Background: Bacteriophytochromes are dimeric histidine kinases, but the functional role of their dimerization interfaces is unclear.
Results: The phytochrome from Deinococcus radiodurans has two dimerization interfaces, which are critical for thermal back reversion and are altered by illumination.
Conclusion: The dimerization interfaces cause strain in the structure.
Significance: A functional role for the dimerization interfaces is proposed.
Keywords: cell signaling, high performance liquid chromatography (HPLC), histidine kinase, molecular dynamics, mutagenesis, photoreceptor, protein conformation, x-ray scattering, phytochrome, dimerization
Abstract
Phytochromes are dimeric photoreceptor proteins that sense red light levels in plants, fungi, and bacteria. The proteins are structurally divided into a light-sensing photosensory module consisting of PAS, GAF, and PHY domains and a signaling output module, which in bacteriophytochromes typically is a histidine kinase (HK) domain. Existing structural data suggest that two dimerization interfaces exist between the GAF and HK domains, but their functional roles remain unclear. Using mutational, biochemical, and computational analyses of the Deinococcus radiodurans phytochrome, we demonstrate that two dimerization interfaces between sister GAF and HK domains stabilize the dimer with approximately equal contributions. The existence of both dimerization interfaces is critical for thermal reversion back to the resting state. We also find that a mutant in which the interactions between the GAF domains were removed monomerizes under red light. This implies that the interactions between the HK domains are significantly altered by photoconversion. The results suggest functional importance of the dimerization interfaces in bacteriophytochromes.
Introduction
Phytochromes are photoreceptors found in plants, fungi, and various microorganisms like cyanobacteria and proteobacteria (1, 2). They modulate their biochemical activity in response to the light environment. Phytochromes exist in two states that absorb red light (the Pr state) and far-red light (the Pfr state). Depending on the species, either Pr or Pfr is the resting state, and the proteins are classed as “prototypical” and “bathy” phytochromes, respectively (3–5). Generally, the proteins have a photosensory module and an output module. The photosensory module usually contains PAS4 (PER, ARNT, SIM) and GAF (cGMP phosphodiesterase, adenylate cyclase, FhlA) domains, which bind a chromophore, and a C-terminal PHY (phytochrome-specific GAF-related) domain. These domains convert an incident light signal into a conformational signal, which is then relayed to control the activity of the output module. The output module varies between species but is a histidine kinase (HK) domain in many bacterial phytochromes (6, 7).
The first step of the photocycle is the photoisomerization of the linear tetrapyrrole chromophore. In bacteriophytochromes this is a biliverdin that is attached to the PAS-GAF domains. The signal is transmitted to the PHY domain by the so-called “tongue” or “hairpin” extension (8–13). Crystal structures of prototypical and bathy phytochromes in their respective Pr and Pfr resting states (8, 9, 12, 14) and crystal structures of the same phytochrome in the dark and after red light illumination show that the PHY-tongue refolds (11). This leads to a shortening of the tongue and a large scale opening of the entire phytochrome photosensory module dimer (11, 12).
The overall arrangement of PAS-GAF-PHY is conserved in all structures published to date (8–10, 14–16). Bacterial phytochromes are generally considered as parallel (head-to-head) dimers (9, 11, 12, 17) even though some phytochrome fragments crystallize as antiparallel (head-to-tail) dimers (8, 10, 15), and head-to-tail arrangement was proposed to be functionally relevant for BphP1 from Rhodopseudomonas palustris (18). The head-to-head arrangement is supported by electron micrographs (12, 19) and small angle x-ray solution scattering data (20). From crystal structures, dimer contacts were identified between the GAF domains (9, 11, 12, 14, 16, 17, 21) and possibly also between the PHY domains (9, 22, 23). Because the structure of a full-length phytochrome is yet to be solved, it is not fully established whether the HK domains have a buried dimer interface. However, electron micrographs (12, 19) and other sensor histidine kinase structures (24–26) suggest that such an interaction exists, at least in one of the photochemical states.
In solution the strength of the dimerization interfaces varies between different phytochromes. The complete photosensory module (PAS-GAF-PHY) and the PAS-GAF fragment of Deinococcus radiodurans form stable dimers in solution (11, 12, 17, 27, 28), but this is not the case in some other species. The photosensory modules of the Agrobacterium tumefaciens Agp1 form a mixture with mainly dimers (29). This is also true for cyanobacterial Cph1 from Synechocystis sp. (8, 15, 30), whereas the photosensory module of Cph2 is predominantly monomeric in solution (10, 31). Plant and cyanobacterial phytochromes are commonly thought to have the principal dimerization interface at the C-terminal region, whereas the N-terminal photosensory module region does not seem to form dimers (14, 32–34). Indeed, the photosensory module of Arabidopsis thaliana phyB behaves as a monomer in solution but crystallizes as a parallel dimer (14). In summary, it is likely that most phytochromes form parallel dimers by interactions between the sister GAF and HK domains. However, the number of exceptions described above makes it worthwhile to put this hypothesis to the test.
Dimerization is an intrinsic feature of phytochromes, but what is its functional role? In A. thaliana, dimerization of the output modules was demonstrated to be necessary for the proper function of the plant phytochrome B (35, 36). Moreover, phytochromes B to E form heterodimers, but the functional role of this flexible quaternary arrangement has yet to be discovered (37, 38). Considering that many bacterial histidine kinases phosphorylate in trans, dimer arrangement of the HK domains may be functionally important for bacterial phytochromes (39–41). However, the role of the dimerization interface in the photosensory module is less clear. While searching for an efficient fluorescent protein, Auldridge et al. (42) found a three-point mutation that completely monomerized the D. radiodurans PAS-GAF fragment. Surprisingly, this also restored photochromicity (42). This hints at that even the dimerization interface in the GAF domain could influence phytochrome function.
Phytochromes can be switched between the Pr and Pfr states by light, but in most cases they thermally dark-revert into one of the two states. The dark reversion rate is likely important for regulating the activity of phytochromes in vivo. Factors that influence this rate include truncation of the HK domain (12, 28, 43). By extensive mutational analysis it has emerged that no single residue alone but, rather, a set of residues controls dark reversion (10, 12, 14, 44). Together, this leads us to hypothesize that the tertiary or quaternary organization of the protein is important for dark reversion. We test this in this study.
It is currently unclear how the activity of the output domain of phytochromes is controlled structurally. Electron microscopy analysis of the full-length bacteriophytochrome from D. radiodurans suggests that the opening in between the PHY domains (11) impacts the relative orientation of the HK output domains (12). Crystallographic snapshots of related dimeric sensor histidine kinases have yielded a number of proposed mechanisms, which often involves asymmetric kinking or bending of the histidine kinase structures (41, 45–48). In these proposals the histidine kinase domains remain dimeric, i.e. the dimer interface is not broken by the light stimulus, neither fully nor partially. This seems reasonable because the hydrophobic interactions that tie together the histidine kinase monomers are presumed to be very stable. However, the opening movement of the PAS-GAF-PHY protein observed by us (11) and the electron micrographs reported by Burgie et al. (12) suggest that a mechanism in which the HK domains separated fully or partially should be tested. Indeed, we find in this study that the dimer interface between the HK domains can be broken by light and conclude that such a mechanism should be considered.
Here, we apply the set of mutations that monomerize PAS-GAF fragment to PAS-GAF-PHY (photosensory module) and the full-length phytochrome from D. radiodurans. Our data show that the phytochrome dimer is stabilized across two interfaces: one between neighboring GAF domains and one between the HK domains but not between the PHY domains. Surprisingly, the HK interface can be broken by red light illumination in the construct with mutations at the GAF/GAF interface. The integrity of both dimerization interfaces is needed for normal thermal (dark) reversion from the Pfr state to the Pr state. The absence of the HK domain (with its dimerization interface) in the PAS-GAF-PHY construct slows down dark reversion, whereas the loss of GAF dimerization completely abolishes it. We conclude that both dimerization interfaces are important for phytochrome function.
Experimental Procedures
Cloning and Protein Purification
The expression plasmids coding for wild-type D. radiodurans fragments (PAS-GAF, PAS-GAF-PHY, full-length) were kindly provided by the laboratories of Prof. R. D. Vierstra and Prof. K. T. Forest and are described elsewhere (12, 17, 27). The three monomerizing mutations were introduced by using the QuikChange Lightning Multi Site-directed Mutagenesis kit (Agilent Technologies, La Jolla, CA) with the following primers: F145S, 5′-CTGCGCAACGCGATGTCAGCGCTCGAAAGTGC-3′; L311E/L314E, 5′-CTCGAATACCTGGGCCGCGAGCTGAGCGAGCAAGTTCAGGTC-3′ (mutated nucleotides are underlined). All sequences were verified using the sequencing facility at the University of Jyväskylä. The phytochrome constructs were expressed and purified as described previously (11, 28, 49). The purified phytochrome fragments in the final size-exclusion chromatography (SEC) buffer of (30 mm Tris·HCl, pH 8.0) were concentrated to 20–40 mg/ml, flash-frozen, and stored at −80 °C. The samples were thawed and filtered with 0.22-μm centrifugal filter units (Amicon Ultrafree, Millipore) immediately before experimental characterization.
Sample Illumination
The samples were illuminated or kept in the dark just before each measurement. The illuminated samples were illuminated with 780 nm (far red, 9-milliwatt output power) or 655 nm (red, 7-milliwatt output power) light-emitting diode lights until photoequilibrium was reached. The samples were kept in the dark unless otherwise indicated.
UV-Visible Spectroscopy
The UV-visible spectra were measured with a PerkinElmer Life Sciences LAMBDA 850 UV-visible spectrophotometer as previously described (11, 28, 49). The samples were diluted with buffer (30 mm Tris·HCl, pH 8.0) to obtain an approximate A280 value of 0.1. All measurements were carried out at ambient conditions (room temperature) and in complete darkness. The dark reversion was measured with Hitachi U-2910 UV-visible spectrophotometer (Hitachi) by sequentially recording the absorption spectrum in the wavelength range of 500–850 nm after the photoequilibrium had been reached under red light illumination. The samples were diluted with buffer (30 mm Tris·HCl, pH 8.0) to obtain approximate A700 value of 1. PAS-GAF and PAS-GAFmon reversion data were recorded at 3-min intervals; longer fragments (PAS-GAF-PHY, PAS-GAF-PHYmon, full-length (FL), and FLmon) were recorded also at 15-min intervals. The UV-visible detection light did not affect the steady-state spectra of the constructs, but the rate of the dark reversion was slightly increased by the detection system (data not shown). However, this effect was minimized by recording data at long time intervals. The effect can be considered identical in all measurements, and therefore, the results are comparable. The exponential fits from normalized dark reversion data were calculated using Equation 1.
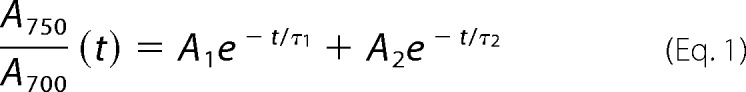 |
Here t is time, A700 and A750 are absorption values at specified wavelength, An is the decay amplitude, and τn is the time constant of the decay component.
SEC with Multi-wavelength Detection
The size-exclusion experiments were conducted as previously described (28). In the experiment Yarra 3u SEC-3000 (300 × 7.80 mm) columns (Phenomenex, Torrance, CA) and a mobile phase of 20 mm Tris, 150 mm NaCl, pH 7.5 with a flow rate of 1 ml/min were used. A protein sample of 5–6 mg/ml concentration was pre-illuminated until photoequilibrium was reached, and 30 μg of protein was injected for each chromatographic run. The elution profiles were detected with multi-wavelength UV-visible detection as described elsewhere (28), and the molecular weights of the phytochrome fragments were determined by comparison to a protein size marker standard (Phenomenex) (28).
Small Angle X-ray Scattering (SAXS)
SAXS data were collected at the beamline BM29 (BioSAXS) at the European Synchrotron Radiation Source, as in Takala et al. (11). All the monomer mutant samples (PAS-GAF-PHYmon and FLmon) as well as full-length wild-type phytochrome were collected in buffer (30 mm Tris·HCl, pH 8.0), whereas wild-type PAS-GAF-PHY in buffer (20 mm Tris·HCl, 150 mm NaCl, pH 7.0) is reported elsewhere (11). The data reduction and analysis was done with the PRIMUS tool (50).
Estimation of the Dimerization Energies
The free energies of dimerization of the PAS-GAF and dimerization histidine phosphotransfer (DHp) fragments were estimated using molecular dynamics simulations. Umbrella sampling was used in combination with the Weighted Histogram Analysis Method (WHAM), both implemented in Gromacs 5.0 (51, 52). For the PAS-GAF subunit (residues 5–325), simulations were based on PDB entry 4Q0J (12), with chromophore parameters adapted from Kaminski et al. (53). For the DHp subunit (residues 505–590), a homology model with the D. radiodurans sequence, based on PDB entry 2C2A (24), created using the SWISS-MODEL server (54), was used as a starting point.
Each starting structure was 1) energy-minimized, 2) solvated with 50 mm NaCl in TIP3P water, 3) equilibrated with position restraints under NVT for 50 ps, and 4) equilibrated with position restraints under NPT conditions for 500 ps. Next, the structures were equilibrated without restraints for 100 ns, and the trajectories were deemed stable on this timescale based on root mean square deviation analysis. Initial conformations for umbrella sampling were generated by pulling the centers of mass of the opposing monomer chains along one of the box vectors from the equilibrium distance at a rate of 5 nm/ns. Conformations separated by 0.1 nm were extracted and used for umbrella sampling covering 2 ns at each window, with a harmonic force coefficient of 1000 kJ/mol/nm2. Analysis was conducted using the Gromacs tool gmx wham, and the built-in bootstrapping feature ensured numerical robustness (with errors <5%).
The CHARMM27 force field (55) was used throughout. All simulations employed a modified Berendsen thermostat at 300 K (56) and the Parrinello-Rahman barostat at 1 atm with time constants of 0.5 and 2.0 ps, respectively. All bonds were constrained using the LINCS algorithm, and a time step of 2 fs was used. Periodic boundary conditions were imposed in three dimensions. Electrostatics was handled using the Particle Mesh Ewald algorithm with 4th order interpolation and a Fourier grid spacing of 0.12 nm. Neighbor searching was done using the Verlet cutoff scheme with a maximum allowed error per particle of 0.005 kJ/mol/ps.
Results
Size-exclusion Chromatography
Three previously describedmutations (F145S, L311E, L314E) (42) were applied to the phytochrome fragments to produce PAS-GAFmon, PAS-GAF-PHYmon, and FLmon (Fig. 1A). The mutations remove stabilizing interactions and induce repulsion between the GAF domains. This leads to monomerization of the PAS-GAF construct (42). We confirmed this using size-exclusion chromatography (Fig. 1B, see panels PAS-GAFmon and PAS-GAF). The same mutations in the photosensory fragment PAS-GAF-PHYmon also led to a monomeric protein (Fig. 1B, PAS-GAF-PHYmon and PAS-GAF-PHY). This indicates that the PHY domains contribute very weakly or not at all to stabilizing the dimers in solution. This is consistent with the absence of PHY-PHY interactions in the published crystal structures of the PAS-GAF-PHY fragment (11, 12). Introducing the same mutations into the full-length phytochrome (FLmon) produced a protein that eluted as a mixture of monomers and dimers in the column (Fig. 1B, FLmon and FL). Thus, a second dimerization interface, formed between the HK domains, must exist.5
FIGURE 1.
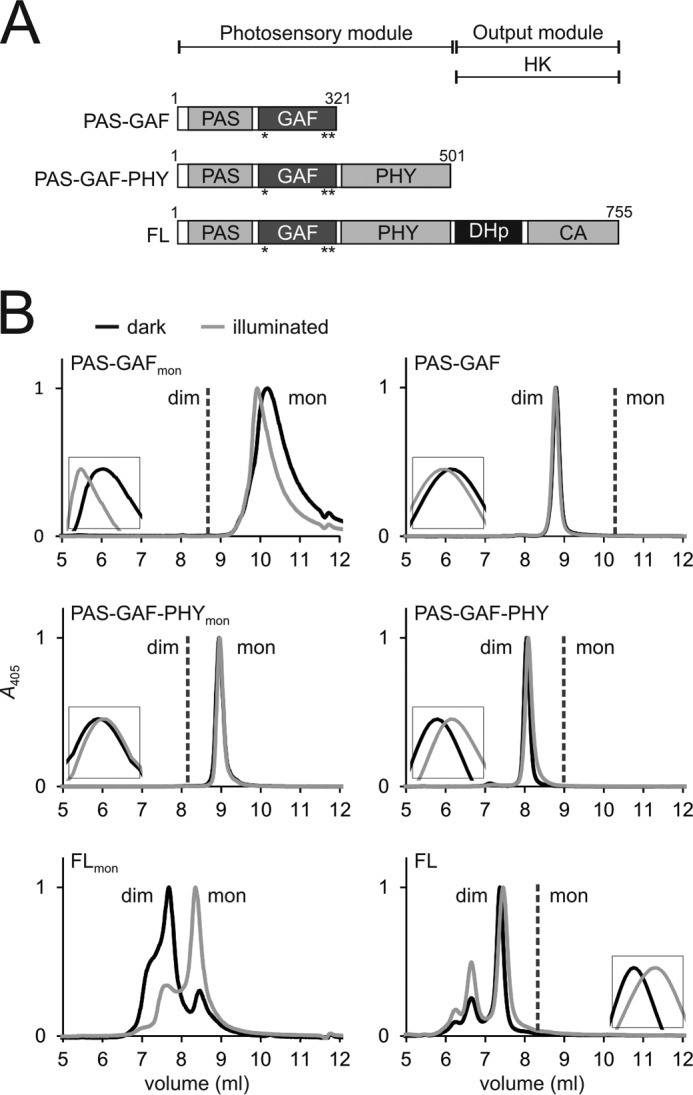
Domain organization and size-exclusion analysis of the phytochrome constructs. A, schematic presentation of the phytochrome fragment used. Mutated residues are indicated as asterisks (*). CA, catalytic ATP binding domain). B, the size exclusion of the monomer mutants is shown in the left panels, and the wild-type constructs are shown in the right panels (28). To consider exclusively holoproteins, the retentions are plotted at a 405-nm wavelength. Samples in the dark (Pr) state are plotted as black lines; the illuminated (Pfr-like) samples are plotted as gray lines. Insets show each retention peak in higher magnification. Whether the construct elutes as monomers (mon) or dimers (dim) is indicated in the figure. The calculated molecular masses are 65 kDa (PAS-GAF-PHYmon) and 220/116 kDa (FLmon). The size of PAS-GAFmon has not been approximated because the sample eluted outside the optimal resolution range of the column. The elution profiles verify that the PAS-GAF-PHY mutant is exclusively monomeric. However, the full-length mutant elutes as a monomer/dimer mixture. The Pr state favors dimers and the Pfr state monomers. Note that the dimeric (28) and monomeric proteins were run at slightly different buffer conditions causing a slight drift of the retention peaks.
Fig. 1B also shows the effect of illumination on the retention of the phytochrome fragments. Although the retention curves of the truncated constructs and the wild-type FL constructs undergo small shifts (Fig. 1B, see the insets), the FLmon construct undergoes a dramatic transition from a dominantly dimeric mixture in Pr to a dominantly monomeric mixture in Pfr. We did not observe any concentration-dependent change in the dimer-monomer ratio in the concentration range from 0.4 μm to 155 μm (data not shown). The wild-type FL protein has two dimerization interfaces between the HK and GAF domains. Because the mutations disturb the latter, this result shows that the interface formed between the sister HK units is significantly altered and weakened in the Pfr state.
Absorption Spectroscopy
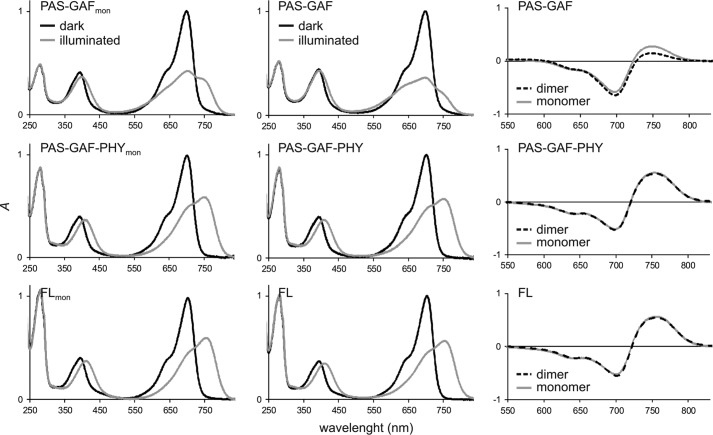
UV-visible absorption measurements reveal that the absorption spectra of the two longer monomeric and dimeric constructs are indistinguishable in Pr and Pfr, respectively, as shown in Fig. 2. Therefore, we can exclude that the mutations directly control the photochromic properties of these proteins. The shortest construct, PAS-GAFmon, is an exception. It has an increased red light absorption in the illuminated state compared with its dimeric counterpart, as already reported by Auldridge et al. (42).
FIGURE 2.
Absorption spectra of the phytochrome constructs. The left and middle panels show the absorption spectra of the monomer mutants (left) and their wild-type counterparts (middle). Dark (Pr) sample spectra are plotted as black lines, and illuminated (Pfr or Pfr-like) sample spectra are plotted as gray lines. The wild-type fragment spectra (17, 28) and the PAS-GAFmon spectra (42) are published elsewhere and are shown here for comparison. The PAS-GAF-PHY and full-length mutants show almost identical spectra with their wild-type counterparts, whereas the PAS-GAF spectra of the illuminated state are different. The right panels show the Pfr-minus-Pr difference spectra of all six constructs. The difference spectra show that the wild-type and mutant spectra are almost identical in the case of PAS-GAF-PHY and full-length phytochrome, whereas they differ in PAS-GAF fragments.
Dark Reversion
Next, we characterized the thermally driven dark reversion time of the constructs by monitoring the time dependence of the ratio of the absorption peaks at 700 and 750 nm (Fig. 3A). The time constants of the reversion were also estimated (Fig. 3B). Among the dimeric fragments, PAS-GAF-PHY has a slower dark reversion toward the Pr state than the FL protein. This was previously attributed to the absence of interactions between the histidine kinase domains (28). The PAS-GAF fragment thermally reverts most rapidly, but it also has an incomplete photocycle (57).
FIGURE 3.
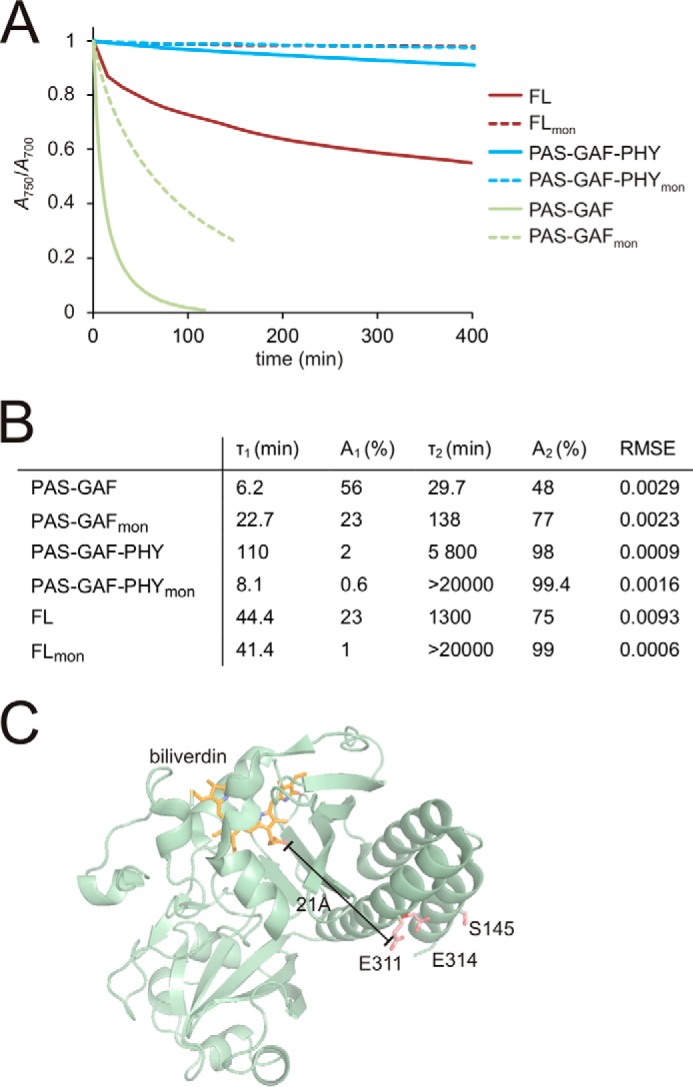
A, thermal (dark) reversion of the phytochrome constructs. To demonstrate the qualitative differences in the dark reversion, the absorption ratio of A750 and A700 values was normalized to 1 in saturating red light illumination and to 0 after illumination with far-red light. The dimeric wild-type fragments and the monomerizing mutants are shown as solid and dotted lines, respectively. The PAS-GAFmon reverts more slowly to the Pr state than its wild-type counterpart. For PAS-GAF-PHYmon and FLmon dark reversion is absent. B, table of dark reversion time constants (τn). Satisfactory fits were obtained by using a sum of two decay components. The obtained values for dimers closely resembled the previously reported ones (28). An, decay amplitude; RMSE, root mean square error. C, distance between mutated residues and the chromophore. The distance between the biliverdin chromophore (orange) and the mutated residues (red) is indicated. The crystal structure of PAS-GAFmon (PDB code 4IJG; Ref. 64) was used for the figure.
Importantly, the monomeric PAS-GAF-PHYmon and FLmon did not dark-revert at all. Also, the monomeric PAS-GAFmon reverted slower to the Pr state than the dimeric PAS-GAF. The mutated residues reside far away (>21 Å) from the chromophore (Fig. 3C), and we have ruled out a direct effect of the mutations on the photochromic properties above. Thus, the dark reversion data imply that both dimerization interfaces are required for efficient dark reversion from the final Pfr state to the Pr state. The absence of the HK/HK interface slows down dark reversion, whereas the absence of the GAF/GAF interface completely suppresses it.
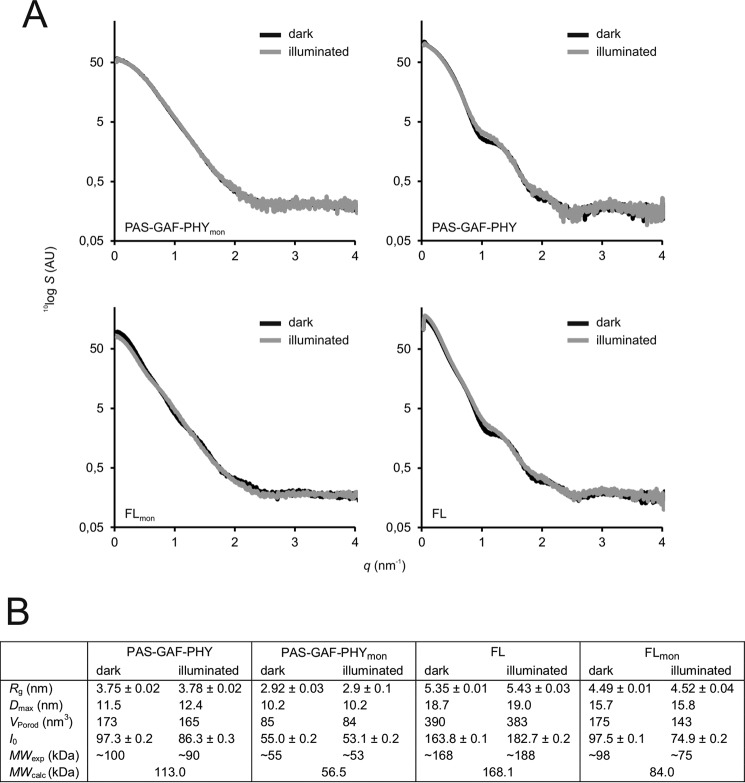
SAXS
Small angle x-ray scattering reveals information about the overall size and shape of proteins. In Fig. 4 we show SAXS data for the PAS-GAF-PHY and FL constructs. The scattering intensity at scattering angle q = 0 (I0) and the structural parameters estimated from the SAXS curves (Fig. 4B) verify that PAS-GAF-PHYmon is a monomer and PAS-GAF-PHY is a dimer. I0 and the molecular weight that is computed from it did not change upon illumination of PAS-GAF-PHYmon, which confirms our findings from size-exclusion chromatography. Furthermore, the scattering patterns for dark and illuminated PAS-GAF-PHYmon appear almost identical within the accuracy of the SAXS measurements (Fig. 4A). Consequently, the structural parameters obtained from the SAXS data are insensitive to illumination (Fig. 4B). Thus, the oligomeric state and shape were not affected by illumination, albeit it cannot be excluded that smaller scale difference signals exist that are not resolved by the standard SAXS measurements. The difference signal of dimeric PAS-GAF-PHY has been considered previously (11).
FIGURE 4.
SAXS of the phytochrome constructs. A, scattering of the phytochrome fragments (S) as a function of scattering angle (q) is plotted on a logarithmic scale. Dark (Pr) state samples are plotted as black lines; illuminated (mostly Pfr) state samples are plotted in gray lines. The scattering data of PAS-GAF-PHY is from Takala et al. (11). B, structural parameters calculated from the SAXS scattering. Rg, radius of gyration; Dmax, maximum particle dimension; VPorod, Porod volume; I0, forward scattering. The molecular weights (MWexp) were estimated from the I0 of BSA using the formula MWexp ≈ MW(BSA) × [I0(sample)/I0(BSA)], where MW(BSA) = 66 kDa. Theoretical molecular weights (MWcalc) were calculated from the protein sequence (58).
The SAXS signal of wild-type FL confirms that it is a dimer in solution. The apparent increase in protein size after illumination as judged from I0 and Rg (Fig. 4B) is explained by light-induced oligomerization as observed in Takala et al. (28) and Fig. 1B. The scattering curves of the dark and illuminated FL sample deviate from each other at a scattering angle (q) of ∼1 nm−1, in similar fashion as in PAS-GAF-PHY (11) (Fig. 4A). For the mutated construct FLmon, the illuminated sample yielded a molecular weight close to the monomeric size. The dark FLmon protein appeared at a significantly larger molecular weight (Fig. 4B), although the increase in size appears to be smaller than in the SEC measurements (see Fig. 1B). This could be due to the difference in salt concentrations in the two experiments. The SEC experiments were performed in 150 mm NaCl, whereas the buffer for SAXS measurement did not contain any salt. We note, however, that the observed increase in size qualitatively confirms the light-induced change in monomer/dimer ratio identified by size-exclusion chromatography (Fig. 1).
Molecular Dynamics Simulation of the Dimerization Interfaces
Molecular Dynamics simulations with umbrella sampling were used to estimate the interaction strengths of the two dimerization interfaces. We calculated the potentials of mean force (PMFs) for the dissociation of the dimeric PAS-GAF and HK domains separately (51). Note that we consider a homology model of the DHp fragment as representative of the whole HK domain (Fig. 5A). The monomers were pulled apart along the vector connecting their centers of mass, as schematically shown by the arrows in Fig. 5A. The resulting PMFs reflect the Gibbs free energies of dissociation and are measures of the dimerization propensity of each interface.
FIGURE 5.
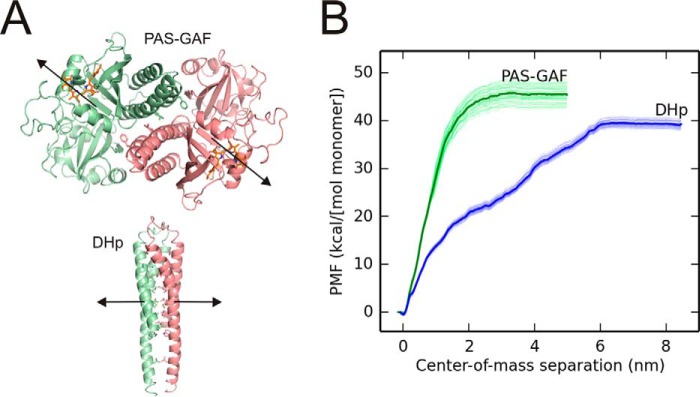
Free energy calculations of the separation of the PAS-GAF and DHp fragments. A, the domain models used for the molecular dynamics simulations, based on crystal structures of D. radiodurans PAS-GAF fragment (PDB entry 4Q0J; Ref. 12) and a DHp fragment from Thermotoga maritima histidine kinase domain (PDB entry 2C2A; Ref. 24). Each monomer is shown in green or red, the approximate direction of the pulling is shown as arrows. The three residues of the PAS-GAF fragment that are mutated in PAS-GAFmon as well as the interacting hydrophobic residues between the DHp helices are presented as sticks. B, the PMF is plotted as a function of the center of mass separation. The separation energies of the PAS-GAF fragment are shown in green and DHp fragment in blue. The thin lines show 50 independent curves from bootstrap analysis. The total binding energies of the both interfaces are approximately similar, although the shapes of the plotted curves differ from each other.
Fig. 5B shows that the GAF and DHp interfaces reach plateaus at separations of 2 and 6 nm, respectively. The plateau for the PAS-GAF fragment appears at slightly higher energy compared with the plateau for the DHp fragment. Even though the computational procedure should correctly estimate the entropy due to fast fluctuations of the protein and water,6 it underestimates the positional and rotational entropy of the monomeric proteins (due to the limited sampling in molecular dynamics simulations) (59). This means that the PMF will decrease for greater separations. The magnitude of this entropic contribution should be very similar for the two protein fragments considered, and we therefore conclude from the molecular dynamics simulations that the relative binding strengths of both dimer interfaces are of similar magnitude, with the PAS-GAF fragment forming a slightly more stable interface.
Fig. 5B also shows the PAS-GAF domains are completely dissociated at a small separation (∼2 nm), whereas the DHp/DHp interface has to be separated by several nanometers before it breaks completely. The former behavior is characteristic of a rigid interface between two compact structures, which is expected for PAS-GAF. The latter observation hints at a more flexible dimerization interface between the long helices of the DHp domains, as might be expected for dissociation of the leucine zipper-like motif that form the interface. Although this structural interpretation is intuitively satisfying, it should be considered with some caution. This is because the shapes of the PMF curves depend on how the trajectories of separation are chosen. The free energy difference between monomer and dimer, however, is a path-independent state function.
Discussion
Three Monomerizing Mutations Reveal Two Dimerization Interfaces
We demonstrate that modification of the hydrophobic interactions in the GAF/GAF interface of the D. radiodurans phytochrome does not only fully monomerize the PAS-GAF fragment as reported by Auldridge et al. (42) but that it also monomerizes the PAS-GAF-PHY fragment. This implies that the PHY domains do not form stabilizing dimer contacts in solution, which is consistent with crystal structures (11, 12) and electron micrographs (12, 19) of the D. radiodurans phytochrome. On the other hand, crystal structures of a phytochrome from Pseudomonas aeruginosa (9, 22) indicate that the sister PHY domains approach each other. Different phytochrome types could have different interaction patterns. Also, the deviating finding could be caused by crystal contacts that force the PHY domains together. We note that the electron micrographs (12, 19) and our data, obtained for the D. radiodurans phytochrome in solution, are not prone to packing artifacts, and we conclude, therefore, that the sister PHY domains do not interact.
Whereas the mutations completely monomerize the PAS-GAF and PAS-GAF-PHY fragments, they result in a mixture of monomers and dimers when applied to the full-length phytochrome. This is because the HK domains stabilize the dimeric form. Thus, our data directly confirm the existence of two dimerization interfaces in the D. radiodurans phytochrome, one between sister GAF domains and one between the HK domains. This is also consistent with our umbrella sampling molecular dynamics simulations, which show that both interfaces support dimer formation.
The Histidine Kinase Domains Can Be Separated by Red-light Illumination
We find that the monomer/dimer ratio of FLmon can be reversibly modified by red and far-red light illumination (Fig. 1). This means that at least when the GAF/GAF interactions are impaired, the HK/HK interface is broken by illumination. We consider this to be caused by a light-induced conformational change in the protein.
For the wild-type phytochrome this finding may imply that the dimers fully or partially separate upon red-light illumination at the HK interface, possibly in a zipper-like motion. The dimeric protein will still be held together by the GAF/GAF interactions. One structural model would be that the GAF/GAF interface acts as a pivot point against which the dimer opening in between the PHY domains can tear the HK domains (partially) apart, although our FLmon data suggest that the HK/HK interface is changed by light even when this pivot point is impaired. This pivot point, however, may be needed for other light-induced changes in the HK domain, as the HK domains are prone to form Pfr-state oligomers in wild-type FL only (28).
The prevailing perception is that the leucine zipper-like motif holds the HK domains together tightly. However, next to the finding in this paper, there is growing evidence that the HK domains partially or fully separate in Pfr. First, the large opening motion of the PAS-GAF-PHY fragment, observed by crystallography and solution x-ray scattering, suggests that a driving force for separating the HK interface exists (11). Second, a large structural change upon illumination is supported by our SAXS data (see Fig. 4B) and the change in SEC retention of the wild-type FL (see the insets in Fig. 1B and see Takala et al. (28)). Finally, cryo-electron micrographs of the Pr and Pfr states (12, 19) are also consistent with this mechanism. We consider none of these evidences to be conclusive by themselves. However, the growing number of items on the list together with our new data suggest that a mechanism in which the sister HK domains separate in the Pfr state should be seriously considered.
Light-driven dimerization or clustering has been widely applied in optogenetic applications (60–62). The mutated FLmon construct is an example of a protein that monomerizes under light exposure, which would make it an interesting candidate for novel optogenetic applications.
Phytochrome Dimerization Does Not Alter the Photochromicity in the Longer Fragments
We confirmed an earlier report that breaking the GAF/GAF dimerization interface changes the absorption properties of the PAS-GAF fragment when illuminated by red light. This is supported by the results where a covalent linkage between the sister GAF domains does not alter the PAS-GAF photocycle (63). The origin of the spectral change is not clear as the mutated residues reside far from the chromophore (Fig. 3C) and they cannot directly interfere with the chromophore binding pocket. However, we do not observe this change for the Pfr spectra of the PAS-GAF-PHYmon and FLmon proteins, which exhibit identical photochromicity to their dimeric counterparts. This indicates that the conformation of the biliverdin and its embedment into the protein is similar in the dimeric and monomeric cases. Whether the details of the photocycle (i.e. intermediates and kinetics, apart from dark reversion) are altered by the monomerizing mutations remains a topic for further investigation.
Both Dimerization Interfaces Are Required for Efficient Dark Reversion
Fig. 3 shows that dark reversion of the phytochrome fragments is critically influenced by the mutations. Earlier, it has been shown that removing the HK domain from the dimeric phytochrome reduces the dark reversion rate (12, 28). In this paper we show that weakening the GAF-GAF interactions in the PAS-GAF-PHYmon and FLmon mutants inhibited dark reversion altogether. Thus, although the HK dimerization interface has an effect on the dark reversion, the GAF-GAF dimerization is even more important for it. Normal dark reversion, therefore, requires that both GAF/GAF and HK/HK interfaces are intact.
This acceleration of dark reversion may be due to strain that is built into the protein between the two interfaces (Fig. 6A). The strain may arise when the switch region in the PHY domain is in the Pfr conformation (11), and it is likely concentrated around the long scaffolding helices. It would pull the dimer into a Pr-like form, thus facilitating the dark reversion of the chromophore surroundings. If one of the dimerization interfaces is lost due to truncations (12, 28) or mutations (this work), the structure is not strained, and dark reversion is significantly slower.
FIGURE 6.
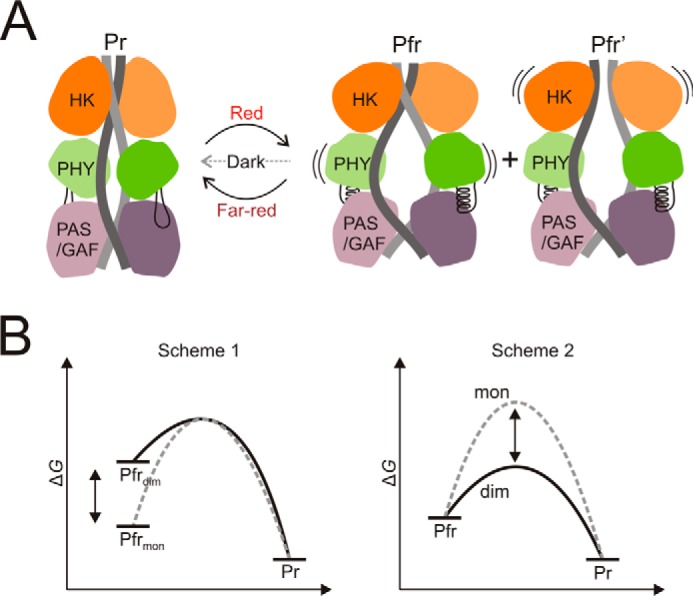
Model scheme of phytochrome dimerization and dark reversion energetics. A, after red light illumination the refolding of the PHY tongue region causes PHY domains to separate and the dimer to open (11, 12). The opening motion along with the two dimerization interfaces creates strain in the protein. This strain is increased by structural changes upon red light illumination, thus facilitating dark reversion. Note that the alternative Pfr form with separated HK domains is denoted as Pfr′, in accordance with Burgie et al. (12). B, hypothetical free energy schemes of dark reversion. Scheme 1, the Pfr state energy of the monomer (mon) and the dimer (dim) differ; Scheme 2, the energies of the transition state for back reversion differ.
The decrease in dark recovery implies that the activation energy for Pfr-to-Pr transition is increased. This could be due to a reduction of the free energy of the Pfr state for the monomeric constructs (Fig. 6B, Scheme 1). Alternatively, it could also be caused by a rise of the energy of the rate-limiting transition state (Fig. 6B, Scheme 2). Both scenarios are possible and distinguishing between them might be an interesting question for future studies. Our data show that the photochromicity of the chromophore is unchanged by the mutations. This means that the relevant structural changes must occur within the protein and not within, or close to, the chromophore.
Model of the Effect of Dimerization on Photocycle
Based on the results presented here and on other results (11, 12), we propose a model for the role of the two dimer interfaces in the photocycle of phytochromes (Fig. 6A). In the Pr state the full-length phytochrome adopts a “closed” state where the neighboring PHY domains reside relatively close to each other. Illumination causes the phytochrome to switch to the Pfr state leading to refolding of the PHY tongue and separation and rotation of the neighboring PHY domains (11, 12). This conformational change modulates the stability of the HK/HK interface. Our data on the mutated proteins demonstrate that the interface breaks. In extension to the wild-type protein, this finding may imply that the HK domains can separate in Pfr, forming a second Pfr form denoted Pfr′ (12) (Fig. 6A). We suggest that separation of the HK domains should be considered as a potential mechanism of modulating the cellular activity of the protein.
Acknowledgments
We thank Prof. R. D. Vierstra and Prof. K. T. Forest for providing the plasmids for wild-type phytochromes. We acknowledge BM29 SAXS beamline access at the European Synchrotron Radiation Facility (ESRF) and its personnel as well as A. Liukkonen and Dr. H. Lehtivuori for assistance in laboratory work and experiments.
This work was supported by Academy of Finland Grant 138063 (to J. A. I.), by a Finnish Cultural Foundation grant (J. A. I.) and Grant 0131067 (to H. T.), Foundation of Strategic Research, Sweden, and the Swedish and European Research Councils, agreement 279944 (to S. W.).
Next to the formation of second interface between the HK domains, our data could also be interpreted as a stabilization of a PHY-PHY dimer interface or formation of a complex interface across both the PHY and HK regions. However, many structures of HK domains show dimerization between HK monomers and PHY-HK interactions are inconsistent with the known structures of the D. radiodurans phytochrome. We, therefore, consider that the new interface is formed between HK domains.
J. Hub, personal communication.
- PAS
- (PER, ARNT, SIM)
- GAF
- cGMP phosphodiesterase, adenylate cyclase, FhlA
- PHY
- phytochrome-specific GAF-related
- DHp
- dimerization histidine phosphotransfer
- FL
- full-length
- HK
- histidine kinase
- PMF
- potentials of mean force
- SEC
- size-exclusion chromatography
- SAXS
- small angle x-ray scattering.
References
- 1. Quail P. H. (2002) Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3, 85–93 [DOI] [PubMed] [Google Scholar]
- 2. Rockwell N. C., Su Y. S., Lagarias J. C. (2006) Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karniol B., Vierstra R. D. (2003) The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. U.S.A. 100, 2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krieger A., Molina I., Oberpichler I., Michael N., Lamparter T. (2008) Spectral properties of phytochrome Agp2 from Agrobacterium tumefaciens are specifically modified by a compound of the cell extract. J. Photochem. Photobiol. B. 93, 16–22 [DOI] [PubMed] [Google Scholar]
- 5. Zienicke B., Molina I., Glenz R., Singer P., Ehmer D., Escobar F. V., Hildebrandt P., Diller R., Lamparter T. (2013) Unusual spectral properties of bacteriophytochrome Agp2 result from a deprotonation of the chromophore in the red-absorbing form Pr. J. Biol. Chem. 288, 31738–31751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burgie E. S., Vierstra R. D. (2014) Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell 26, 4568–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Auldridge M. E., Forest K. T. (2011) Bacterial phytochromes: more than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88 [DOI] [PubMed] [Google Scholar]
- 8. Essen L. O., Mailliet J., Hughes J. (2008) The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. U.S.A. 105, 14709–14714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang X., Kuk J., Moffat K. (2008) Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc. Natl. Acad. Sci. U.S.A. 105, 14715–14720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anders K., Daminelli-Widany G., Mroginski M. A., von Stetten D., Essen L. O. (2013) Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. J. Biol. Chem. 288, 35714–35725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takala H., Björling A., Berntsson O., Lehtivuori H., Niebling S., Hoernke M., Kosheleva I., Henning R., Menzel A., Ihalainen J. A., Westenhoff S. (2014) Signal amplification and transduction in phytochrome photosensors. Nature 509, 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burgie E. S., Wang T., Bussell A. N., Walker J. M., Li H., Vierstra R. D. (2014) Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion. J. Biol. Chem. 289, 24573–24587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stojković E. A., Toh K. C., Alexandre M. T., Baclayon M., Moffat K., Kennis J. T. (2014) FTIR spectroscopy revealing light-dependent refolding of the conserved tongue region of bacteriophytochrome. J. Phys. Chem. Lett. 5, 2512–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burgie E. S., Bussell A. N., Walker J. M., Dubiel K., Vierstra R. D. (2014) Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. U.S.A. 111, 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mailliet J., Psakis G., Feilke K., Sineshchekov V., Essen L. O., Hughes J. (2011) Spectroscopy and a high-resolution crystal structure of Tyr-263 mutants of cyanobacterial phytochrome Cph1. J. Mol. Biol. 413, 115–127 [DOI] [PubMed] [Google Scholar]
- 16. Bellini D., Papiz M. Z. (2012) Dimerization properties of the RpBphP2 chromophore-binding domain crystallized by homologue-directed mutagenesis. Acta Crystallogr. D Biol. Crystallogr. 68, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 17. Wagner J. R., Zhang J., Brunzelle J. S., Vierstra R. D., Forest K. T. (2007) High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J. Biol. Chem. 282, 12298–12309 [DOI] [PubMed] [Google Scholar]
- 18. Bellini D., Papiz M. Z. (2012) Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 20, 1436–1446 [DOI] [PubMed] [Google Scholar]
- 19. Li H., Zhang J., Vierstra R. D., Li H. (2010) Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 10872–10877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Evans K., Grossmann J. G., Fordham-Skelton A. P., Papiz M. Z. (2006) Small angle X-ray scattering reveals the solution structure of a bacteriophytochrome in the catalytically active Pr state. J. Mol. Biol. 364, 655–666 [DOI] [PubMed] [Google Scholar]
- 21. Karniol B., Wagner J. R., Walker J. M., Vierstra R. D. (2005) Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem. J. 392, 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang X., Kuk J., Moffat K. (2009) Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc. Natl. Acad. Sci. U.S.A. 106, 15639–15644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang X., Ren Z., Kuk J., Moffat K. (2011) Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature 479, 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marina A., Waldburger C. D., Hendrickson W. A. (2005) Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 24, 4247–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamada S., Sugimoto H., Kobayashi M., Ohno A., Nakamura H., Shiro Y. (2009) Structure of PAS-linked histidine kinase and the response regulator complex. Structure 17, 1333–1344 [DOI] [PubMed] [Google Scholar]
- 26. Casino P., Rubio V., Marina A. (2009) Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139, 325–336 [DOI] [PubMed] [Google Scholar]
- 27. Wagner J. R., Brunzelle J. S., Forest K. T., Vierstra R. D. (2005) A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 [DOI] [PubMed] [Google Scholar]
- 28. Takala H., Lehtivuori H., Hammarén H., Hytönen V. P., Ihalainen J. A. (2014) Connection between absorption properties and conformational changes in Deinococcus radiodurans phytochrome. Biochemistry 53, 7076–7085 [DOI] [PubMed] [Google Scholar]
- 29. Noack S., Michael N., Rosen R., Lamparter T. (2007) Protein conformational changes of Agrobacterium phytochrome Agp1 during chromophore assembly and photoconversion. Biochemistry 46, 4164–4176 [DOI] [PubMed] [Google Scholar]
- 30. Strauss H. M., Schmieder P., Hughes J. (2005) Light-dependent dimerisation in the N-terminal sensory module of cyanobacterial phytochrome 1. FEBS Lett. 579, 3970–3974 [DOI] [PubMed] [Google Scholar]
- 31. Anders K., von Stetten D., Mailliet J., Kiontke S., Sineshchekov V. A., Hildebrandt P., Hughes J., Essen L. O. (2011) Spectroscopic and photochemical characterization of the red-light sensitive photosensory module of Cph2 from Synechocystis PCC 6803. Photochem. Photobiol. 87, 160–173 [DOI] [PubMed] [Google Scholar]
- 32. Wagner D., Koloszvari M., Quail P. H. (1996) Two small spatially distinct regions of phytochrome B are required for efficient signaling rates. Plant Cell 8, 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sineshchekov V., Koppel' L., Esteban B., Hughes J., Lamparter T. (2002) Fluorescence investigation of the recombinant cyanobacterial phytochrome (Cph1) and its C-terminally truncated monomeric species (Cph1Delta2): implication for holoprotein assembly, chromophore-apoprotein interaction, and photochemistry. J. Photochem. Photobiol. B. 67, 39–50 [DOI] [PubMed] [Google Scholar]
- 34. Esteban B., Carrascal M., Abian J., Lamparter T. (2005) Light-induced conformational changes of cyanobacterial phytochrome Cph1 probed by limited proteolysis and autophosphorylation. Biochemistry 44, 450–461 [DOI] [PubMed] [Google Scholar]
- 35. Matsushita T., Mochizuki N., Nagatani A. (2003) Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424, 571–574 [DOI] [PubMed] [Google Scholar]
- 36. Oka Y., Matsushita T., Mochizuki N., Suzuki T., Tokutomi S., Nagatani A. (2004) Functional analysis of a 450-amino acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell 16, 2104–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharrock R. A., Clack T. (2004) Heterodimerization of type II phytochromes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 11500–11505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clack T., Shokry A., Moffet M., Liu P., Faul M., Sharrock R. A. (2009) Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21, 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang Y., Inouye M. (1991) Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 88, 11057–11061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marina A., Mott C., Auyzenberg A., Hendrickson W. A., Waldburger C. D. (2001) Structural and mutational analysis of the PhoQ histidine kinase catalytic domain: insight into the reaction mechanism. J. Biol. Chem. 276, 41182–41190 [DOI] [PubMed] [Google Scholar]
- 41. Casino P., Miguel-Romero L., Marina A. (2014) Visualizing autophosphorylation in histidine kinases. Nat. Commun. 5, 3258. [DOI] [PubMed] [Google Scholar]
- 42. Auldridge M. E., Satyshur K. A., Anstrom D. M., Forest K. T. (2012) Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J. Biol. Chem. 287, 7000–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoon J. M., Hahn T. R., Cho M. H., Jeon J. S., Bhoo S. H., Kwon Y. K. (2008) The PHY domain is required for conformational stability and spectral integrity of the bacteriophytochrome from Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 369, 1120–1124 [DOI] [PubMed] [Google Scholar]
- 44. Zhang J., Stankey R. J., Vierstra R. D. (2013) Structure-guided engineering of plant phytochrome B with altered photochemistry and light signaling. Plant Physiol. 161, 1445–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang C., Sang J., Wang J., Su M., Downey J. S., Wu Q., Wang S., Cai Y., Xu X., Wu J., Senadheera D. B., Cvitkovitch D. G., Chen L., Goodman S. D., Han A. (2013) Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol. 11, e1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Diensthuber R. P., Bommer M., Gleichmann T., Möglich A. (2013) Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure 21, 1127–1136 [DOI] [PubMed] [Google Scholar]
- 47. Mechaly A. E., Sassoon N., Betton J. M., Alzari P. M. (2014) Segmental helical motions and dynamical asymmetry modulate histidine kinase autophosphorylation. PLoS Biol. 12, e1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ferris H. U., Coles M., Lupas A. N., Hartmann M. D. (2014) Crystallographic snapshot of the Escherichia coli EnvZ histidine kinase in an active conformation. J. Struct. Biol. 186, 376–379 [DOI] [PubMed] [Google Scholar]
- 49. Lehtivuori H., Rissanen I., Takala H., Bamford J., Tkachenko N. V., Ihalainen J. A. (2013) Fluorescence properties of the chromophore binding domain of bacteriophytochrome from Deinococcus radiodurans. J. Phys. Chem. B. 117, 11049–11057 [DOI] [PubMed] [Google Scholar]
- 50. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 51. Hub J. S., de Groot B. L., van der Spoel D. (2010) g_wham: free-weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput. 6, 3713–3720 [Google Scholar]
- 52. Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M. R., Smith J. C., Kasson P. M., van der Spoel D., Hess B., Lindahl E. (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kaminski S., Daminelli G., Mroginski M. A. (2009) Molecular dynamics simulations of the chromophore binding site of Deinococcus radiodurans bacteriophytochrome using new force field parameters for the phytochromobilin chromophore. J. Phys. Chem. B. 113, 945–958 [DOI] [PubMed] [Google Scholar]
- 54. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 55. Bjelkmar P., Larsson P., Cuendet M. A., Hess B., Lindahl E. (2010) Implementation of the CHARMM force Field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J. Chem. Theory Comput. 6, 459–466 [DOI] [PubMed] [Google Scholar]
- 56. Bussi G., Donadio D., Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 [DOI] [PubMed] [Google Scholar]
- 57. Wagner J. R., Zhang J., von Stetten D., Günther M., Murgida D. H., Mroginski M. A., Walker J. M., Forest K. T., Hildebrandt P., Vierstra R. D. (2008) Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J. Biol. Chem. 283, 12212–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wilkins M. R., Gasteiger E., Bairoch A., Sanchez J. C., Williams K. L., Appel R. D., Hochstrasser D. F. (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552 [DOI] [PubMed] [Google Scholar]
- 59. Zhang B. W., Brunetti L., Brooks C. L., 3rd (2011) Probing pH-dependent dissociation of HdeA dimers. J. Am. Chem. Soc. 133, 19393–19398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Taslimi A., Vrana J. D., Chen D., Borinskaya S., Mayer B. J., Kennedy M. J., Tucker C. L. (2014) An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 5, 4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tischer D., Weiner O. D. (2014) Illuminating cell signalling with optogenetic tools. Nat. Rev. Mol. Cell Biol. 15, 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pathak G. P., Strickland D., Vrana J. D., Tucker C. L. (2014) Benchmarking of optical dimerizer systems. ACS Synth. Biol. 3, 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bornschlögl T., Anstrom D. M., Mey E., Dzubiella J., Rief M., Forest K. T. (2009) Tightening the knot in phytochrome by single-molecule atomic force microscopy. Biophys. J. 96, 1508–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bhattacharya S., Auldridge M. E., Lehtivuori H., Ihalainen J. A., Forest K. T. (2014) Origins of fluorescence in evolved bacteriophytochromes. J. Biol. Chem. 289, 32144–32152 [DOI] [PMC free article] [PubMed] [Google Scholar]