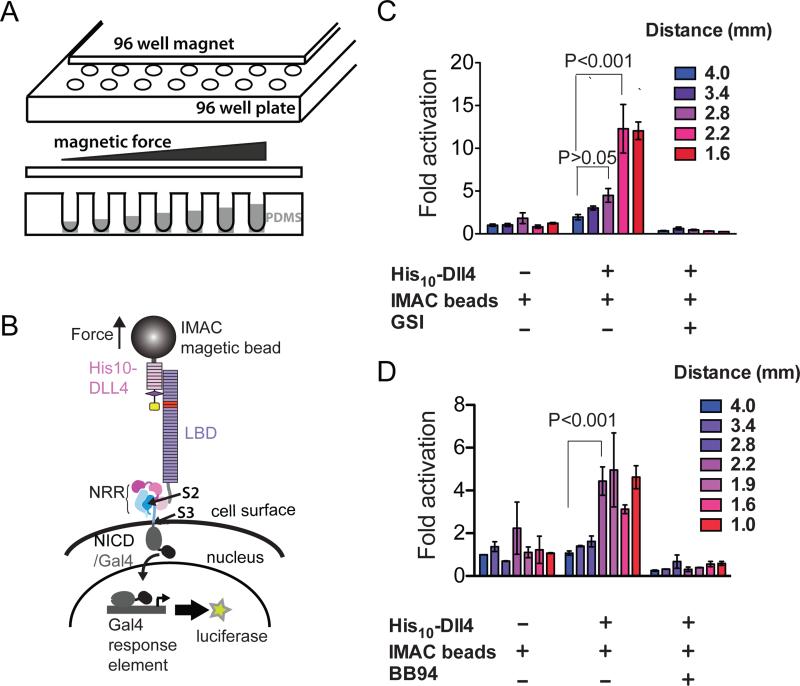
We next wished to determine whether force is required for the induction of a Notch signal when ligands bind to Notch receptors on cells. Because studies using genetically encoded or surface tethered force sensors have shown that signaling proteins such as integrins (Morimatsu et al., 2013; Wang and Ha, 2013), cadherins (Borghi et al., 2012), and vinculin (Grashoff et al., 2010) respond to applied force in the 1-10 pN range, we developed a high-throughput magnetic tweezers assay to apply a wide range of pN-scale forces to Notch receptors on the cell-surface. Our method uses magnetic tweezers in 96-well format, and applies force to cell-surface receptor molecules bound to ligands on paramagnetic beads (Figure 2A). By controlling the distance between the cells and the magnet, it is possible to vary the force applied to cells as a function of their well position on the plate. In order to present the cells at different distances from the magnet, we dispensed different amounts of PDMS polymer into the culture chambers, creating a “terraced” configuration of wells of different depths across the plate. The range of forces sampled in a given experiment is specified simply by varying the heights of the terraces, the size of the beads and the characteristics of the magnet. For example, when 1 μm beads are used and the distance of the magnet from the cells ranges from 0.15 to 0.5 mm, the applied force estimated from force calibration using phage lambda DNA ranges from 0.5 to 2.5 pN (see Figure S2 for 96-well magnet calibration).
Figure 2. Response of cell-surface Notch receptors to applied force using a multiplexed magnetic tweezers assay.
(A) Experimental design. A plate containing 96 cylindrical magnets is positioned over a 96 well plate of cells in order to apply force to magnetic beads tethered to Notch molecules on the cell-surface. The distance between the cells and the magnet is varied by using the polymer PDMS to create terraces of different heights. (B) Assay schematic. Cells expressing Notch1 receptors in which the ankyrin-repeat domain has been replaced by the Gal4 DNA-binding domain (Malecki et al., 2006) are stimulated by magnetic beads loaded with the ligand DLL4, followed by measurement of luciferase reporter gene activity. (C, D) Luciferase reporter gene activity in response to various treatments as a function of the distance from the magnet. U2OS cells expressing Notch1-Gal4 receptors were incubated with magnetic beads alone or beads loaded with the ligand DLL4 in the absence or presence of a gamma secretase inhibitor (GSI) (C) or metalloprotease inhibitor BB94 (D). Luciferase reporter gene activity is reported relative to the response of cells to beads alone at a distance of 4 mm from the magnet. Error bars represent the standard error of the mean of triplicate measurements, and statistical significance was determined with a two-way ANOVA followed by a post-hoc Bonferroni's test. 96-well magnet calibration is provided in Figure S2.
To probe the force dependence of Notch activation using this assay, we cultured cells expressing Notch1-Gal4 chimeric receptors (Malecki et al., 2006) in wells of different depths, and treated the cells with paramagnetic beads loaded with the ligand DLL4. Force was applied to the beads by placing a 96-well magnet over the plate, and luciferase reporter-gene activity was measured six hours later (Figure 2B). When cells expressing the Notch1-derived receptors are incubated with DLL4-loaded magnetic beads, a statistically significant signal is induced only when the magnet is less than or equal to 2.2 mm from the beads, and is suppressed in the presence of gamma-secretase or metalloprotease inhibitors (Figures 2C,D). [This magnet distance exerts a force of about 1.4 pN based on in vitro calibration with lambda DNA; Figure S2.] Given the many differences between the cell-based and in vitro proteolysis assays, it is not surprising that the amount of force sufficient to induce Notch proteolysis differs between the two experiments. In particular, the sustained delivery of force (over several hours) to receptors on cells combined with intrinsic protein dynamic motions promoting conformational opening likely results in irreversible capture of transiently open states by proteolysis at reduced forces, and accounts for the lower force requirement in cells. Other factors, such as the influence of the membrane or its microenvironment, the ligand-binding domain of the receptor, or the clustering of receptors in response to bead-tethered ligand, may also contribute. Regardless, the key finding is that force must be applied to bead-tethered ligands in order to induce the canonical proteolytic steps responsible for Notch activation.
Robust Notch signals in synthetic systems
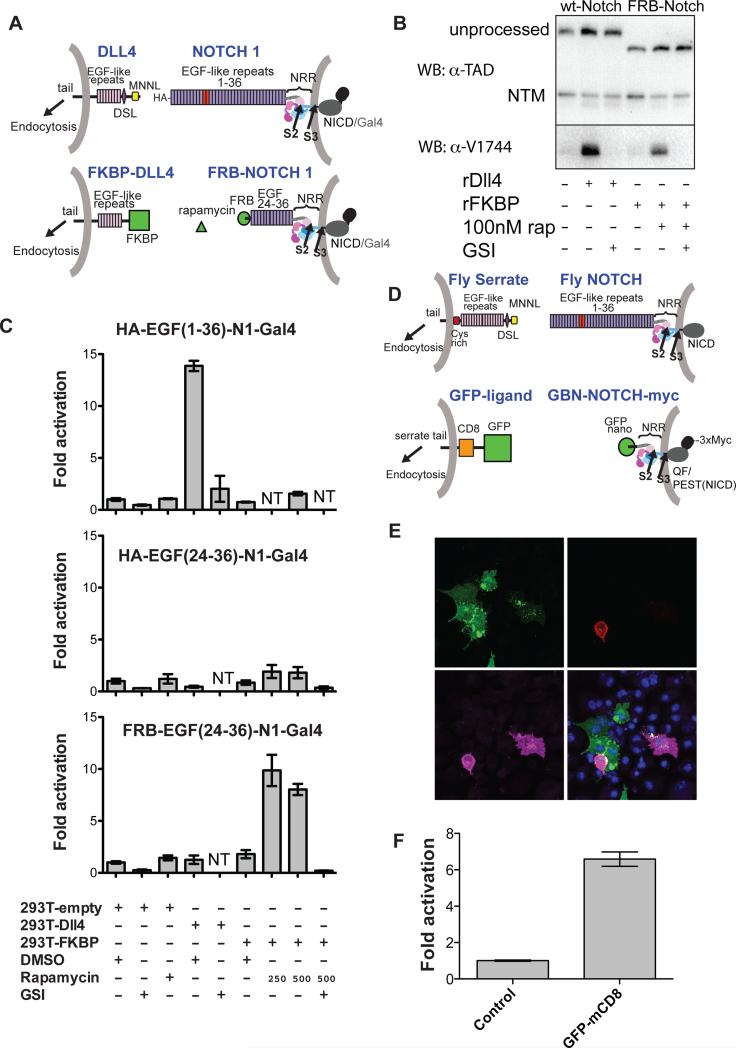
To explore whether or not a signal-sending cell can directly deliver sufficient force to induce NRR proteolysis, we created “synthetic” ligand-receptor signaling systems that substitute the native binding interaction between Notch1 and DLL4 with non-native interacting pairs to tether signal-sending and receiving cells together. These systems dispense with native interaction domains and thus eliminate the possibility that formation of a native ligand-receptor complex allosterically lowers the barrier to proteolysis of the NRR.
In the first system, we tethered sending and receiving cells using the FRB domain of mTor and the FK506 binding protein (FKBP), which interact to form a stable complex only in the presence of rapamycin (Figure 3A). The chimeric DLL4 ligand molecules substitute FKBP in place of the Notch-binding MNNL and DSL domains, but retain the rest of their extracellular region, as well as the transmembrane region and cytoplasmic tail, which contains the sequences that target the ligand for endocytosis. The Notch-derived molecules substitute the FRB domain of mTor in place of EGF-like repeats 1-23 that encompass the ligand binding region, but retain repeats 24-36, the NRR, the transmembrane region, and the Notch1-Gal4 intracellular fusion for monitoring luciferase reporter activity with a Gal4 response element as above (Figure 3A).
Figure 3. Development and evaluation of two synthetic Notch signaling systems.
(A) Schematic comparing the natural human Notch-ligand signaling system (top; EGF repeats 11-12 in red) to a synthetic signaling system placing NRR proteolysis under rapamycin-inducible control (bottom). Here, FKBP replaces the N-terminal portion of DLL4, and FRB replaces EGF-like repeats 1-23 of Notch1. The Notch1 ankyrin domain is also replaced with Gal4, as above (Malecki et al., 2006). (B) Western blots monitoring receptor proteolysis. U2OS cells stably expressing wild-type or FRB-Notch1 were grown in the presence of the DLL4 ectodomain or FKBP immobilized on plastic tissue culture dishes in the absence or presence of rapamycin (250 nM) and/or a GSI (Compound E, 400 nM). Blots were probed with an antibody directed against an epitope of intracellular Notch1 (α-TAD), or the α-V1744 antibody to S3-cleaved Notch1 (Cell Signaling). (C) Cell-based reporter gene assay. U2OS cells stably transfected with the indicated Notch variants were co-cultured with 293T cells transiently transfected with the indicated ligands. Luciferase activity for each U2OS line is reported relative to co-culture with 293T cells transfected with empty vector. Error bars reflect the standard error of readings performed in triplicate. Additional control experiments are provided in Figure S3. (D) Schematic illustrating design of a GFP - GFP-binding nanobody (GBN) synthetic ligand-receptor pair. Full-length fly Serrate and Notch are shown for reference. The artificial ligand consists of GFP, CD8 and the Serrate-derived tail. The ectodomain of the Notch-derived molecule consists of the GFP binding nanobody (GBN) and the NRR, and the intracellular domain contains the QF transcription factor, the Notch PEST domain, and a triple Myc tag. (E) Co-culture assay. S2R+ cells expressing GFP-mcd8-Ser as ligand (green, upper left panel) were co-cultured with cells expressing GBN-FlyNotch(NRR)-QF-3XMyc (GBN-N-QFMyc). Receptor is stained with anti-myc antibody (magenta, lower left panel). The tdTomato reporter signal is red (upper right panel). DNA was stained with DAPI (blue, lower right panel). (F) QAS luciferase readout of cell-mixing experiment. Luciferase reporter gene activity for the GBN-Notch cell line is reported relative to co-culture with control cells. Error bars represent the standard error of measurements performed in quadruplicate. Additional supporting data are provided in Figure S3.
We first tested the fidelity of this synthetic ligand-receptor pair and compared its signaling activity with normal ligand-receptor complexes using a well-established “plated ligand” assay, in which cells expressing receptors of interest are cultured in dishes coated with a ligand ectodomain. Plated DLL4 (or plated anti-HA) stimulates protetolytic activation of the intact HA-Notch1-Gal4 fusion protein, but not the FRB-Notch1 chimera; in contrast, plated FKBP, when in the presence of rapamycin, induces proteolytic activation of the FRB-Notch1 chimera, but not the standard Notch1-Gal4 fusion (Figure 3B, S3A).
We next tested whether this synthetic system signals in a co-culture assay, in which ligand-expressing cells are used to stimulate a signal in receptor-expressing cells (Figure 3C). Control experiments confirm that DLL4 expressing cells induce a reporter response in the cells expressing full-length HA-tagged Notch1, but not in cells expressing a truncated HA-tagged receptor, or the chimeric FRB-Notch receptor. In contrast, cells expressing the FKBP chimeric ligand only activate signaling in cells expressing the FRB-Notch chimeric receptor in a rapamycin-dependent fashion (Figure 3C). This signaling activity is sensitive to a gamma secretase inhibitor and to the metalloprotease inhibitor BB94, indicating that activating proteolysis of the NRR at S2 and subsequent S3 cleavage can be triggered in the absence of native receptor-ligand interactions. Similar results are obtained when the experiment is performed with Notch molecules lacking all 36 EGF repeats (Figure S3B).
We also created a second chimeric signaling system derived from Drosophila proteins that pairs an anti-GFP nanobody and the QF transcriptional activator under Notch NRR control with a Serrate-derived protein that substitutes GFP (followed by CD8) in place of the normal Serrate ectodomain (Figure 3D). Thus, the entire ligand binding domain of Notch and the entire Notch-binding region of the ligand have been removed. Nevertheless, this system induces expression of tomato-GFP (under control of a QF-responsive element) only in nanobody-driven responder cells that are in direct contact with GFP-expressing ligand cells (Figure 3E, S3C), and signals in co-culture assays (Figure 3F). The robust signaling observed in two synthetic systems utilizing non-native modes of protein-protein interaction shows that a pair of interacting moieties sufficient to i) bring signal sending and receiving cells into contact and ii) withstand rupture under the force required to expose S2 is all that is needed to induce NRR proteolysis and transduce a signal. Though the native ligand-receptor interaction may alter the energy landscape associated with conformational exposure of the S2 site of the NRR, the synthetic systems show conclusively that an allosteric effect of ligand binding is not necessary for S2 cleavage to occur.
Endocytosis is required for S2 site exposure
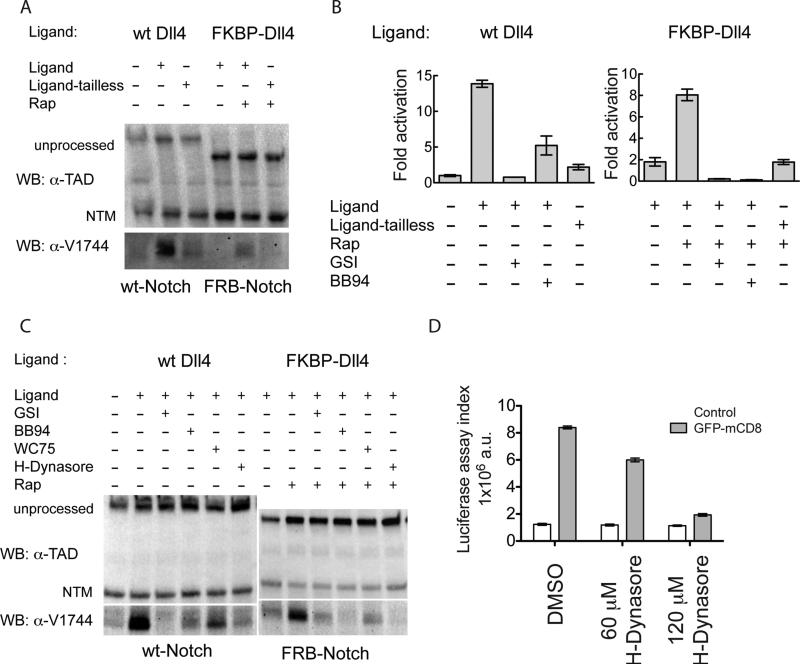
To address whether or not endocytosis of the ligand is required for proteolytic activation of the receptor, we blocked ligand endocytosis in the mammalian and rapamycin-dependent co-culture assays using two different approaches: i) deletion of the cytoplasmic tail of the ligand (which is required for endocytosis-dependent activation) and ii) treatment of ligand-expressing cells with hydroxydynasore, a small-molecule endocytosis inhibitor (McCluskey et al., 2013). The response to both interventions in the rapamycin-based synthetic signaling system mirrors that of Notch1 responding cells to DLL4-expressing signal-sending cells. Tailless ligands, which are transported to the cell surface as well as ligands with intact cytoplasmic tails (Figure S4), attenuate production of the gamma-secretase cleaved product (Figure 4A) and reporter gene expression (Figure 4B). Similarly, treatment of ligand cells with hydroxydynasore suppresses accumulation of the S3-cleaved product in both wild-type Notch-DLL4 and synthetic signaling systems (Figure 4C). Production of the S3-cleaved product is comparably attenuated in both systems by treatment with a gamma-secretase inhibitor, the metalloprotease inhibitor BB94, or the anti-Notch1 inhibitory antibody WC75, which binds specifically to the Notch1 NRR and stabilizes the autoinhibited conformation. Similar decreases in signaling activity occur upon hydroxydynasore treatment in the Drosophilia GFP nanobody-GFP synthetic system (Figure 4D). Together, these data show that endocytosis of ligands artificially tethered to receptor molecules promotes proteolytic activation of Notch signaling in a fashion that remains dependent on a conformational change in the NRR permissive for S2 and S3 cleavages. Importantly, tethering alone without ligand endocytosis is insufficient for activation. We conclude that a step dependent on ligand endocytosis is required for signal-sending cells to deliver sufficient mechanical force to the receptor to induce the proteolytic cascade responsible for receptor activation and downstream signaling events.
Figure 4.
Western blot analysis of natural and synthetic Notch receptor signaling in co-culture assays. (A) Effect of ligand-tail deletion on signaling. Wild-type and synthetic Notch receptors were co-cultured with full-length or tail-deleted cognate ligands (Ligand-tailless) and in the absence or presence of rapamycin (250 nM), as indicated. Blots were probed with an antibody directed against an epitope of intracellular Notch1 (α-TAD), or the α-V1744 antibody to S3-cleaved Notch1 (Cell Signaling). (B) Cell-based reporter gene assay probing Notch activation in co-culture experiments. 293T cells were signal-sending cells, and U2OS cells were signal-receiving cells. 293T cells were transfected with plasmids encoding wild-type Dll4, tailless Dll4, FKBP-Dll4, FKBP-Dll4-tailless or empty vector in the presence or absence of rapamycin (250 nM), Compound E (GSI, 400nM), or BB94 (20 μM). U2OS cells were transfected in 96-well format with plasmids encoding HA-Notch1-Gal4 (left), or FRB-Notch1-Gal4 (right) along with a luciferase reporter plasmid containing the Gal4 response element and an internal control plasmid expressing Renilla luciferase. 24 h after transfection, the 293T cells were added to the U2OS cells. Luciferase activity relative to the Renilla control was determined 24 h later. Fold activation is relative to U2OS cells transfected with HA-Notch1-Gal4 and co-cultured with empty-vector transfected 293T cells. Error bars represent the standard error of triplicate measurements. (C) Effect of various drug or antibody treatments on signaling by wild-type or synthetic receptors when co-cultured with cognate ligands. Blots were probed with α-TAD or the α-V1744 antibody as in (A). (D) Effect of hydroxydynasore in the fly synthetic signaling assay. Ligand expressing cells and untransfected control cells were first treated with the indicated concentration of H-Dynasore for 30 min. Receptor and ligand (or control) cells were then mixed together in a 1:5 ratio. Fresh drug was added to maintain the desired concentration, and luciferase activity was determined 6 h later. Trypan Blue staining after 10 hours of co-culture showed no difference in viability between DMSO and drug treatment (not shown). Additional control experiments are provided in Figure S4.
Discussion
The goal of these studies was to gain insight into the still elusive mechanism of ligand-induced proteolysis of Notch receptors. Previous X-ray structures of the Notch NRR “activation switch” show that a major conformational change must occur in order to unmask the solvent inaccessible S2 processing site for metalloprotease cleavage. A leading model (Parks et al., 2000) proposes that endocytosis of ligands applies a pulling force to bound Notch receptors, thereby exposing the S2 proteolytic site. In this model, the NRR would then be a mechanosensitive switch responding to this pulling force. A number of indirect lines of evidence are consistent with the mechanotransduction model, and the requirement for a specialized pathway for endocytosis of ligands in the signal-sending cells is well established (Musse et al., 2012).
As appealing as a mechanotransduction model might be, however, it has remained unclear i) whether or not the force required to induce proteolytic sensitivity in vitro or in vivo lies in a physiologically accessible force regime, ii) whether allostery is required to lower the barrier to proteolysis, and iii) whether or not the force is delivered by ligand endocytosis. AFM studies in which the Notch2 NRR was pulled under high loading rates showed that multiple unfolding transitions occur in the 100 pN range, but how these findings relate to a physiological context is unclear (Stephenson and Avis, 2012). Studies using plated ligands conjugated to tension-gated tethers (TGT), which sense forces imposed on cellular receptors based on rupturing short DNA duplexes, led to the conclusion that Notch activation occurs at forces under 12 pN, but could not establish whether or not applied force was needed at all because of the limits of DNA duplexes as force sensors (Wang and Ha, 2013). The use of plate-bound ligands as activators also does not address whether or not signal-sending cells are capable of supplying an activating force.
Here, we developed assays to determine how the proteolytic sensitivity of site S2 varies as a function of applied force both in vitro and in cells under physiologically relevant conditions. The in vitro magnetic tweezers assay revealed that the isolated activation switch undergoes a transition from protease resistance to sensitivity between 3.5 and 5.4 pN of force. Typical rate constants of cleavage were roughly 6 × 103 M−1s−1, in line with reported catalytic efficiencies for the isolated metalloprotease domain of Adam17 (Caescu et al., 2009). Cell-surface Notch1 receptors also exhibit mechanosensitivity upon application of force via ligand tethered magnetic beads, as the presence of ligand-coated beads alone is not sufficient to induce Notch activation.
The low forces required to relieve Notch autoinhibition show that the NRR is a highly mechanosensitive switch. How do these forces compare to the forces required for unfolding of other mechanosensors, or for other biological force-dependent events? The A2 domain of von Willebrand Factor, a mechanosensor that undergoes proteolysis in response to shear stress, unfolds with a transition at 8 pN of force (Zhang et al., 2009). Similarly, the binding of vinculin to talin relies on unfolding in the talin R3 domain over a force range of ~2-5 pN (del Rio et al., 2009; Yao et al., 2014). In addition, the force required for S2 exposure is comparable to the 3-4 pN force generated by a myosin motor taking a step on actin (Finer et al., 1994) and stall forces measured for kinesin (4-6 pN) and dynein (1 pN) (Blehm et al., 2013). Importantly, the force required to relieve autoinhibition of the activation switch is lower than the forces measured by optical tweezers to rupture ligand-receptor interactions and in line with the measured stall force generated by endocytosis of DLL1 (Meloty-Kapella et al., 2012; Shergill et al., 2012) and with the force experienced by EGFR during endocytosis (Stabley et al., 2011). Our data using synthetic signaling systems now show that allostery is not required to render the NRR sensitive to proteolytic activation. Moreover, these experiments directly link ligand-receptor engagement to proteolytic site exposure of the NRR in a step that depends on ligand endocytosis, though whether or not endocytosis itself supplies the pulling force remains to be determined.
Our studies investigating the responsiveness of the Notch1 NRR to force also raise a number of new questions about the mechanosensitive behavior of Notch receptors. What degree of domain movement is required to relieve autoinhibition? Is the barrier to mechanical exposure of the metalloprotease site in Notch1 influenced by the EGF-repeat region, or is the mechanosensitive property of the entire receptor completely encoded within the NRR? How do lateral interactions among Notch receptors in the membrane affect receptor mechanosensitivity? And how does the intrinsic sensitivity to force vary among the various Notch receptors, both in isolation, in response to disease-associated mutations, different ligands, or mechanical forces generated in the cellular microenvironment (e.g. by blood flow or muscle contraction)?
The methods developed here to investigate the role of Notch signaling should have wide utility for exploring the consequences of Notch signal transduction under precise chemical and temporal control and for investigation of other mechanosensitive processes in biology. The synthetic GFP-nanobody and rapamycin-dependent signaling systems open up new possibilities for controlling and reporting on Notch activation in a defined cellular context. The approaches can be used to investigate the kinetics of metalloprotease recruitment, receptor proteolysis, as well as events downstream of receptor cleavage. The assays can also report on whether or not two cells contact each other in vivo. Finally, the cell-based magnetic tweezers assay should facilitate new studies of other biological processes that may rely on mechanical force for the induction of signaling, such as ephrin-ephrin receptor signaling (Salaita et al., 2010), atypical cadherin complexes of the inner ear (Sotomayor et al., 2012), and other transmembrane signaling events.
Experimental Procedures
Materials
A complete description of constructs, recombinant proteins, and cell lines is provided in the Supplemental Experimental Procedures.
Single-Molecule Magnetic Tweezer Experiments
Briefly (see Supplemental Experimental Procedures), single-molecule experiments were performed using custom microfluidic flow cells with glass coverslips as described previously (Tanner and van Oijen, 2010). Two stacked 6 mm cube magnets were attached to a mount containing a micrometer in order to control the distance from the magnet to the flow cell. Biotinylated NRRs or peptides are delivered into the flowcell with a syringe pump and captured in the flowcell with streptavidin. After delivery of magnetic beads followed by extensive washing, buffer with Adam17 (1 μM), and ZnCl2 (4 μM) was added. Movies were recorded using Metavue or MicroManager in one-second increments for up to 30 minutes. The total number of beads in each frame (10× objective) was counted using a built-in algorithm in ImageJ. For NRR experiments, Adam17 was loaded into the flow cell at ~1 pN force, and the magnet subsequently lowered to the appropriate distance corresponding to the desired applied force. The magnet calibration is described in Supplemental Experimental Procedures.
96-well magnetic tweezer assays
PDMS Components A and B (Sylgard 184, Dow Corning) were added to a 50 ml falcon tube in a ratio of 10:1, and were mixed by slow rotation over 30 minutes. The mixture was centrifuged at 4000 g for 5 min, and then dispensed with an Eppendoff digital repeat pipetter using the slowest setting to ensure reproducible dispensing. The PDMS was dispensed into 96 well TC-coated plates in volume “steps” from 40-120 μl, and was cured overnight at 37 C. Before cells were plated, the wells were bathed in 70 μl of fibronectin (10 μg/mL in PBS; Sigma) for 1 hour at 37 C. U2OS cell lines stably expressing Flag-Notch1-Gal4 were then reverse transfected with luciferase reporter plasmids as above, treated with 1 μM doxycycline to induce protein expression, and plated onto the PDMS-modified wells. After 24-48 hr, cells were incubated in DMEM with or without 500 nM recombinant Dll4 ectodomain (R&D Systems). After 20 min, an excess of 1 μm IMAC magnetic beads in DMEM (Dynal) was added, the plate with the 96-well configuration of magnets was placed over the cells (Alpaqua), and the level of luciferase reporter activity was determined six hours later using a Promega Dual Luciferase kit.
Chimeric Notch/ligand experiments
Co-culture experiments, human cell lines. On Day 1, Notch1-Gal4 fusion constructs and reporter plasmids were reverse transfected into U2OS cells in 96 well format as above. Ligand molecules were reverse transfected separately into 293T cells in 6 well plates (2 μg ligand/well) using Lipofectamine 2000. On Day 2, ligand-transfected cells were resuspended in fresh DMEM with 10% FBS, drugs were added as indicated, and the 293T cells were plated on top of the Notch-expressing cells. On day 3, the luciferase reporter activity was determined as above.
Co-culture experiments, Drosophila cell lines. On day one, S2R+ cells were transfected in six-well dishes with 400 ng total DNA/well using Effectene Transfection Reagent (QIAGEN). Receptor positive cells were generated by transfection of 396 ng DNA of QUAT::tdTomato and 4 ng ubi::GBN-flyNotch(NRR)-QF-3XMyc. Ligand positive cells were generated by transfection of 100 ng Actin::Gal4 together with 300 ng UAST::GFP-mcd8-Ser or UAS-GFP-mcd8-Dl. On day three, receptor and ligand positive cells were each washed in fresh culture medium to remove transfection reagents and dislodged from dishes by pipetting. Half of the receptor positive cells were mixed with ligand positive cells, while the other half were mixed with the same number of S2R+ cells without transfection (control). The cell mixture was transferred into a 1.5 ml Eppendorf tube and slowly rotated at room temperature for 1hr to allow the ligand and receptor positive cells to bind to each other. Then the cell mixture was plated back into a new 6-well dish and cultured for one additional day before assay. For immunofluorescence imaging, the cell mixture was plated on cover-glass bottom chamber slides (Lab-Tek) coated with Concanavalin-A. Cells were fixed in 4% formaldehyde, stained with mouse anti-Myc antibody (1:400, 9E10, Santa Cruz Biotech) followed by Alexa 647 goat anti-mouse IgG (1:500, Invitrogen), and observed after mounting in a Zeiss LSM 780 confocal microscope using a 63X/N.A. 1.4 oil objective. Western blot methods are provided in Supplemental Experimental Procedures.
Statistical analysis of reporter assays. Error bars in reporter assays represent SEM of triplicate or quadruplicate measurement. Statistical analysis to assess significance (p values) was performed with GraphPad Prism software using two-way analysis of variance (ANOVA) followed by a Bonferroni post-hoc test.
Supplementary Material
Highlights.
- Developed single molecule assays to monitor Notch proteolysis under force
- Exposure of the Notch1 masked proteolytic site occurs in a physiological force regime
- Dll4-induced activation of Notch1 requires the application of mechanical force
- Non-native tethering can substitute for receptor-ligand complexes in signaling
Acknowledgements
We thank Benedikt Bauer and Anthony Nguyen for help in optimizing single molecule experiments and Adam17 expression, Scott Ficarro for mass spectrometry, and Kelly Arnett and Brian McMillan for helpful discussions. WRG was supported by an AHA SDG grant. LH was supported by a Damon Runyon Postdoctoral Fellowship. This work was supported by NIH grants P01 CA119070 (JJL, JCA, and SCB), R01 CA092433 (JCA and SCB), HHMI, and the NIH (NP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blehm BH, Schroer TA, Trybus KM, Chemla YR, Selvin PR. In vivo optical trapping indicates kinesin's stall force is reduced by dynein during intracellular transport. Proceedings of the National Academy of Sciences. 2013;110:3381–3386. doi: 10.1073/pnas.1219961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proceedings of the National Academy of Sciences. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israël A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Caescu CI, Jeschke GR, Turk BE. Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem J. 2009;424:79–88. doi: 10.1042/BJ20090549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science (New York, NY) 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113:4381–4390. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey A, Daniel JA, Hadzic G, Chau N, Clayton EL, Mariana A, Whiting A, Gorgani NN, Lloyd J, Quan A, et al. Building a Better Dynasore: The Dyngo Compounds Potently Inhibit Dynamin and Endocytosis. Traffic. 2013;14:1272–1289. doi: 10.1111/tra.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloty-Kapella L, Shergill B, Kuon J, Botvinick E, Weinmaster G. Notch Ligand Endocytosis Generates Mechanical Pulling Force Dependent on Dynamin, Epsins, and Actin. Dev Cell. 2012;22:1299–1312. doi: 10.1016/j.devcel.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 2013;13:3985–3989. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Musse AA, Meloty-Kapella L, Weinmaster G. Notch ligand endocytosis: Mechanistic basis of signaling activity. Seminars in Cell & Developmental Biology. 2012 doi: 10.1016/j.semcdb.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols JT, Miyamoto A, Olsen SL, D'Souza B, Yao C, Weinmaster G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007;176:445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Salaita K, Nair PM, Petit RS, Neve RM, Das D, Gray JW, Groves JT. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science (New York, NY) 2010;327:1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shergill B, Meloty-Kapella L, Musse AA, Weinmaster G, Botvinick E. Optical Tweezers Studies on Notch: Single-Molecule Interaction Strength Is Independent of Ligand Endocytosis. Dev Cell. 2012;22:1313–1320. doi: 10.1016/j.devcel.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotomayor M, Weihofen WA, Gaudet R, Corey DP. Structure of a force-conveying cadherin bond essential for inner-ear mechanotransduction. Nature. 2012;492:128–132. doi: 10.1038/nature11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabley DR, Jurchenko C, Marshall SS, Salaita KS. Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nat Meth. 2011;9:64–67. doi: 10.1038/nmeth.1747. [DOI] [PubMed] [Google Scholar]
- Stephenson NL, Avis JM. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proceedings of the National Academy of Sciences. 2012;109:E2757–E2765. doi: 10.1073/pnas.1205788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner NA, van Oijen AM. Visualizing DNA replication at the single-molecule level. Meth Enzymol. 2010;475:259–278. doi: 10.1016/S0076-6879(10)75011-4. [DOI] [PubMed] [Google Scholar]
- Wang X, Ha T. Defining Single Molecular Forces Required to Activate Integrin and Notch Signaling. Science (New York, NY) 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science (New York, NY) 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Yao M, Goult BT, Chen H, Cong P, Sheetz MP, Yan J. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Halvorsen K, Zhang C-Z, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science (New York, NY) 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.