Background: Lysophospholipids activate P-type plasma membrane H+-ATPase proton pump by an unknown mechanism.
Results: In contrast to fungal plasma membrane H+-ATPase, plant H+-ATPase is activated by lysophospholipids directly via a mechanism involving both terminal domains of the pump.
Conclusion: Plant plasma membrane H+-ATPase responds specifically to lysophospholipids.
Significance: Lysophospholipids may function in plant signaling.
Keywords: H+-ATPase, lysophospholipid, plasma membrane, post-transcriptional regulation, proton pump
Abstract
Eukaryotic P-type plasma membrane H+-ATPases are primary active transport systems that are regulated at the post-translation level by cis-acting autoinhibitory domains, which can be relieved by protein kinase-mediated phosphorylation or binding of specific lipid species. Here we show that lysophospholipids specifically activate a plant plasma membrane H+-ATPase (Arabidopsis thaliana AHA2) by a mechanism that involves both cytoplasmic terminal domains of AHA2, whereas they have no effect on the fungal counterpart (Saccharomyces cerevisiae Pma1p). The activation was dependent on the glycerol backbone of the lysophospholipid and increased with acyl chain length, whereas the headgroup had little effect on activation. Activation of the plant pump by lysophospholipids did not involve the penultimate residue, Thr-947, which is known to be phosphorylated as part of a binding site for activating 14-3-3 protein, but was critically dependent on a single autoinhibitory residue (Leu-919) upstream of the C-terminal cytoplasmic domain in AHA2. A corresponding residue is absent in the fungal counterpart. These data indicate that plant plasma membrane H+-ATPases evolved as specific receptors for lysophospholipids and support the hypothesis that lysophospholipids are important plant signaling molecules.
Introduction
Plasma membrane (PM)3 H+-ATPases of fungi and plants generate the electrochemical proton-motive force across the PM that energizes the secondary active transport of nutrients across the membrane (1). PM H+-ATPases belong to a large family of ion transport proteins termed P-type ATPases, which include ion pumps, such as animal Na+/K+-ATPases and Ca2+-ATPases (2, 3). Common features of the catalytic machinery of P-type ATPases are a transmembrane ion-binding domain and three cytoplasmic domains, including a phosphorylation domain, a nucleotide-binding domain, and an actuator domain. Phosphorylation and dephosphorylation of a conserved aspartate residue within the phosphorylation domain trigger movements of the cytoplasmic domains that are coupled with the opening and closing of discrete ion-binding sites within the membrane and ion translocation across the membrane.
The activities of many P-type ATPases are tightly regulated by accessory inhibitory proteins or alternatively cis-acting autoinhibitory domains. For instance, the P2A sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) is inhibited by the accessory proteins phospholamban and sarcolipin, small membrane proteins with a single transmembrane segment (4). Sarcolipin uncouples the SERCA pump (5) by trapping it in the Ca2+-binding conformation (6, 7). Autoinhibition by a regulatory domain is observed in a closely related family of P-type ATPases, the P2B calmodulin-activated Ca2+-ATPases, which occur in both plants and animals (8–10). These ATPases possess an autoinhibitory domain that interacts with calmodulin to relieve autoinhibition.
In yeast and plant PM H+-ATPases, pump activation by loss of autoinhibition is induced by distinct extracellular stimuli, such as changes in carbon source availability (11, 12) and in plants blue light (13) and pathogens (14, 15). In plant PM H+-ATPases, both the N and C termini constitute autoinhibitory domains, but the C-terminal regulatory domain is more extensive (16, 17). Activation following neutralization of this domain causes a change in the coupling ratio of the pump with a large increase in proton pumping and a small increase in ATP hydrolytic activity (13, 16, 18, 19). Furthermore, the activated pump has an increased Vmax, an increased affinity for ATP, and a shift in pH optimum from slightly acidic to neutral, which suggests an increased apparent H+ affinity (20).
The C-terminal domain of the PM H+-ATPase AHA2 from the model plant Arabidopsis thaliana is divided into two assumed autoinhibitory segments of predicted α-helical nature denoted as Region I between ∼80 and 60 residues from the terminus and Region II between ∼50 and 30 residues from the terminus (21). Studies of the related Nicotiana plumbaginifolia (tobacco) pump PMA2 confirmed this intricate organization of the C-terminal regulatory domain (22). Transformation of the PM H+-ATPase molecule from the low to the high activity state in vivo is proposed to involve structural rearrangement of the terminal domains without the need for their actual separation from the rest of the protein. A well described mechanism for the in vivo activation of the PM H+-ATPase protein involves phosphorylation of the C-terminal threonine residue (Thr-947 in the penultimate position of AHA2), which allows for 14-3-3 binding (23–25). Several additional phosphorylation sites in the C-terminal domain have been identified that either activate or inhibit the enzyme independently of 14-3-3 binding (26). The structure of a C-terminal truncated form of Arabidopsis AHA2 has been solved to 3.6 Å (27), and the co-crystal structure of the 52-residue C-terminal fragment of tobacco PMA2 in complex with 14-3-3 protein has been published (27, 28). However, these structures do not reveal how the terminal domains regulate pump function.
Phospholipids are important regulators of P-type ATPases. PM Ca2+-ATPases are strongly activated by acidic phospholipids (29) and contain a binding site for acidic phospholipids that interfere with regulation by a calmodulin-binding terminal domain (30–32). A phospholipid-binding site is apparent in crystal structures of SERCA (33–35), and it has been suggested that this site is conserved among distantly related P-type ATPases (36). Lysophosphatidylcholine (lyso-PC) is a detergent-like molecule produced from cleavage of phosphatidylcholine by the enzyme phospholipase A2 and has been proposed to serve as a signaling molecule in plants (37–39). Plant PM H+-ATPase is strongly activated by micromolar concentrations of lyso-PC, which cannot be explained by the unmasking of latent ATP-binding sites alone (40, 41). However, the specificity and mechanism of activation are unknown.
A long term goal of our research is to determine the full-length structure of PM H+-ATPase in its autoinhibited state. To purify membrane protein for crystallization trials, it is essential to solubilize the protein from its native membrane using detergents. During this work, we observed a partial activation of the plant PM H+-ATPase by some detergents but none that was as strong and with such a clear threshold as lysophospholipids. Strikingly, lysophospholipids had no effect on a fungal counterpart of the plant pump, Saccharomyces cerevisiae Pma1p. The effect of lysophospholipids on the plant pump was direct as it did not involve phosphorylation of Thr-947 and was completely abolished by substitution of a single hydrophobic residue in the C terminus of the pump. The identification of the plant PM H+-ATPase as a lysophospholipid sensor supports the hypothesis that signaling lipids are involved in regulating PM electrochemical gradients in plants.
Experimental Procedures
Chemicals
For the detergent screen, DSOL-MK was acquired from Anatrace, Maumee, OH (a list of detergents used can be found in the product literature). Dioleoylphosphatidylcholine was acquired from Avanti Polar Lipids, Alabaster, AL. 1-Hexadecanoyl-sn-glycero-3-phosphoglycerol (lyso-PG), 1-dodecanoyl-sn-glycerol-3-phosphocholine (lyso-PC 12:0) and 1-tetradecanoyl-sn-glycerol-3-phosphocholine (lyso-PC 14:0) were from Anatrace. 1-Hexadecanoyl-sn-glycero-3-phosphocholine (lyso-PC 16:0), 1-hexadecanoyl-sn-glycero-3-phosphate, 1-hexadecanoyl-sn-glycero-3-phospho-l-serine, 1-hexadecyl-2-hydroxy-sn-glycero-3-phosphocholine, hexadecylphosphocholine (miltefosine), and 1-octadecyl-2-O-methyl-glycero-3-phosphocholine (edelfosine) were acquired from Avanti Polar Lipids. n-Dodecyl β-d-maltoside (DDM) was acquired from Glycon GMBH, Luckenwalde, Germany. All other chemicals were acquired from Sigma-Aldrich.
Strains and Plasmids Used
The RS72 yeast strain (42), which harbors the native PM H+-ATPase, PMA1, under the control of a galactose promoter was used. Plasmids containing AHA2 under the control of the PMA1 promoter were described previously (17, 20, 43). The pma1,pma2 knock-out strain of S. cerevisiae YAK2 (MAT, ade2–101, leu2Δ1, his3-Δ200, ura3–52, trp1Δ63, lys2–801 pma1Δ::HIS3, pma2-Δ::TRP1; Ref. 44) was used to express native PMA1 from the same expression plasmid as that used for AHA2. Glucose-metabolizing cells producing yeast PM H+-ATPases in the activated state and glucose-starved cells producing PM H+-ATPases in the basal state were prepared as described (45). Plasmids carrying AHA2 with deletions corresponding to 20 or 25 amino acids in the C terminus as well as single or double point mutations were generated by standard procedures using polymerase chain reactions and verified by DNA sequencing. Other constructs were as described previously (17, 20, 43). Strains were transformed as described (43).
Complementation Assay
The ability of plant PM H+-ATPase variants to complement the deficiency in PMA1 during growth on glucose was performed as described previously (21).
Isolation of Membranes
Total membranes expressing AHA2 PM H+-ATPase were isolated as described previously (28). Yeast microsomal membranes in both activation states were isolated as described previously (28) except that 2 mm sodium molybdate was present in all buffers used for purification to inhibit phosphatases. Yeast plasma membranes were isolated as described (11).
SDS-PAGE
Sodium dodecylsulfate polyacrylamide gel electrophoresis was performed according to standard techniques.
Treatment with Detergent
Membranes were diluted to 10 mg/ml in a buffer (G20MKED) containing 20% glycerol, 50 mm MES-KOH (pH 6.5), 50 mm KCl, 1 mm EDTA, 1 mm DTT, 2 μg/ml pepstatin A, and 0.2 μm phenylmethylsulfonyl fluoride (PMSF) and mixed with an equal volume of G20MKED with typically 3% detergent, corresponding to a 3:1 mass ratio of detergent to protein. For titrations with DDM, the concentration of detergent varied from 0.08 to 3%. The samples were incubated for 30 min at 4 °C with agitation. For the ATPase assay, the mixture was diluted in G20MKED and incubated with an equal volume containing a 2.5 mass ratio of dioleoylphosphatidylcholine in G20MKED for 8 min immediately before use. Solubilization was assessed by ultracentrifugation at 150,000 × g. The supernatant was removed, the pellet was resuspended in an equal volume of G20MKED, and both fractions were assayed by SDS-PAGE. The activity of the solubilized fractions was assayed as described below.
ATPase Assay
ATPase activity was determined essentially as described previously (20) with the following modifications. The assay was performed in a final volume of 60 μl in a 96-well microplate and typically contained 0.2–0.4 μg of membranes. Each assay was performed in triplicate or quadruplicate. Absorbance was measured using a SpectraMax M5 microplate reader. The final reaction mixture contained 20 mm MOPS adjusted to pH 7.0 with N-methyl-d-glucamine or 20 mm MES adjusted to pH 5.9 for yeast Pma1p, 5 mm NaN3, 0.25 mm NaMoO4, 25 mm KNO3, 8 mm MgSO4, 2 mm phosphoenolpyruvate, 0.7 milliunit/μl pyruvate kinase from rabbit muscle (Sigma-Aldrich), and 3 mm ATP. To determine ATP kinetics, ATP concentrations were varied between 0.085 and 3 mm. Lysophospholipids were assayed at concentrations ranging from 3 to 800 μm. To evaluate the effect of DDM and lyso-PCs on ATP kinetics, the protein was preincubated for 30 min at 4 °C with dioleoylphosphatidylcholine and with or without 1.5% DDM as described above. Then 0 or 25 μm lyso-PC 16:0 (final concentration) was added directly before the start of the assay, and the same ATP concentrations as above were used.
Protein Determination
Protein concentrations were estimated using the Bradford method (46) with bovine serum albumin as standard.
Statistical Analysis
All figures were generated using GraphPad Prism® 6.0 and analyzed using non-linear regression Michaelis-Menten kinetic tools. The standard errors are calculated as S.E.
Results
Effect of Detergents on the Activation State of AHA2
For structural studies of full-length PM H+-ATPases with correct positioning of autoinhibitory domains, it is essential that the low affinity state of the pump be preserved during protein purification. An important step in membrane protein purification is solubilization with detergents. We therefore tested whether the solubilization procedure as such affected the activation parameters of the PM H+-ATPase AHA2. To identify detergents that do not influence the activity state of AHA2, we evaluated the effect of various detergents on the activation and solubilization state of AHA2. Eighty detergents were tested, and of those, 19 were able to solubilize AHA2 without denaturing it (data not shown). All solubilizing detergents were found to increase both Vmax and ATP affinity (i.e. lowered the Km for ATP) of AHA2. Among the 19 solubilizing detergents, 18 had a relatively modest effect on the ATPase activity of AHA2 (as exemplified by DDM in Table 1 and Fig. 1, A and B), whereas lyso-PC stood out as having a potent effect with a marked threshold level (Table 1 and Fig. 1).
TABLE 1.
Kinetic parameters for the plasma membrane H+-ATPase AHA2 and derived deletion mutants measured at 30 °C in the presence and absence of DDM and lyso-PC, respectively (n = 3–5 biological replicates; ±S.E.)
| DDM | Lyso-PC | Specific activity at 3 mm ATPa | Vmaxb | Km(ATP) | |
|---|---|---|---|---|---|
| μmol Pi/mg/min | % | mm | |||
| AHA2 | − | − | 0.87 ± 0.06 | 100 | 1.20 ± 0.10 |
| + | − | 1.46 ± 0.03 | 127 ± 14 | 0.51 ± 0.11 | |
| − | + | 3.37 ± 0.27 | 287 ± 6.5 | 0.23 ± 0.01 | |
| + | + | 2.78 ± 0.19 | 240 ± 5.2 | 0.25 ± 0.01 | |
| aha2ΔN10 | − | − | 1.54 ± 0.04 | 100 | 0.71 ± 0.02 |
| + | − | 0.97 ± 0.02 | 51 ± 2.2 | 0.32 ± 0.03 | |
| − | + | 1.57 ± 0.02 | 82 ± 1.9 | 0.21 ± 0.01 | |
| + | + | 0.95 ± 0,01 | 50 ± 1.3 | 0.23 ± 0.02 | |
| aha2ΔC92 | − | − | 1.07 ± 0.05 | 100 | 0.13 ± 0.01 |
| + | − | 0.43 ± 0.01 | 29 ± 0.6 | 0.17 ± 0.01 | |
| − | + | 1.18 ± 0.05 | 118 ± 1.2 | 0.18 ± 0.01 | |
| + | + | 0.46 ± 0.01 | 34 ± 0.5 | 0.20 ± 0.01 |
a Specific activity between deletions cannot be compared because expression level may vary.
b 100% is defined as Vmax without DDM and lyso-PC present for given construct.
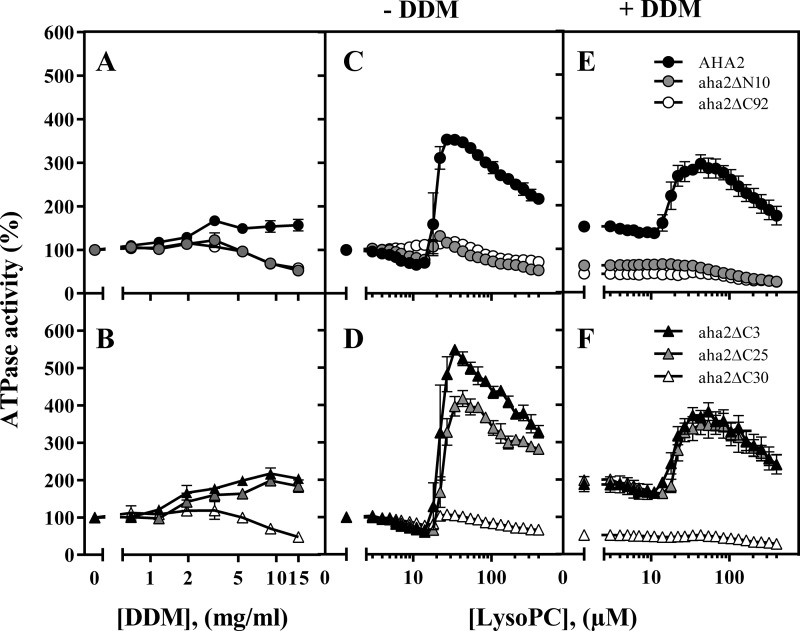
FIGURE 1.
Activation of the PM H+-ATPase by DDM and lyso-PC 16:0 depends on the presence of intact terminal domains. The ATPase activities of membranes isolated from cells expressing either WT AHA2 (●); a variant with an N-terminal truncation (aha2ΔN10; gray circles); a variant with a C-terminal truncation encompassing the entire regulatory domain (aha2Δ92; ○); or variants with three, 25, or 30 amino acids removed from the C terminus (aha2Δ3/25/30; ▴, gray triangles, and ▵, respectively) were analyzed with increasing concentrations of DDM (A and B) or lyso-PC (C, D, E, and F) ±1.5% DDM. Concentrations of DDM refer to concentrations in the preincubation mixture with a membrane protein concentration of 5 mg/ml. The mixture was diluted to 0.4 mg/ml protein and mixed 1:1 with 1 mg/ml dioleoylphosphatidylcholine before assaying the ATPase activity at 30 °C with 3 mm ATP. LysoPC refers to the concentration directly in the assay. An activity of 100% corresponds to the specific activity at 3 mm ATP without lyso-PC (see Table 2 for quantification) (n = 3–5 biological replicates; ±S.E. (error bars)).
Effect of DDM on AHA2 Activity
To learn more about the effect of detergents on the plant PM H+-ATPase, we used DDM as a representative for the group of detergents with a modest effect on the kinetic properties of AHA2. DDM treatment specifically increased the ATP hydrolytic activity of membranes expressing the full-length enzyme by ∼50% (Fig. 1A). The effect peaked at a DDM concentration of ∼3 mg/ml, corresponding to a detergent to protein ratio of 3:5, and remained stable to a concentration of 15 mg/ml, corresponding to a 3:1 ratio.
As the observed detergent effect could be indirect (for instance, the result of unmasking latent ATP-binding sites and/or increasing fluidity of the membranes), we tested the effect of DDM on N- and C-terminally truncated versions of AHA2 reported previously to be in an activated state (17, 20). If DDM affects AHA2 activity indirectly, then we would expect these mutants to be equally affected and thus activated further by DDM. However, truncated mutants lacking 10 N-terminal and 92 C-terminal residues, respectively, did not respond with an increase in activity to a DDM concentration of 3 mg/ml, and at higher concentrations, the activity decreased (Fig. 1A). This suggests that DDM has at least two effects on AHA2, namely, a stimulatory effect, which only occurs when both autoinhibitory termini are intact, and an inhibitory effect, which likely results from delipidation of the protein, because it affects all variants equally.
We subsequently assayed the ability of DDM to stimulate mutant versions of AHA2 carrying shorter truncations at the C terminus (Fig. 1B). Deletion of the three C-terminal-most amino acid residues removes the conserved threonine residue (Thr-947) that when phosphorylated forms the 14-3-3 binding motif. This mutant responded to DDM in the same way as the wild type (Fig. 1B), suggesting that the activation is direct and not mediated by protein kinase-mediated phosphorylation and subsequent binding of 14-3-3 protein. The stimulatory effect of DDM on AHA2 activity was still evident following deletion of 25 amino acids from the C terminus (aha2ΔC25; Fig. 1B), but surprisingly, removal of an additional five residues (to produce aha2ΔC30) completely abolished the stimulatory effect of DDM (Fig. 1B). This pointed to the presence of specific amino acid residues in the C terminus of AHA2 that were essential for the detergent effect.
Effect of Lysophosphatidylcholine on the Activity State of AHA2
To compare the effect of DDM with that of lyso-PC, we next tested the effect of lyso-PC on AHA2 at a broader concentration range than tested previously (40, 41). The ATPase activity decreased slightly as the concentration of lyso-PC increased until a sharp increase occurred at ∼25–30 μm lyso-PC, corresponding to a concentration of ∼4 nmol of lyso-PC/μg of membrane protein. Lyso-PC treatment increased the specific ATP hydrolytic activity of membranes expressing AHA2 by ∼400% (Fig. 1C), which is more than 6 times that observed for DDM. At higher concentrations, we observed a successive decrease in activity, consistent with the role of lyso-PC as a detergent, as higher concentrations could be expected to delipidate the protein.
Strikingly, the pump lacking either its full C terminus or the outermost part of the N terminus (aha2ΔC92 and aha2ΔN10, respectively) completely lost its ability to respond to lyso-PC (Fig. 1C). Whereas a mutant lacking 25 amino acids from the C terminus (aha2ΔC25; Fig. 1D) could be activated by lyso-PC, the effect was abolished in a mutant lacking a further five C-terminal residues (aha2ΔC30; Fig. 1D). Notably, it was the same stretch of five amino acid residues that appeared essential for the effects of both DDM and lyso-PC.
Another interesting observation was that a mutant lacking only three amino acids from its C terminus responded more strongly (550%) to lyso-PC than did wild-type AHA2 (400%; Fig. 1C). A similar pronounced effect was seen after mutation of the penultimate residue (Thr-947) (Fig. 2, A–D). This result is in line with the finding that a subset of the AHA2 expressed in yeast is post-translationally activated in the heterologous host by phosphorylation at Thr-947 (43). Accordingly, when this residue is deleted, a larger population of recombinant AHA2 is present in the basal state, and the population as a whole can respond more strongly to lyso-PC.
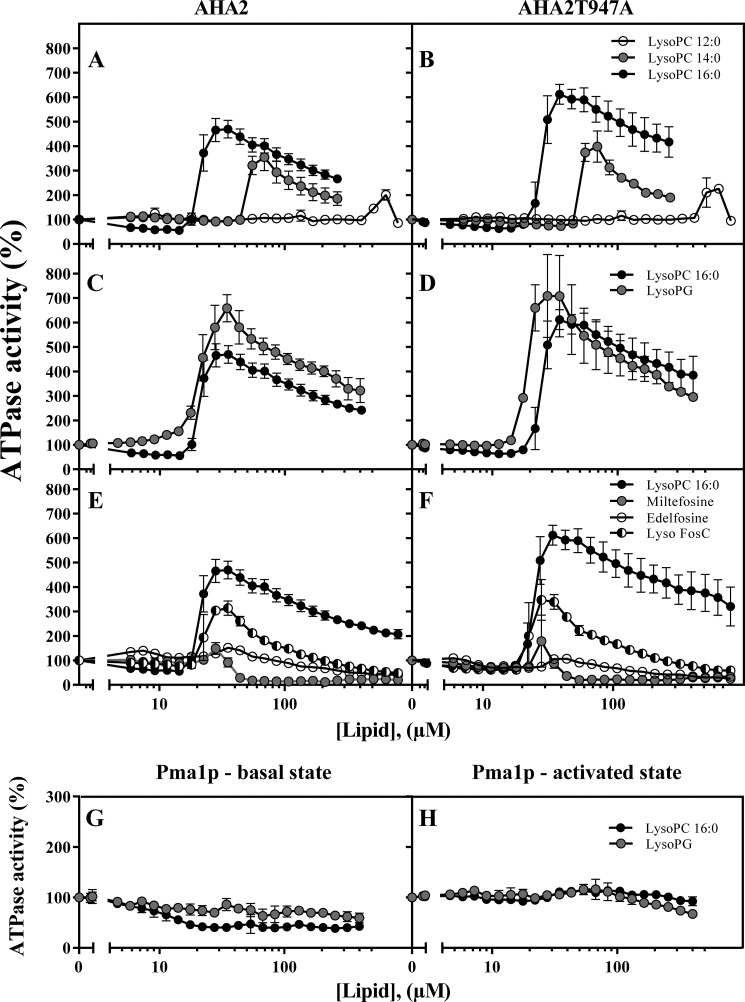
FIGURE 2.
Lysophospholipid activation of PM H+-ATPase. The ability of lysophospholipids and their analogues to activate the ATP hydrolytic activity of the wild-type plant PM H+-ATPase AHA2 (A, C, and E), a T947A mutant (B, D, and F), and the yeast plasma membrane PM H+-ATPase Pma1p (G and H) was tested at 3 mm ATP. For wild-type AHA2 and the T947A mutant, 0.3 μg of microsomal membrane protein was used per sample, and for Pma1p, 0.15 μg of protein from purified yeast plasma membranes expressing Pma1p as the only H+-ATPase was used. An activity of 100% corresponds to the specific activity at 3 mm ATP without lipid. For quantification of AHA2 and T947A kinetic parameters, see Table 2. Specific activity at 3 mm ATP for Pma1p in the basal state was 2.3 ± 0.05 μmol of Pi/mg/min and in the activated state was 9.6 ± 0.23 μmol of Pi/mg/min. A and B, test of the effect of acyl chain length of lyso-PC on wild-type AHA2 and T947A (lyso-PC 12:0, ○; lyso-PC 14:0, gray circles; lyso-PC 16:0, ●). C and D, test of the lysophospholipid headgroup on the wild type and T947A (lyso-PC 16:0, ●; lyso-PG 16:0, gray circles). E and F, test of the glycerol backbone on the wild type and T947A (lyso-PC 16:0, ●; miltefosine 16:0, gray circles; edelfosine 18:0, ○; 1-hexadecyl-2-hydroxy-sn-glycero-3-phosphocholine (Lyso FosC) 16:0, ◐). G and H, test of the lysophospholipid headgroup on Pma1p (lyso-PC 16:0, ●; lyso-PG 16:0, gray circles) (n = 2–3 biological replicates; ±S.E. (error bars)).
DDM and Lyso-PC Alter AHA2 Kinetics in a Non-additive Manner
To understand the effect of the interplay between detergents and lyso-PC on the kinetic properties of the pump, we analyzed the ATP hydrolytic activity of AHA2 at various ATP concentrations and in the absence and presence of DDM and lyso-PC (Table 1). When the full-length enzyme was treated with DDM, it exhibited an increase in apparent ATP affinity (i.e. the Km changed from 1.2 to 0.5 mm ATP; Table 1) and a slight increase in Vmax. The addition of lyso-PC lowered the Km to 0.2 mm ATP and increased Vmax over 3-fold regardless of whether DDM was present (Table 1 and Fig. 1, E and F) or not (Table 1 and Fig. 1, C and D). A C-terminally truncated pump (aha2ΔC92) exhibited a high apparent affinity for ATP (Km = 0.2 mm; Table 1), which was not affected by the addition of either DDM or lysoPC (Fig. 1, C and D). The N-terminally truncated pump (aha2ΔN10) had a somewhat lower Km for ATP (Km = 0.7 mm; Table 1), but application of DDM increased the ATP affinity further (Km = 0.3 mm) and of lyso-PC even more so (Km = 0.2 mm; Table 1). Taken together, we conclude that (i) DDM and lyso-PC alter important kinetic properties of the PM H+-ATPase, (ii) the effect of lyso-PC is stronger than that of DDM, and (iii) these effects are not cumulative, but the effect of DDM is superseded by treatment with lyso-PC. Furthermore, the effect of both compounds is abolished by the removal of a five-residue element in the C-terminal domain but at least partially retained in a mutant lacking the N-terminal domain.
Effect of Terminal Deletions on the Ability of AHA2 to Function in Vivo
To learn how lyso-PC-insensitive N- and C-terminal deletion and C-terminal mutants of AHA2 function in vivo, we tested their ability to complement a yeast pma1 mutant (Fig. 3). In this assay, the essential PM H+-ATPase PMA1 was placed under a galactose-inducible promoter to ensure that PMA1 was expressed on galactose medium only, whereas the transgenic AHA2 was expressed constitutively. The full-length AHA2 molecule supported yeast growth on glucose medium on which PMA1 is not expressed (Fig. 3), whereas deletion of the three C-terminal-most residues (resulting in aha2ΔC3) abolished growth on this medium (Fig. 3). This was due to removal of Thr-947, which in yeast is recognized by an endogenous protein kinase to allow for binding of activating endogenous 14-3-3 protein (43), as mutation of this residue to Ala abolished the ability of the pump to complement the yeast mutant (Fig. 3). Removal of up to 25 C-terminal residues had no effect on the inability of the pump to complement pma1. Deletion of 30 C-terminal residues, however, allowed the pump to complement pma1 when pumping against an external pH of 5.5 (Fig. 3). To remove the effect of endogenous activation following phosphorylation of Thr-947, we combined the N-terminal deletion with mutation of the penultimate Thr residue to produce aha2ΔN10T947A and found that the resulting pump was unable to complement pma1 (Fig. 3). This suggests that the role of the N terminus is to inhibit protein kinase-mediated phosphorylation of Thr-947.
FIGURE 3.
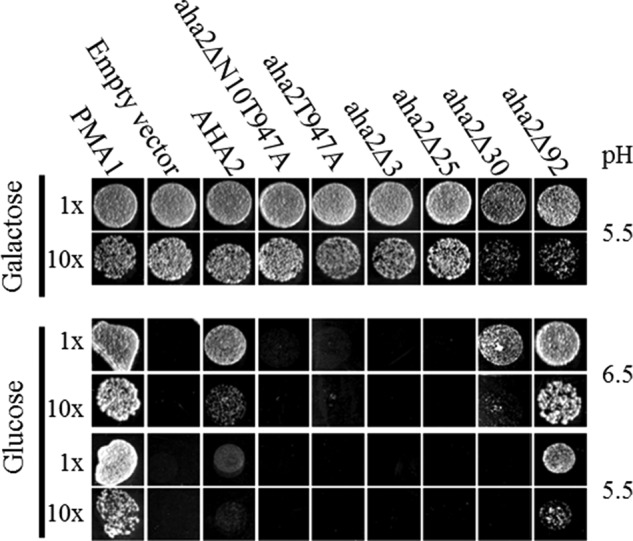
N- and C-terminal deletions activate the plant PM H+-ATPase in a yeast growth assay. A yeast strain with the native PM H+-ATPase placed under a galactose-inducible promoter was transformed with plasmids bearing wild-type or mutant plant PM H+-ATPases under the native promoter. Growth was assayed on minimal medium plates with either glucose or galactose at the given pH values and recorded after 3 days. Droplets contained initially around 103 cells.
A Single Point Mutation in the C Terminus Abolishes Lyso-PC Activation
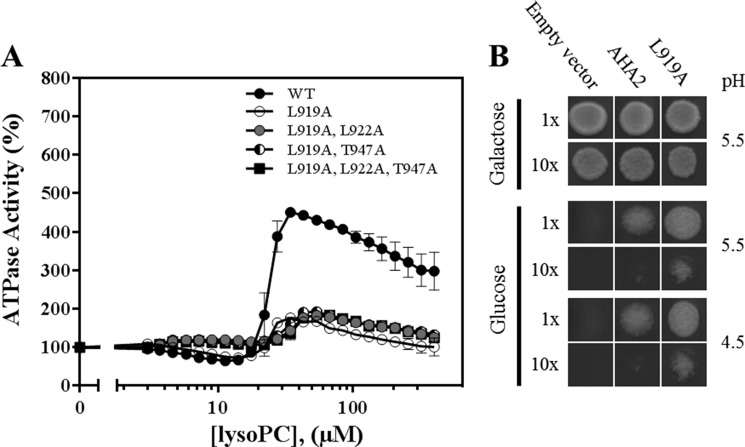
To narrow down the number of residues involved in lyso-PC activation, each residue between aha2ΔC25 and aha2ΔC30, corresponding to residues 918–923, was substituted with alanine. The effect of lyso-PC was tested on all mutants (Table 2). Of the six mutations tested, only L919A and L922A displayed significantly altered kinetic profiles (Table 2). The L922A mutant showed an increased stimulatory response to lyso-PC resembling that of the T947A mutant. By contrast, the L919A mutant was constitutively in the high activity state and responded only marginally to lyso-PC (Fig. 4A). Double or triple mutants of L919A in combination with T947A and L922A resembled the single L919A mutant with respect to Km and lyso-PC insensitivity (Table 2 and Fig. 4). From this, we conclude that the L919A substitution dominates over the L922A and T947A substitutions and thus plays a major role in AHA2 autoinhibition. When introduced into yeast, the L919A substitution complemented pma1 both when introduced as a single mutant and in combination with T947A and even when the external pH was as low as 4.5 (Fig. 4B). This demonstrates that the stimulatory effect of the Leu-919 substitution is direct and does not involve phosphorylation of Thr-947.
TABLE 2.
Kinetic parameters for the plasma membrane H+-ATPase AHA2 and C-terminal single, double, and triple mutants and lyso-PC activation measured at 30 °C with 3 mm ATP (n = 3 biological replicates; ±S.E.)
| H+-ATPase variant | Specific activity at 3 mm ATPa | Km(ATP) | Lyso-PC activation, max.b |
|---|---|---|---|
| μmol Pi/mg/min | mm | % | |
| AHA2 | 0.97 ± 0.11 | 1.20 ± 0.10 | 450 ± 2 |
| T947A | 0.62 ± 0.01 | 1.92 ± 0.30 | 612 ± 42 |
| R918A | 0.95 ± 0.07 | 1.48 ± 0.24 | 508 ± 1 |
| L919A | 1.97 ± 0.22 | 0.32 ± 0.08 | 177 ± 1 |
| R920A | 1.22 ± 0.09 | 1.42 ± 0.20 | 418 ± 5 |
| E921A | 1.23 ± 0.05 | 1.40 ± 0.30 | 433 ± 15 |
| L922A | 0.80 ± 0.09 | 1.82 ± 0.20 | 585 ± 45 |
| H923A | 1.21 ± 0.07 | 1.72 ± 0.20 | 469 ± 4 |
| L919A,T947A | 1.38 ± 0.13 | 0.36 ± 0.10 | 192 ± 8 |
| L919A,L922A | 1.71 ± 0.23 | 0.24 ± 0.02 | 182 ± 4 |
| L919A,L922A,T947A | 1.31 ± 0.12 | 0.34 ± 0.06 | 185 ± 8 |
a Specific activity between mutations cannot be compared because expression level may vary.
b Activity at 100% corresponds to the specific activity at 3 mm ATP without lyso-PC.
FIGURE 4.
Point mutations of Leu-919 activate the plant PM H+-ATPase in a yeast growth assay and in vitro. A, AHA2 C-terminal single, double, or triple mutants from purified microsomes were assayed with increasing concentrations of lyso-PC 16:0. WT (●), L919A (○), L919A,L922A (gray circles), L919A,T947A (◐), and L919A,L922A,T947A (■). A total of 0.3 μg of microsomes was used per sample, and 100% activity corresponds to the specific activity at 3 mm ATP without lyso-PC (see Table 2 for quantification) (n = 3 biological replicates; ± S.E. (error bars)). B, a yeast strain with the native H+-ATPase placed under a galactose-inducible promoter was transformed with plasmids bearing wild-type plant H+-ATPases or the L919A mutant under the yeast pma1 promoter. Growth was assayed on minimal medium plates with either glucose or galactose at the given pH values and recorded after 3 days.
Specificity of the Lysophospholipid Effect
Lyso-PC is characterized by having a choline headgroup, a glycerol backbone, and a single acyl chain. To test the importance of each component for the lipid effect, we titrated both wild-type AHA2 and the T947A mutant with a series of lipids with variations in headgroup, glycerol backbone, and acyl chain length (Table 3 and Fig. 2).
TABLE 3.
Overview of lysophospholipids and analogues tested
Red boxes indicate the modified group in relation to lyso-PC 16:0. Lyso-PA, 1-hexadecanoyl-sn-glycero-3-phosphate; lyso-PS, 1-hexadecanoyl-sn-glycero-3-phospho-l-serine; lysoFosC, 1-hexadecyl-2-hydroxy-sn-glycero-3-phosphocholine.
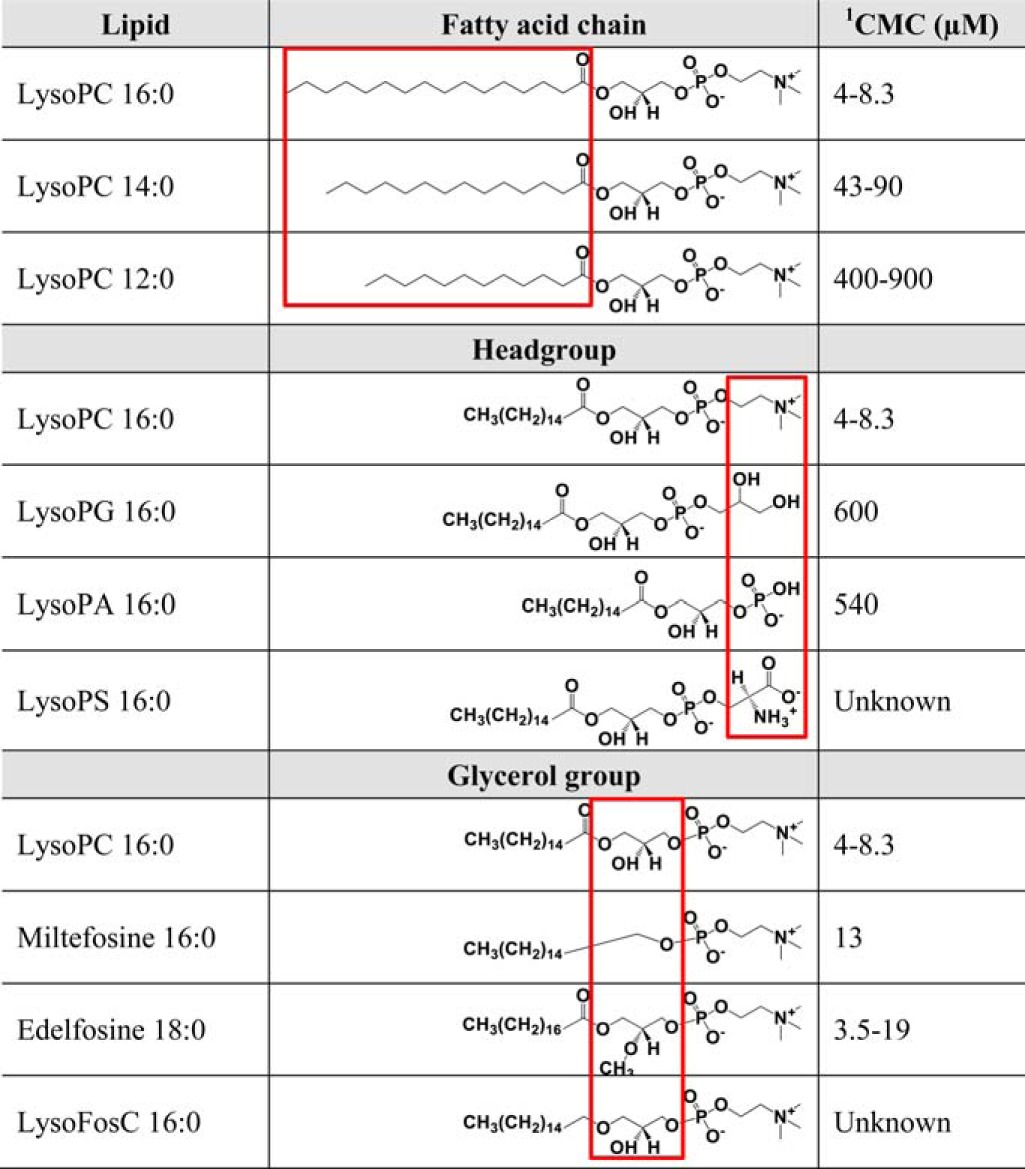
1 Critical micelle concentrations (CMC) were obtained from the suppliers (Avanti Polar Lipids and Anatrace).
The activating effect was approximately the same when choline in the headgroup was substituted with glycerol to produce lyso-PG (Fig. 2, C and D). Activation by lyso-PG was at a similar threshold level (around 15 μm) as for lyso-PC (Fig. 2, C and D). Lysophosphatidic acid and lysophosphatidylserine were also tested, but these lipids precipitated with the concentrations of Mg2+ used in the assay.
Modification of the glycerol backbone by removing the carbonyl oxygen to produce lysofos choline ether (Fig. 2, E and F, Lyso FosC) reduced the potency to about 50% of that of lyso-PC, whereas other modifications of the backbone, such as introduction of a methyl group (edelfosine; Fig. 2, E and F) or removing it completely (miltefosine; Fig. 2, E and F), abolished the effect. Decreasing the acyl chain length from C16 to C14 and C12 reduced the strength of the effect by more than a third (Fig. 2, A and B), and the threshold levels for activation of AHA2 increased significantly to 30 and 170 μm, respectively. We conclude that the chemical nature of the glycerol backbone and length of the acyl chain are critically important for the lysophospholipid effect, whereas the headgroup does not appear to determine specificity.
Lysophospholipids Do Not Activate the Yeast PM H+-ATPase
To establish whether lysophospholipids activate all PM H+-ATPases, we tested their effect on Pma1p, the PM H+-ATPase of the yeast S. cerevisiae. Like AHA2, Pma1p has a C-terminal regulatory domain and, depending on the phosphorylation status of this domain, exists in either a basal or activated state (45). However, the C-terminal domain of the fungal PM H+-ATPase is considerably shorter than that of the plant pump, and no sequence stretch with homology to Region II in the plant pump is present (Fig. 5A). We expressed PMA1 from the same expression plasmid as that used for AHA2 and in a pma1,pma2 background and isolated membranes harboring Pma1p from yeast grown in the absence or presence of glucose, which triggers the formation of the basal and activated states, respectively (11). When the ATP hydrolytic activity of Pma1p was titrated with lyso-PC or lyso-PG, no activating effect was observed for either lipid (Fig. 2, G and H). Rather, they inhibited activity with the basal state of Pma1p being more sensitive than the activated state. This indicates that lyso-PC activation is a specific feature that evolved in plant PM H+-ATPase but not in its fungal counterpart.
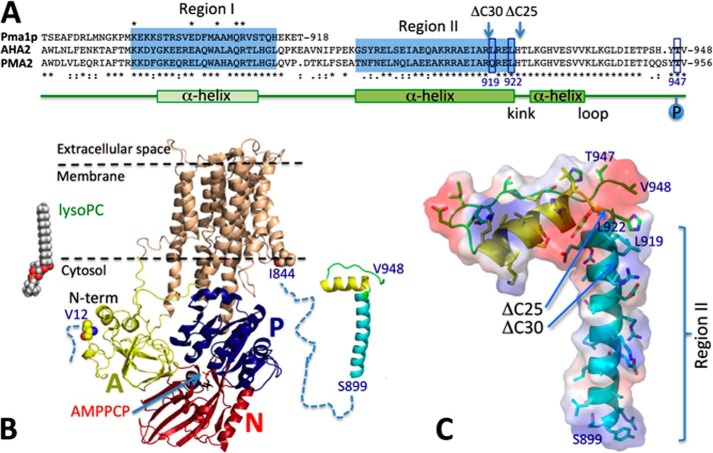
FIGURE 5.
Working model for lyso-PC activation of the plant PM H+-ATPase. A, alignment of fungal S. cerevisiae Pma1p with Arabidopsis AHA2 and tobacco PMA2 C-terminal regions with the structure of PMA2 indicated below (predicted in the case of Region I). Autoinhibitory Regions I and II are indicated by blue boxes. AHA2 residues Leu-919 and Leu-922, which play opposing roles in autoinhibition, and the phosphorylation site Thr-947 are indicated. Asterisks below sequence indicate full conservation in plants; asterisks above sequence indicate full conservation across species; colons indicate conservation between groups of strongly similar properties; periods indicate conservation between groups of weakly similar properties. B, known structural elements involved in the regulation of the plant PM H+-ATPase. A, actuator domain; N, nucleotide-binding domain; P, phosphorylation domain. Left, the structure of lyso-PC 16:0 is inserted for size comparison with the PM H+-ATPase. How lyso-PC interacts with the core PM H+-ATPase and/or terminal domains is not known. Middle, a ribbon model of the crystal structure of a truncated AHA2 PM H+-ATPase (residues 12–844) (Protein Data Bank code 3B8C) is shown. The terminal modeled residues are highlighted, and the cytosolic domains are noted. In the crystal structure, AMPPCP is present as a non-hydrolyzable ATP analogue. Right, attached with a dotted line to the core structure is a homology model of the 50 C-terminal residues of AHA2 based on the crystal structure of the corresponding region of PMA2 in complex with 14-3-3 protein (Protein Data Bank code 2O98). How the C-terminal domain interacts with the core protein is not known. C, space-filling structure of the homology model of the 50 C-terminal residues of AHA2 shown in B with key amino acid residues indicated. The position of truncations corresponding to Δ25 and Δ30 are shown. Leu-919 and Leu-922 are seen on the same face of the α-helix constituting Region II (in cyan). Electrostatic potential as determined by PyMOL is shown as a surface: white, no charge; blue, positive charge; red, negative charge. Region II has a hydrophobic patch (left side of the helix). The α-helix in yellow is part of the 14-3-3-binding site and has a partially hydrophobic surface (top), which in the basal non-activated state might be docked in a hydrophobic environment, such as the membrane, to protect Thr-947 from phosphorylation.
Discussion
Lyso-PC, a naturally occurring detergent-like metabolite in eukaryotic membranes, was reported previously to activate the plant PM H+-ATPase (40, 41). In this work, we expanded on these observations and conclude that the effect of lysophospholipids is specific for plant PM H+-ATPases, is dependent on the glycerol backbone and the length of the acyl chain, is direct, and involves the concerted action of both terminal domains of the pump. These results refine our understanding of the structural organization of autoinhibited PM H+-ATPases and reveal a mechanistic role for lysophospholipids as signaling molecules in plants.
Effects of Detergents on AHA2 Involve the Terminal Domains
Strikingly, the effects of surfactants were critically dependent on the presence of the terminal domains of the pump. The stimulation by DDM, which represented that of most detergents, was unrelated to phosphorylation of Thr-947 because the effect persisted in mutants lacking this residue. This implies that the effect of DDM is not the result of facilitating exposure of Thr-947 to a protein kinase in the preparation. What then could be the mechanism of activation? As the effect of DDM disappeared when 10 N-terminal residues and more than 25 C-terminal residues of AHA2 were truncated, the most likely explanation is that DDM and other detergents in some way loosen the interactions between the terminal autoinhibitory domains and/or the rest of the protein.
Lysophospholipids stood out among detergent-like molecules in that they produced a substantially stronger and concentration-dependent activating effect that mimicked the effect of deleting both of the autoinhibitory regions (Regions I and II) in the C-terminal domain (as in aha2ΔC92). This finding suggests that lysophospholipids neutralize both Regions I and II. Whether DDM affects these regions is unclear; however, the fact that lysophospholipids can further stimulate pump activity even after DDM treatment suggests that the interactions between autoinhibitory regions and the core of the pump protein might be retained even in the presence of DDM. Taken together, DDM induces a general detergent effect associated with partial activation, whereas lysophospholipids cause a specific effect resulting in full activation.
Lysophospholipid Activation of the PM H+-ATPase Involves a C-terminal Hydrophobic Anchor Stretch
By carrying out sequential truncations at the C-terminal end combined with point mutations at single residues, we identified Leu-919 as a residue that when mutated to Ala causes AHA2 to respond only marginally to lyso-PC. The L919A mutant was identified previously as a so-called Class B mutant, a name coined for substitutions that confer partial complementation of yeast pma1 (21). Some Class B mutants have been shown to associate with 14-3-3 protein (21, 47), but we found that the L919A mutant is insensitive to lysophospholipids even in a T947A background, which demonstrates that the effect is not through facilitating exposure of Thr-947 for phosphorylation and subsequent 14-3-3 binding but more likely is direct. We also identified Leu-922 further downstream of Leu-919 as a residue that when substituted with Ala results in a mutant that, like T947A, has a stronger response to lyso-PC than does wild-type AHA2. In this regard, the effects of mutating Leu-919 and Leu-922 were opposite to each other. Both residues are highly conserved in angiosperms, but interestingly, Leu-919 is substituted with a Glu in tobacco PMA2 (Fig. 5A). It remains to be seen whether this particular isoform is lysophospholipid-responsive.
A minimal model for the role of these residues in autoinhibition is that Leu-919 promotes an interaction that keeps the C-terminal domain in place relative to the rest of the molecule, whereas Leu-922 opposes this interaction (Fig. 5, B and C). We found that the pump was activated by substitution of the hydrophobic Leu-919 with Ala, another hydrophobic residue, whereas substituting the nearby charged residues Arg-920 and Glu-921 with Ala had no effect, suggesting that the interaction is highly hydrophobic or alternatively involves the backbone carbonyl oxygen of Leu-919. A direct interaction of Leu-919 with lysophospholipids has not been shown in this study, but this possibility is in accordance with our observation that the charged headgroup of lyso-PC does not determine the specificity of the lipid effect.
Role of the N-terminal Domain in Regulating PM H+-ATPases
What could be the mechanistic role of the N-terminal domain in regulating PM H+-ATPases? Removal of the N terminus results in increased phosphorylation of Thr-947 (17), suggesting that the N terminus modulates the accessibility of protein kinases to Thr-947. In this work, we showed that mutation of this residue to Ala abolishes the ability of the pump to complement pma1 and further that truncation of the N terminus does not restore this function. This finding supports the notion that the N terminus controls the ability of Thr-947 to become phosphorylated.
A Concerted Action between Autoinhibitory Regions May Affect Activation Kinetics
A peculiar phenomenon we observed in this work is that AHA2 is activated by lysophospholipids within a narrow concentration range. Similar abrupt activation kinetics in response to an increase in modulator concentration have been reported for the autoinhibited Ca2+-ATPase ACA8 (48). This P-type ATPase is activated by calmodulin binding to two autoinhibitory regions in response to an increase in Ca2+ concentration. Mathematical modeling of Ca2+/calmodulin action on ACA8 demonstrates that when two intramolecular autoinhibitors are present, rather than just one, activation becomes almost instantaneous above a certain threshold value of Ca2+ (48). In accordance with this model, a concerted action between several autoinhibitory regions (the N terminus and Regions I and II of the C-terminal domain), which is neutralized by lysophospholipids, could explain why lipid activation of the PM H+-ATPase AHA2 by lysophospholipids is instantaneous above a certain threshold value.
Is the Effect of Lysophospholipids Related to Their Detergent Properties?
Besides being implicated as a signaling molecule in both plants and animals, lyso-PC is known to lyse cell membranes even at micromolar concentrations (49). Can a molecule that permeabilizes cell membranes be involved in energizing the cell membrane? Interestingly, the lytic properties of lyso-PC depend strongly on the lipid environment in which it is present, and in combination with sterols, phosphatidylethanolamines, or fatty acids, lyso-PC may even stabilize a bilayer structure (50). Notably, a bilayer structure is preserved when lyso-PC is present at an equimolar ratio with free fatty acids, a ratio that is the result of phospholipase A2 action, which allows for the specific effects of lyso-PC other than lysis to take place. Further studies are needed to clarify the interplay between the solubilizing effect of lysophospholipids and their selective activating effect on the PM H+-ATPase in planta. Several binding sites for lipids have been identified in crystal structures of the model P-type ATPase SERCA (35); of these, four do not appear to serve merely as crystal contacts and could represent tightly bound annular lipids. Of special note is the binding site near the calcium entrance pathway modeled as a phosphatidylethanolamine-binding site in the structures. The presence of a specific lysophospholipid-binding site in AHA2, which could be related to a lipid-binding site in SERCA, may also explain the selective activation of AHA2 and lack of activation of yeast Pma1p.
Possible Physiological Role of Lysophospholipid-mediated Activation of PM H+-ATPase
Lysophospholipids have been suggested to function as signaling molecules in plants as they appear to play a role during symbiotic encounters between plants and microorganisms (38) and in plant-pathogen interactions (51, 52). Root growth is inhibited during phosphate starvation in mutant plants lacking phospholipase A (AtPLAIVA), an enzyme that degrades phosphatidylglycerol and phosphatidylcholine to lyso-PG and lyso-PC (53), a phenotype that also is seen when aha2 plants are exposed to nutrient limitation (47, 54). In leaves, phospholipase A2 activation has been implicated in light-induced opening of the stomatal pores through which plants regulate carbon dioxide uptake and water loss (39), a process that is strictly dependent on activation of the PM H+-ATPase (13, 15). In this context, our results add to the growing body of evidence suggesting that lysophospholipids regulate primary active proton transport across the plant plasma membrane. In conclusion, our results suggest a molecular mechanism for the phosphorylation-independent activation of the plant PM H+-ATPase that requires the concerted action of its terminal domains and renders the pump protein responsive to lysophospholipids.
This work was supported by the Danish Strategic Research Council (FungalFight) and the Danish National Research Foundation (Pumpkin). The authors declare that they have no conflicts of interest with the contents of this article.
This work is dedicated to the memory of Alex Green Wielandt.
- PM
- plasma membrane
- SERCA
- sarco(endo)plasmic reticulum Ca2+-ATPase
- lyso-PC
- lysophosphatidylcholine
- lyso-PG
- hexadecanoyl-sn-glycero-3-phosphoglycerol
- lyso-PC 12:0
- 1-dodecanoyl-sn-glycerol-3-phosphocholine
- lyso-PC 14:0
- 1-tetradecanoyl-sn-glycerol-3-phosphocholine
- lyso-PC 16:0
- 1-hexadecanoyl-sn-glycero-3-phosphocholine
- DDM
- n-dodecyl β-d-maltoside
- AMPPCP
- adenylyl 5′-(β,γ-methylene)diphosphonate.
References
- 1. Palmgren M. G. (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845 [DOI] [PubMed] [Google Scholar]
- 2. Axelsen K. B., Palmgren M. G. (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46, 84–101 [DOI] [PubMed] [Google Scholar]
- 3. Palmgren M. G., Nissen P. (2011) P-type ATPases. Annu. Rev. Biophys. 40, 243–266 [DOI] [PubMed] [Google Scholar]
- 4. Traaseth N. J., Shi L., Verardi R., Mullen D. G., Barany G., Veglia G. (2009) Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc. Natl. Acad. Sci. U.S.A. 106, 10165–10170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sahoo S. K., Shaikh S. A., Sopariwala D. H., Bal N. C., Periasamy M. (2013) Sarcolipin protein interaction with sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA) is distinct from phospholamban protein, and only sarcolipin can promote uncoupling of the SERCA pump. J. Biol. Chem. 288, 6881–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Winther A. M., Bublitz M., Karlsen J. L., Møller J. V., Hansen J. B., Nissen P., Buch-Pedersen M. J. (2013) The sarcolipin-bound calcium pump stabilizes calcium sites exposed to the cytoplasm. Nature 495, 265–269 [DOI] [PubMed] [Google Scholar]
- 7. Toyoshima C., Iwasawa S., Ogawa H., Hirata A., Tsueda J., Inesi G. (2013) Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state. Nature 495, 260–264 [DOI] [PubMed] [Google Scholar]
- 8. Geisler M., Axelsen K. B., Harper J. F., Palmgren M. G. (2000) Molecular aspects of higher plant P-type Ca2+-ATPases. Biochim. Biophys. Acta 1465, 52–78 [DOI] [PubMed] [Google Scholar]
- 9. Penniston J. T., Enyedi A. (1998) Modulation of the plasma membrane Ca2+ pump. J. Membr. Biol. 165, 101–109 [DOI] [PubMed] [Google Scholar]
- 10. Brini M., Calì T., Ottolini D., Carafoli E. (2013) The plasma membrane calcium pump in health and disease. FEBS J. 280, 5385–5397 [DOI] [PubMed] [Google Scholar]
- 11. Serrano R. (1983) In vivo glucose activation of the yeast plasma membrane ATPase. FEBS Lett. 156, 11–14 [DOI] [PubMed] [Google Scholar]
- 12. Niittylä T., Fuglsang A. T., Palmgren M. G., Frommer W. B., Schulze W. X. (2007) Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell. Proteomics 6, 1711–1726 [DOI] [PubMed] [Google Scholar]
- 13. Kinoshita T., Shimazaki K. (1999) Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J. 18, 5548–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marre E. (1979) Fusicoccin: a tool in plant physiology. Annu. Rev. Plant Physiol. 30, 273–288 [Google Scholar]
- 15. Elmore J. M., Coaker G. (2011) The role of the plasma membrane H+-ATPase in plant-microbe interactions. Mol. Plant 4, 416–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Palmgren M. G., Sommarin M., Serrano R., Larsson C. (1991) Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J. Biol. Chem. 266, 20470–20475 [PubMed] [Google Scholar]
- 17. Ekberg K., Palmgren M. G., Veierskov B., Buch-Pedersen M. J. (2010) A novel mechanism of P-type ATPase autoinhibition involving both termini of the protein. J. Biol. Chem. 285, 7344–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Venema K., Palmgren M. G. (1995) Metabolic modulation of transport coupling ratio in yeast plasma membrane H+-ATPase. J. Biol. Chem. 270, 19659–19667 [DOI] [PubMed] [Google Scholar]
- 19. Baunsgaard L., Venema K., Axelsen K. B., Villalba J. M., Welling A., Wollenweber B., Palmgren M. G. (1996) Modified plant plasma membrane H+-ATPase with improved transport coupling efficiency identified by mutant selection in yeast. Plant J. 10, 451–458 [DOI] [PubMed] [Google Scholar]
- 20. Regenberg B., Villalba J. M., Lanfermeijer F. C., Palmgren M. G. (1995) C-terminal deletion analysis of plant plasma membrane H+-ATPase: yeast as a model system for solute transport across the plant plasma membrane. Plant Cell 7, 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Axelsen K. B., Venema K., Jahn T., Baunsgaard L., Palmgren M. G. (1999) Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2: mapping of residues that when altered give rise to an activated enzyme. Biochemistry 38, 7227–7234 [DOI] [PubMed] [Google Scholar]
- 22. Speth C., Jaspert N., Marcon C., Oecking C. (2010) Regulation of the plant plasma membrane H+-ATPase by its C-terminal domain: what do we know for sure? Eur. J. Cell Biol. 89, 145–151 [DOI] [PubMed] [Google Scholar]
- 23. Jahn T., Fuglsang A. T., Olsson A., Brüntrup I. M., Collinge D. B., Volkmann D., Sommarin M., Palmgren M. G., Larsson C. (1997) The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 9, 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fuglsang A. T., Visconti S., Drumm K., Jahn T., Stensballe A., Mattei B., Jensen O. N., Aducci P., Palmgren M. G. (1999) Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem. 274, 36774–36780 [DOI] [PubMed] [Google Scholar]
- 25. Maudoux O., Batoko H., Oecking C., Gevaert K., Vandekerckhove J., Boutry M., Morsomme P. (2000) A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 275, 17762–17770 [DOI] [PubMed] [Google Scholar]
- 26. Rudashevskaya E. L., Ye J., Jensen O. N., Fuglsang A. T., Palmgren M. G. (2012) Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments. J. Biol. Chem. 287, 4904–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pedersen B. P., Buch-Pedersen M. J., Morth J. P., Palmgren M. G., Nissen P. (2007) Crystal structure of the plasma membrane proton pump. Nature 450, 1111–1114 [DOI] [PubMed] [Google Scholar]
- 28. Ottmann C., Marco S., Jaspert N., Marcon C., Schauer N., Weyand M., Vandermeeren C., Duby G., Boutry M., Wittinghofer A., Rigaud J. L., Oecking C. (2007) Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining x-ray crystallography and electron cryomicroscopy. Mol. Cell 25, 427–440 [DOI] [PubMed] [Google Scholar]
- 29. Niggli V., Adunyah E. S., Carafoli E. (1981) Acidic phospholipids, unsaturated fatty acids, and limited proteolysis mimic the effect of calmodulin on the purified erythrocyte Ca2+-ATPase. J. Biol. Chem. 256, 8588–8592 [PubMed] [Google Scholar]
- 30. Brodin P., Falchetto R., Vorherr T., Carafoli E. (1992) Identification of two domains which mediate the binding of activating phospholipids to the plasma-membrane Ca2+ pump. Eur. J. Biochem. 204, 939–946 [DOI] [PubMed] [Google Scholar]
- 31. Meneghelli S., Fusca T., Luoni L., De Michelis M. I. (2008) Dual mechanism of activation of plant plasma membrane Ca2+-ATPase by acidic phospholipids: evidence for a phospholipid binding site which overlaps the calmodulin-binding site. Mol. Membr. Biol. 25, 539–546 [DOI] [PubMed] [Google Scholar]
- 32. Brini M., Di Leva F., Ortega C. K., Domi T., Ottolini D., Leonardi E., Tosatto S. C., Carafoli E. (2010) Deletions and mutations in the acidic lipid-binding region of the plasma membrane Ca2+ pump: a study on different splicing variants of isoform 2. J. Biol. Chem. 285, 30779–30791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Obara K., Miyashita N., Xu C., Toyoshima I., Sugita Y., Inesi G., Toyoshima C. (2005) Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+. Proc. Natl. Acad. Sci. U.S.A. 102, 14489–14496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Olesen C., Picard M., Winther A. M., Gyrup C., Morth J. P., Oxvig C., Møller J. V., Nissen P. (2007) The structural basis of calcium transport by the calcium pump. Nature 450, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 35. Drachmann N. D., Olesen C., Møller J. V., Guo Z., Nissen P., Bublitz M. (2014) Comparing crystal structures of Ca2+-ATPase in the presence of different lipids. FEBS J. 281, 4249–4262 [DOI] [PubMed] [Google Scholar]
- 36. Baldridge R. D., Xu P., Graham T. R. (2013) Type IV P-type ATPases distinguish mono- versus diacyl phosphatidylserine using a cytofacial exit gate in the membrane domain. J. Biol. Chem. 288, 19516–19527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laxalt A. M., Munnik T. (2002) Phospholipid signalling in plant defence. Curr. Opin. Plant Biol. 5, 332–338 [DOI] [PubMed] [Google Scholar]
- 38. Drissner D., Kunze G., Callewaert N., Gehrig P., Tamasloukht M., Boller T., Felix G., Amrhein N., Bucher M. (2007) Lyso-phosphatidylcholine is a signal in the arbuscular mycorrhizal symbiosis. Science 318, 265–268 [DOI] [PubMed] [Google Scholar]
- 39. Seo J., Lee H. Y., Choi H., Choi Y., Lee Y., Kim Y. W., Ryu S. B., Lee Y. (2008) Phospholipase A2β mediates light-induced stomatal opening in Arabidopsis. J. Exp. Bot. 59, 3587–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Palmgren M. G., Sommarin M., Ulvskov P., Jorgensen P. L. (1988) Modulation of plasma membrane H+-ATPase from oat roots by lysophosphatidylcholine, free fatty acids and phospholipase A2. Physiol. Plant. 74, 11–19 [Google Scholar]
- 41. Palmgren M. G., Sommarin M. (1989) Lysophosphatidylcholine stimulates ATP dependent proton accumulation in isolated oat root plasma membrane vesicles. Plant Physiol. 90, 1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cid A., Perona R., Serrano R. (1987) Replacement of the promoter of the yeast plasma membrane ATPase gene by a galactose-dependent promoter and its physiological consequences. Curr. Genet. 12, 105–110 [DOI] [PubMed] [Google Scholar]
- 43. Jahn T. P., Schulz A., Taipalensuu J., Palmgren M. G. (2002) Post-translational modification of plant plasma membrane H+-ATPase as a requirement for functional complementation of a yeast transport mutant. J. Biol. Chem. 277, 6353–6358 [DOI] [PubMed] [Google Scholar]
- 44. de Kerchove d'Exaerde A., Supply P., Dufour J. P., Bogaerts P., Thinés D., Goffeau A., Boutry M. (1995) Functional complementation of a null mutation of the yeast Saccharomyces cerevisiae plasma membrane H+-ATPase by a plant H+-ATPase gene. J. Biol. Chem. 270, 23828–23837 [DOI] [PubMed] [Google Scholar]
- 45. Lecchi S., Nelson C. J., Allen K. E., Swaney D. L., Thompson K. L., Coon J. J., Sussman M. R., Slayman C. W. (2007) Tandem phosphorylation of Ser-911 and Thr-912 at the C terminus of yeast plasma membrane H+-ATPase leads to glucose-dependent activation. J. Biol. Chem. 282, 35471–35481 [DOI] [PubMed] [Google Scholar]
- 46. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 47. Fuglsang A. T., Borch J., Bych K., Jahn T. P., Roepstorff P., Palmgren M. G. (2003) The binding site for regulatory 14-3-3 protein in plant plasma membrane H+-ATPase: involvement of a region promoting phosphorylation-independent interaction in addition to the phosphorylation-dependent C-terminal end. J. Biol. Chem. 278, 42266–42272 [DOI] [PubMed] [Google Scholar]
- 48. Tidow H., Poulsen L. R., Andreeva A., Knudsen M., Hein K. L., Wiuf C., Palmgren M. G., Nissen P. (2012) A bimodular mechanism of calcium control in eukaryotes. Nature 491, 468–472 [DOI] [PubMed] [Google Scholar]
- 49. Weltzien H. U. (1979) Cytolytic and membrane-perturbing properties of lysophosphatidylcholine. Biochim. Biophys. Acta 559, 259–287 [DOI] [PubMed] [Google Scholar]
- 50. Jain M. K., van Echteld C. J., Ramirez F., de Gier J., de Haas G. H., van Deenen L. L. (1980) Association of lysophosphatidylcholine with fatty acids in aqueous phase to form bilayers. Nature 284, 486–487 [DOI] [PubMed] [Google Scholar]
- 51. Viehweger K., Dordschbal B., Roos W. (2002) Elicitor-activated phospholipase A2 generates lysophosphatidylcholines that mobilize the vacuolar H+ pool for pH signaling via the activation of Na+-dependent proton fluxes. Plant Cell 14, 1509–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao J., Devaiah S. P., Wang C., Li M., Welti R., Wang X. (2013) Arabidopsis phospholipase Dβ1 modulates defense responses to bacterial and fungal pathogens. New Phytol. 199, 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rietz S., Dermendjiev G., Oppermann E., Tafesse F. G., Effendi Y., Holk A., Parker J. E., Teige M., Scherer G. F. (2010) Roles of Arabidopsis patatin-related phospholipases A in root development are related to auxin responses and phosphate deficiency. Mol. Plant 3, 524–538 [DOI] [PubMed] [Google Scholar]
- 54. Mlodzińska E., Kłobus G., Christensen M. D., Fuglsang A. T. (2015) The plasma membrane H+-ATPase AHA2 contributes to the root architecture in response to different nitrogen supply. Physiol. Plant. 154, 270–282 [DOI] [PubMed] [Google Scholar]