Abstract
Appropriate repair of DNA lesions and the inhibition of DNA repair activities at telomeres are critical to prevent genomic instability. By fuelling the generation of genetic alterations and by compromising cell viability, genomic instability is a driving force in cancer and aging1, 2. Here we identify MAD2L2 (also known as MAD2B or REV7) through functional genetic screening as a novel factor controlling DNA repair activities at mammalian telomeres. We show that MAD2L2 accumulates at uncapped telomeres and promotes non-homologous end-joining (NHEJ)-mediated fusion of deprotected chromosome ends and genomic instability. MAD2L2 depletion causes elongated 3′ telomeric overhangs, implying that MAD2L2 inhibits 5′ end-resection. End-resection blocks NHEJ while committing to homology-directed repair (HDR) and is under control of 53BP1, RIF1 and PTIP3. Consistent with MAD2L2 promoting NHEJ-mediated telomere fusion by inhibiting 5′ end-resection, knockdown of the nucleases CTIP or EXO1 partially restores telomere-driven genomic instability in MAD2L2-depleted cells. Control of DNA repair by MAD2L2 is not limited to telomeres. MAD2L2 also accumulates and inhibits end-resection at irradiation (IR)-induced DNA double-strand breaks (DSBs) and promotes end-joining of DSBs in multiple settings, including during immunoglobulin class switch recombination (CSR). These activities of MAD2L2 depend on ATM kinase activity, RNF8, RNF168, 53BP1 and RIF1, but not on PTIP, REV1 and REV3, the latter two acting with MAD2L2 in translesion synthesis (TLS)4. Together our data establish MAD2L2 as a critical contributor to the control of DNA repair activity by 53BP1 that promotes NHEJ by inhibiting 5′ end-resection downstream of RIF1.
As the processes underlying telomere-driven genomic instability are not completely understood we performed a functional genetic screen to identify telomere-induced genomic instability regulators (TIGIRs). The TIGIR-screen relies on well-controlled reversible inactivation of telomere component TRF2 by expressing temperature-sensitive TRF2-I468A (TRF2ts) in Trf2−/−;p53−/− mouse embryo fibroblasts (MEFs)5. At the permissive temperature (32°C) TRF2ts MEFs have intact TRF2-mediated telomere protection, but at non-permissive temperatures (37-39°C) TRF2ts is inactive, causing ATM kinase activation, accumulation of DNA damage response (DDR) proteins and NHEJ-dependent ligation at chromosome ends5. In the TIGIR-screen (Fig. 1a) prolonged TRF2-inactivation causes TRF2ts cells to irreversibly arrest or die due to severe chromosome end fusion that drives cells into genomic crisis6-8. However, diminished telomere fusion, such as upon DNA-ligaseIV- or RNF8-deficiency, allows survival and proliferation despite telomere uncapping9-11.
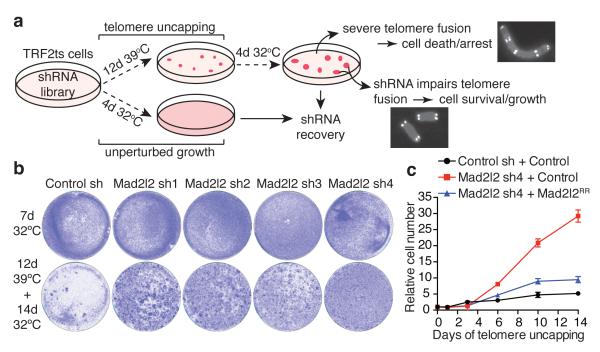
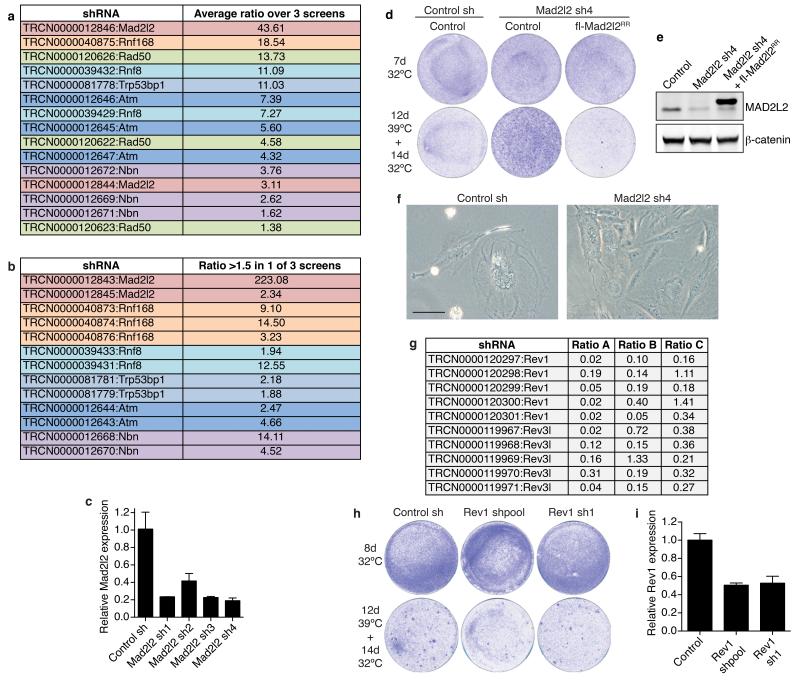
Figure 1. A functional genetic screen identifies Mad2l2 as a critical factor in telomere-driven genomic instability.
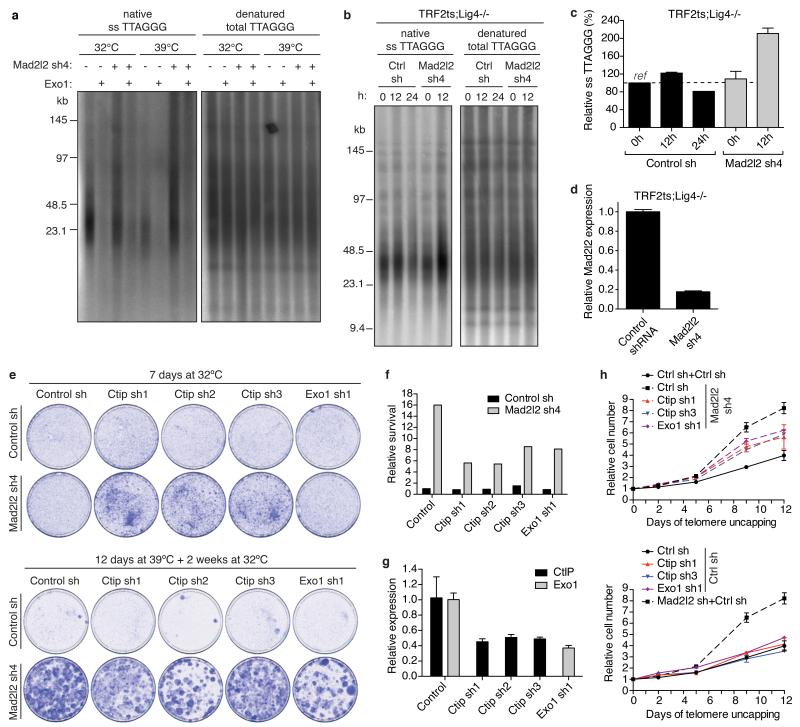
a, Outline of TIGIR-screen to identify factors controlling telomere-driven genomic instability. b, Survival assays of TRF2ts MEFs infected with control or Mad2l2 shRNAs, stained after growth as indicated. c, Growth curves at 39°C of TRF2ts cells transduced with control or Mad2l2 sh4 shRNAs and complemented with empty control or RNAi-resistant Flag-Mad2l2RR (quadruplicate ± s.d.).
In a triplicate TIGIR-screen we assayed 1976 short-hairpin RNAs (shRNAs), targeting 391 genes linked to DDR, for shRNAs that allow TRF2ts cells to survive 12 days of telomere uncapping. Among shRNA-targets significantly enriched in at least 2 of 3 screens were multiple factors previously shown to control telomere fusion (Extended Data Fig. 1a,b), including ATM, NBS1, RAD50, 53BP1 and RNF811-15. Remarkably, the most prominent screen hit, also independently recovered in a genome-wide TIGIR-screen (our unpublished results), was MAD2L2 (Extended Data Fig. 1a,b). MAD2L2 has no known function at telomeres but acts as a non-catalytic interaction partner of REV1 and REV3 in TLS4. Knockdown of Mad2l2 with independent shRNAs markedly increased survival of TRF2ts MEFs subjected to telomere uncapping, which was abolished by complementation with exogenous shRNA-resistant human MAD2L2 (Fig. 1b,c, Extended Data Fig. 1c-f). Interestingly, we failed to detect a similar activity for REV1 or REV3 (Extended Data Fig. 1g-i).
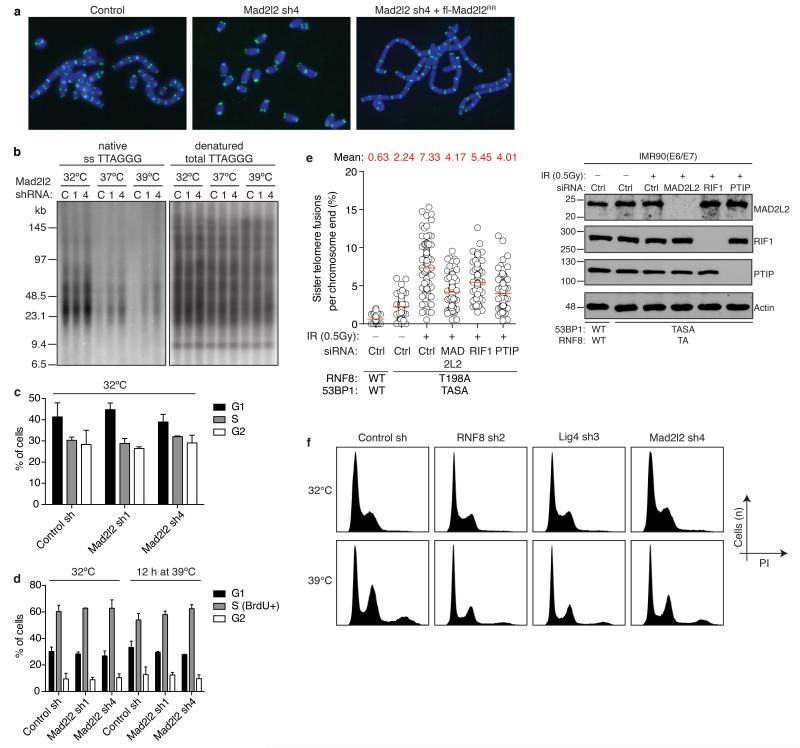
Indeed, enhanced survival of MAD2L2-depleted cells upon long-term TRF2 inactivation was due to diminished telomere fusion. Mad2l2 knockdown significantly reduced telomere fusions, which was prevented by complementation with exogenous MAD2L2 (Fig. 2a, Extended Data Fig. 2a). Telomeres end with a single-stranded 3′ G-overhang that undergoes DNA-LigaseIV/NHEJ-dependent degradation upon loss of TRF2-mediated telomere protection9, 10. Analysis of telomeric 3′ G-overhangs showed that MAD2L2 depletion prevents overhang loss at 48 hours of TRF2 inactivation, further confirming that MAD2L2 is essential for efficient processing of deprotected telomeres by NHEJ (Fig. 2b, Extended Data Fig. 2b). The defect in NHEJ-mediated telomere fusion in Mad2l2-knockdown cells was not explained by changes in cell-cycle progression (Fig. 1b: 32°C panels, Extended Data Fig. 1e, 2c,d). Moreover, MAD2L2 is essential for sister-telomere fusion upon activation of DNA repair in mitosis16 (Extended Data Fig. 2e). In line with MAD2L2 promoting telomere fusion, chromosomal rearrangement and mis-segregation6-8, MAD2L2 depletion prevented tetraploidy and aneuploidy upon telomere uncapping (Fig. 2c, Extended Data Fig. 2f).
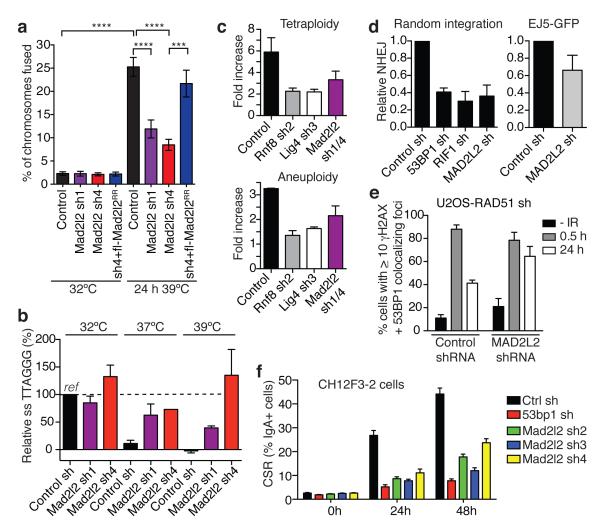
Figure 2. Mad2l2 facilitates telomeric G-overhang degradation, NHEJ-mediated telomere fusion, repair and CSR.
a, Chromosome fusion in shRNA-transduced TRF2ts MEFs (n=2-4, ± s.e.m., ****p< 0.0001, ***p=0.0002, Kruskal Wallis test, ANOVA). b, Telomeric single-strand G-overhang quantification in TRF2ts MEFs at 32°C and after 48 h at 37°C or 39°C. (n=4 except Mad2l2 sh4 at 37°C: n=1, ± s.e.m.). c, Tetraploidy and aneuploidy upon telomere uncapping in TRF2ts MEFs (n=2, ± s.d.). d, NHEJ-mediated repair in U2OS cells analysed by random plasmid integration (left, n=3) or an EJ5-GFP reporter (right, n=6). Error bars: s.d.. e, MAD2L2 depletion impairs resolution of DDR foci after IR (2Gy) in HR-deficient U2OS cells, indicating defective NHEJ (n=2, ± s.e.m.). f, CSR in shRNA-transduced CH12F3-2 cells (n=2, ± s.d.).
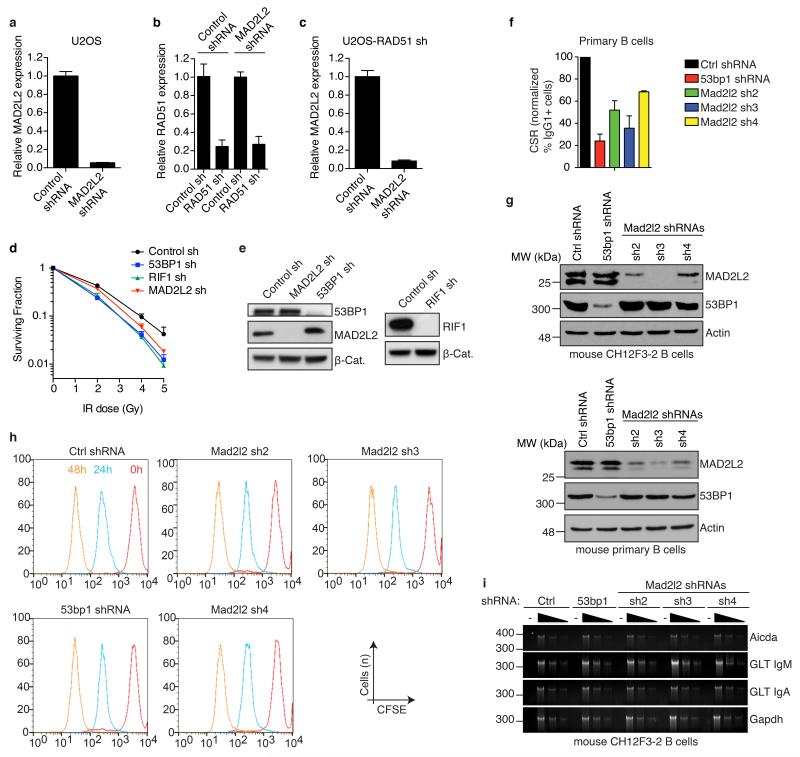
To address if the requirement of MAD2L2 for NHEJ extends beyond telomeres we investigated whether MAD2L2 depletion affects end-joining in other settings. Knockdown of human MAD2L2 significantly impaired NHEJ-mediated repair of I-Sce1-induced DNA DSBs, NHEJ-mediated random-plasmid integration, resolution of IR-induced DSBs in HR-deficient cells and clonogenic survival upon IR, similar to depletion of the NHEJ promoting factors 53BP1 and RIF1 (Fig. 2d,e, Extended Data Fig. 3a-e). Moreover, CSR, a physiological process depending on joining of DNA DSBs by NHEJ17, was significantly impaired in MAD2L2-depleted cells (Fig. 2f, Extended Data Fig. 3f-i). This CSR defect was not due to defects in cell proliferation or expression of AID mRNA, IgM and IgA germ line transcripts. Thus, MAD2L2 promotes NHEJ-mediated repair in multiple contexts.
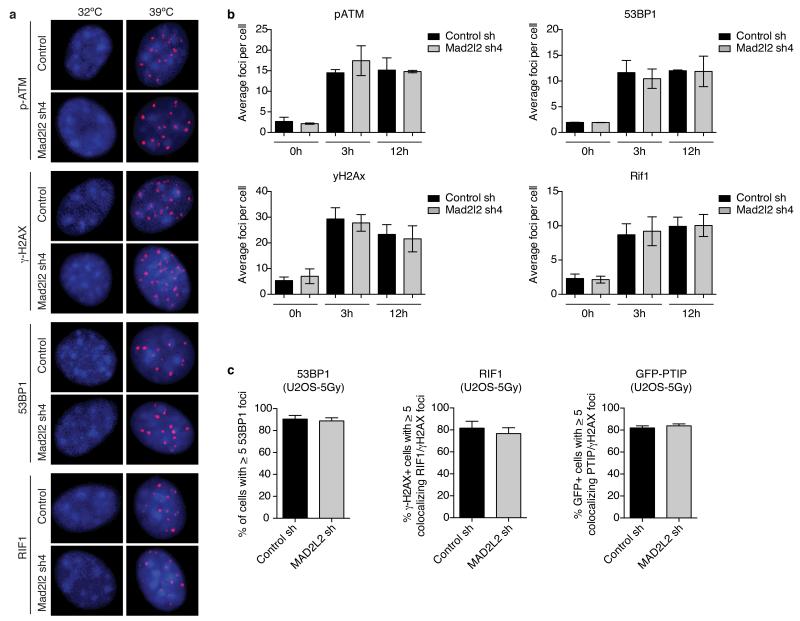
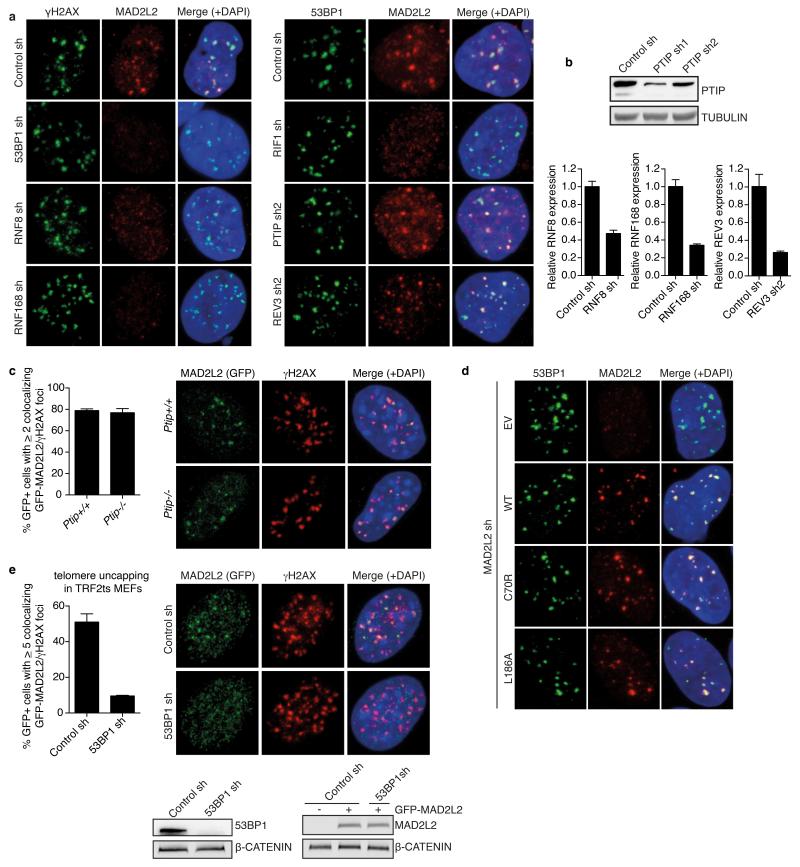
To investigate how MAD2L2 promotes NHEJ, we first addressed its localization. MAD2L2 accumulated both at TRF2-deprotected telomeres and IR-induced DSBs (Fig. 3a), implying a direct activity of MAD2L2 at these sites. Next we examined activation of the DDR. At 3, 6 and 12 hours of TRF2 inactivation both control and MAD2L2-depleted cells activated ATM, phosphorylated H2AX, KAP1 and CHK2, and formed p-ATM and γ-H2AX foci to a similar extent (Fig. 3b,c, Extended Data Fig. 4a,b). Moreover, accumulation of 53BP1, RIF1 and PTIP to uncapped telomeres, as well as IR-induced DSBs, was unperturbed in MAD2L2-depleted cells (Fig. 3c, Extended Data Fig. 4a-c). Thus, MAD2L2 is not required for recognition of uncapped telomeres or DSBs as damage, nor for initiation of DDR signalling. However, probably reflecting aberrant processing of deprotected telomeres, we noticed increased DDR signalling at 24 and 48 hours of TRF2 inactivation in MAD2L2-depleted cells, visible as elevated H2AX and KAP1 phosphorylation, and H2AX ubiquitylation (Fig. 3b).
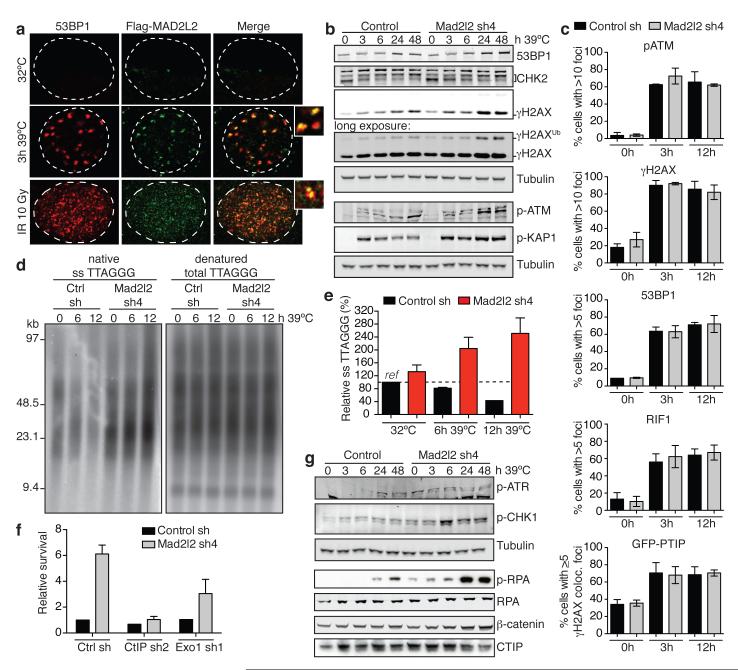
Figure 3. Mad2l2 inhibits end-resection and promotes telomere-induced genomic instability in a CTIP and EXO1 dependent manner.
a, Flag-MAD2L2 and 53BP1 localisation in TRF2ts MEFs at 32°C and 39°C, and in p53−/− MEFs after IR. b, Immunoblotting for DDR factors in TRF2ts MEFs upon telomere uncapping at 39°C. c, p-ATM, γ-H2AX, 53BP1, RIF1 and PTIP-GFP foci in TRF2ts MEFs grown for 0, 3 or 12 h at 39°C (n=2, ± s.d.). d-e, Representative example (d) and quantification (e) of telomeric single-strand G-overhang analysis in TRF2ts MEFs grown as specified (n=3, ± s.e.m.). f, Quantification of survival assays of TRF2ts MEFs shRNA-transduced as indicated (n=2, ± s.e.m.). g, Immunoblotting for p-ATR(S428), p-CHK1(S345), p-RPA32(S4/S8), RPA34 and CTIP in TRF2ts MEFs upon telomere uncapping at 39°C.
Next we examined the terminal structure of telomeres in more detail. Analysis of 3′ single-strand G-overhangs at 6 and 12 hours of TRF2 inactivation revealed that in absence of MAD2L2, telomeres gain significantly longer 3′ single-stranded overhangs. At 12 hours, Mad2l2-knockdown cells contained 2,5-fold higher, 3′-exonuclease sensitive, overhang signals than control cells without uncapping and nearly 6-fold higher overhang signals than control cells at 12 hours of telomere uncapping (Fig. 3d,e, Extended Data Fig. 5a). Overhang increase upon MAD2L2 depletion was observed in both wild-type and DNA-LigaseIV-deficient TRF2ts cells (Extended Data Fig. 5b-d), indicating it is not a passive consequence of lack of NHEJ-mediated overhang degradation, but represents an active role of MAD2L2 in inhibiting 5′ end-resection. 5′ DNA end-resection is a powerful obstruction to NHEJ by creating DNA-ends unsuitable for ligation18, explaining why MAD2L2 depletion impairs NHEJ-mediated repair.
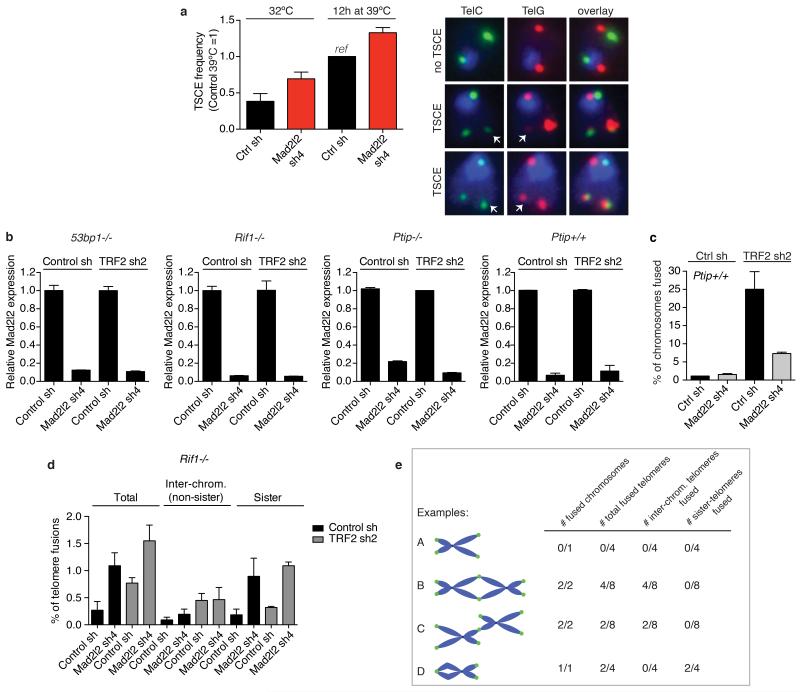
5′ End-resection involves the combined activities of CTIP, MRN, EXO1, BLM and DNA2, and is blocked by 53BP1 and its interaction partners RIF1 and PTIP3, 13, 19-24. In line with MAD2L2 promoting NHEJ by impeding 5′ end-resection, co-depletion of CTIP or EXO1 with MAD2L2 partially reversed the increased survival of Mad2l2-knockdown cells undergoing prolonged telomere uncapping (Fig. 3f, Extended Data Fig. 5e-h). Moreover, consistent with the generation of longer single-stranded overhangs that block NHEJ and prime for HDR, Mad2l2-knockdown cells showed elevated phosphorylation of the ATR target CHK1 and of the single-strand DNA-binding protein RPA (Fig. 3g). In addition, MAD2L2-depleted cells showed increased HDR at telomeres by elevated telomere sister-chromatid exchanges (TSCE, Extended Data Fig. 6a).
Because the effects of MAD2L2 depletion on end-resection, telomere fusion and CSR resemble those of 53BP1, RIF1 and, to certain extent, PTIP inactivation3, 13, 19-24, we asked whether MAD2L2 is epistatic with 53BP1, RIF1 or PTIP. Similarly, we addressed the requirement of REV1 and REV3 for the ability of MAD2L2 to control end-resection and telomere fusion. Unlike in wild-type or PTIP-deficient MEFs, MAD2L2 depletion did not reduce fusion of TRF2-deprotected telomeres in 53BP1- or RIF1-deficient MEFs (Fig. 4a, Extended Data Fig. 6b-e). However, MAD2L2-L186A and MAD2L2-C70R, respectively defective in REV1 or REV3 interaction25, 26, were still capable of promoting telomere fusion and telomere-driven genomic instability (Fig. 4b, Extended Data Fig. 7a,g,i). Moreover, MAD2L2 depletion resulted in elevated RPA phosphorylation upon IR in wild-type, Ptip−/−, Rev1−/− or Rev3−/− MEFs, but not in 53bp1−/− or Rif1−/− MEFs (Fig. 4c). This indicates that MAD2L2 controls end-resection and fusion of deprotected telomeres in a 53BP1 and RIF1 dependent, but PTIP, REV1 and REV3 independent manner. Unanticipated, MAD2L2-depleted Rif1−/−MEFs showed an elevated background of chromosome end fusion due to telomere association between sister-chromatids (Fig. 4a, Extended Data Fig. 6d,e). The molecular nature of these sister-telomere associations remains to be determined, but they occur irrespective of TRF2 inhibition and potentially reflect replication problems27.
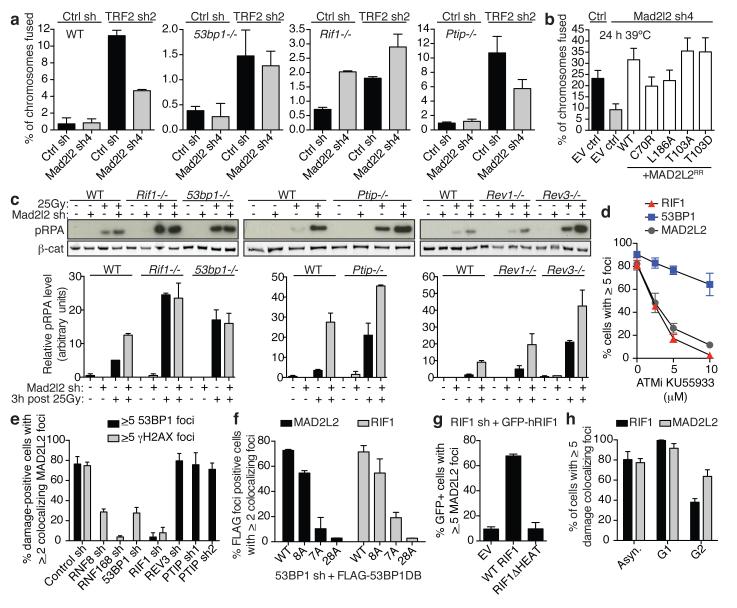
Figure 4. Mad2l2 localizes to DSBs, inhibits end-resection and promotes telomere-NHEJ in a 53BP1 and RIF1 dependent manner.
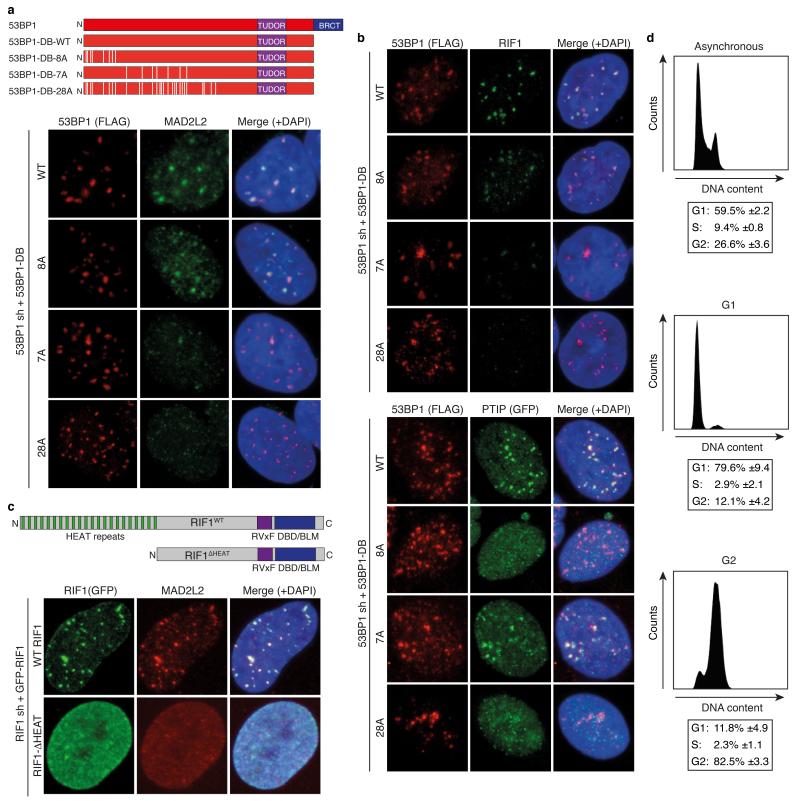
a, Chromosome fusion upon TRF2 inhibition in MEFs (n=2, ± s.e.m.). WT: controls to 53bp1−/− and Rif1−/− MEFs. See Extended Data Fig. 6c for Ptip+/+ MEFs. b, Chromosome fusion in TRF2ts MEFs (Error bars: s.e.m.). c, pRPA levels in MEFs (n=2, ± s.e.m.). d, IR-induced MAD2L2, RIF1 and 53BP1 foci in ATM inhibitor-treated U2OS cells (n=3, ± s.e.m.). e, MAD2L2 foci in irradiated shRNA-transduced U2OS cells (n=2, ± s.d.). f, MAD2L2 and RIF1 foci in irradiated U2OS cells expressing 53BP1-DB alleles with wild-type or substituted S/TQ sites (schematic in Extended Data Fig. 9a; n=2, ± s.d.). g, MAD2L2 foci in irradiated U2OS cells expressing wild-type RIF1 or RIF1 lacking heat repeats (n=2, ± s.d.). h, MAD2L2 and RIF1 foci in irradiated asynchronous, G1 or G2 synchronized RPE cells (n=3, ± s.d.).
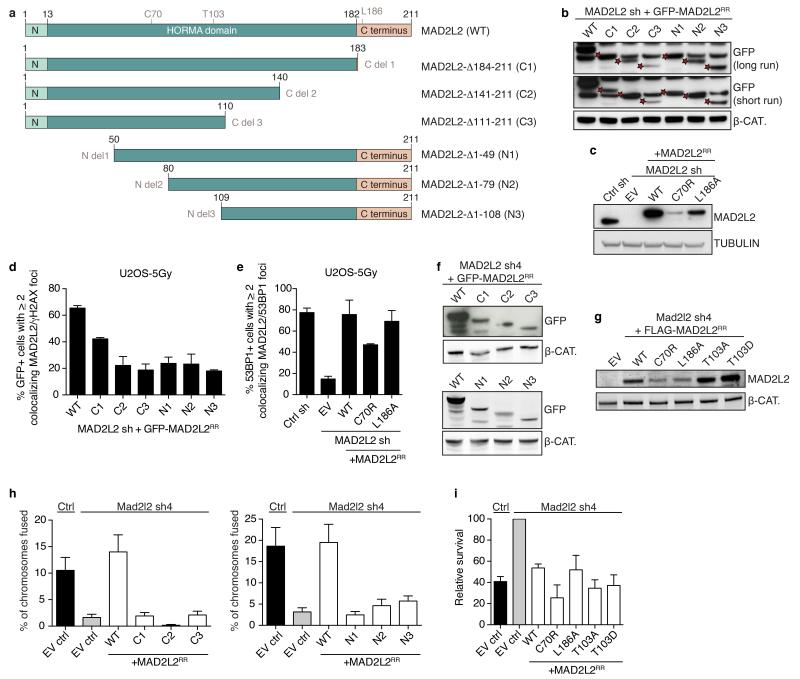
To further address the dependencies for MAD2L2 function in DNA repair, we investigated the requirements for MAD2L2 accumulation to DSBs and NHEJ activity at telomeres. Both DSB accumulation and the ability to promote telomere-driven genomic instability by MAD2L2 require an intact HORMA domain and C-terminus, while potential S/TQ phosphorylation on T103 appears dispensable (Fig. 4b, Extended Data Fig. 7). Interestingly, while MAD2L2 accumulation to IR-induced DDR foci (IRIFs) was independent of PTIP or interaction of MAD2L2 with REV1 or REV3, it clearly depended on ATM kinase activity, RNF8, RNF168, 53BP1 and RIF1 (Fig. 4d,e, Extended Data Fig. 7c,e, 8). More specifically, MAD2L2 IRIF accumulation required the HEAT repeats of RIF1 and the same ATM/ATR phosphorylation sites in 53BP1 that promote RIF1 IRIFs, while those that promote PTIP IRIFs were dispensable21-24 (Fig. 4f,g, Extended Data Fig. 9a-c). Similarly, MAD2L2 localization to uncapped telomeres required 53BP1 (Extended Data Fig. 8e). In line with dependency on RIF1, whose localization to DSBs is strongly reduced in G2, MAD2L2 foci were also diminished in G2, although less than RIF1 foci (Fig. 4h, Extended Data Fig. 9d). The latter could relate to a proposed role for MAD2L2 in late steps of HDR28.
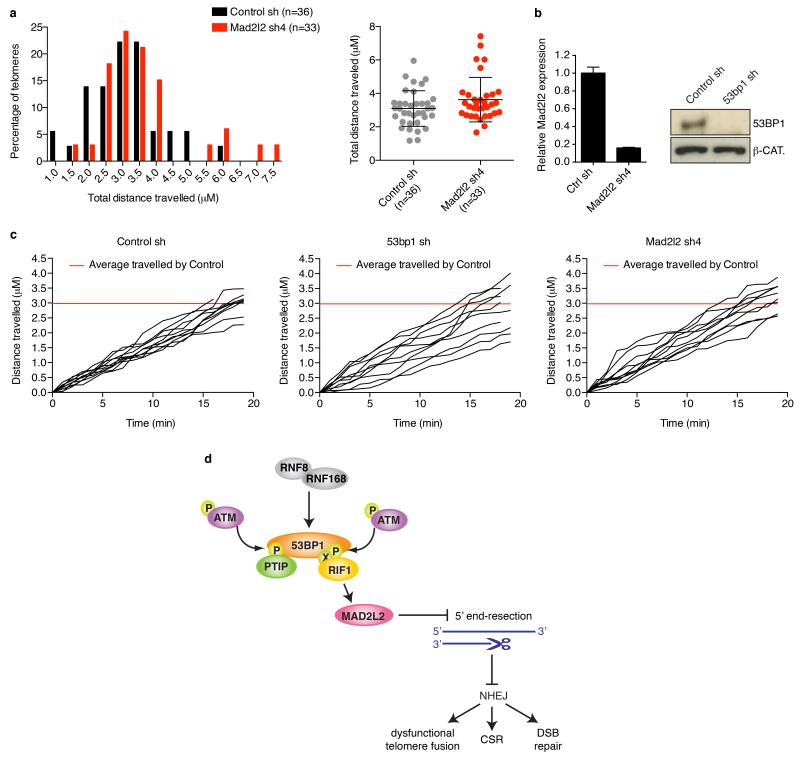
In addition to inhibiting end-resection, 53BP1, but not RIF1, promotes NHEJ by increasing mobility of uncapped telomeres13, 20. Consistent with MAD2L2 acting via end-resection control downstream of RIF1, MAD2L2-deficiency did not impair mobility of uncapped telomeres (Extended Data Fig. 10a-c).
Altogether, our data identify MAD2L2 as an important regulator of DNA repair pathway choice that promotes NHEJ at telomeres and DSBs in multiple settings, by inhibiting CTIP and EXO1 dependent 5′ end-resection downstream of RIF1 (Extended Data Fig. 10d). Whether MAD2L2 affects regulatory acetylation or phosphorylation of CTIP18, or blocks other end-resection factors, awaits further investigation.
MAD2L2 and REV3 together compose Polζ and act with REV1 in TLS to facilitate DNA replication past blocking lesions4. Based on reduced gene conversion and impaired DSB, RPA and RAD51 foci resolution in REV1- and Polζ-depleted cells, it has been proposed that the ability of REV1/Polζ to alleviate replication stalling might also be important during HDR repair-synthesis28. Although requiring further characterization, together with the data presented here, this suggests differential roles for MAD2L2 in DSB repair. As shown here, MAD2L2 acts downstream of 53BP1/RIF1 in a REV1/REV3-independent manner to promote NHEJ by inhibiting resection, mostly in G1. In addition, MAD2L2, as part of Polζ, might promote HDR by facilitating replication of damaged DNA formed after resection in G2.
The contribution of MAD2L2 to DNA repair at uncapped telomeres and DSBs suggests that aberrant MAD2L2 expression could have pathological consequences by compromising genome integrity. In this respect it is interesting that in particular MAD2L2, more than REV1 or REV3, is reported overexpressed in multiple cancers and has prognostic significance (ref29 and Oncomine database). Furthermore, our finding that MAD2L2 is essential for CSR identifies MAD2L2 as a potential disease-susceptibility gene for human primary immunodeficiencies.
Methods
TIGIR-screen
Trf2−/−;p53−/−;TRF2ts mouse embryo fibroblasts (in short TRF2ts MEFs) were infected in triplicate with PLKO lentiviral shRNA pools consisting of 1,976 shRNAs selected from the Mouse TRC 1.0 Collection. Cells were infected with a multiplicity of infection below 1 and drug selected on 4 μg/ml puromycin. After completion of selection, TRF2ts MEFs were plated in duplicate on 15 cm dishes at 150.000 cells per dish and allowed to adhere at 32°C. One set of plates was kept at 32°C for 4 additional days, the other set of plates was subjected to 12 days of telomere uncapping at 39°C, followed by recovery for 4 additional days at 32°C. Cells were harvested for genomic DNA isolation using a DNeasy Blood & Tissue Kit (Qiagen) and shRNA inserts were recovered from genomic DNA by 2 sequential PCR amplifications (PCR1 and PCR2). In PCR1 6 individual PCR reactions were performed on 325 ng genomic DNA using for each sample a common reverse primer P7_PLKO1_R (5′-CAAGCAGAAGACGGCATACGAGATTTCTTTCCCCTGCACTGTACCC-3′) and 1 of 6 forward primers with a unique index (underlined): Illuseq_01_PLKO1_f (5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTCGTGATCTTGTGGAAAGG ACGAAACACCGG-3′), Illuseq_02_PLKO1_f (5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTACATCGCTTGTGGAAAGG ACGAAACACCGG-3′), Illuseq_03_PLKO1_f (5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTGCCTAACTTGTGGAAAGG ACGAAACACCGG-3′), Illuseq_04_PLKO1_f (5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTTGGTCACTTGTGGAAAGG ACGAAACACCGG-3′), Illuseq_05_PLKO1_f (5’ACACTCTTTCCCTACACGACGCTCTTCCGATCTCACTGTCTTGTGGAAA GGACGAAACACCGG-3′), Illuseq_06_PLKO1_f (5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTATTGGCCTTGTGGAAAGG ACGAAACACCGG-3′). In PCR2, 2,5 μl DNA from each PCR1 reaction was amplified using primers P5_Illuseq (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTT CCGATCT-3′) and P7 (5′-CAAGCAGAAGACGGCATACGAGAT-3′). PCR products were pooled per condition, purified using a Qiagen PCR purification kit (Qiagen), analyzed on a bioanalyzer for quality and concentration, and subjected to deep-sequencing on a Illumina Hiseq2000 genome analyzer at the NKI Genomics Core Facility.
Cell culture, growth assays, flow-cytometry
Trf2−/−;p53−/−;TRF2ts MEFs (TRF2ts MEFs) and Trf2−/−;p53−/−;Lig4−/−;TRF2ts MEFs (TRF2ts;Lig4−/− MEFs) were generated as described before11. All cells were grown in DMEM with 100 U penicillin, 0.1 mg ml−1 streptomycin, 2 mM l-glutamine and 10% FBS. TRF2ts MEFs were maintained at the permissive temperature of 32°C and only grown at 37°C or 39°C to induce telomere uncapping through inactivation of TRF2. All other cells were grown at 37°C. Wild-type, 53bp1−/−, Rif1−/−, Ptip+/+ and Ptip−/− MEFs were SV40 immortalized.
For colony survival assays, TRF2ts MEFs were plated at 150,000 cells per 15 cm dish or 45,000 per 10 cm dish, allowed to adhere and kept at 32°C or placed at 39°C. To evaluate potential toxicity or growth rate differences, plates were fixed with 10% formalin and stained with 0.1% crystal violet after growth for 1 week at 32°C. To evaluate survival upon prolonged telomere uncapping plates were fixed and stained after growth for 12 days at 39°C, followed by 0 or 2 weeks recovery at 32°C. Quantifications of survival assays reflect the relative survival after growth for 12 days at 39°C and 2 weeks recovery at 32°C, corrected for plating efficiency and growth at 32°C and assessed by crystal violet extraction as described below.
To address cell survival under telomere uncapping conditions in short term growth assays, TRF2ts MEFs were plated in quadruplicate at 5,000 cells per well on a 12-well plate (Fig. 1c) or in duplicate at 2,500 cells per well on a 24-wells plate (Extended Data Fig. 5h), allowed to adhere overnight at 32°C and then placed for 12-14 days at 39°C. During this time period of non-permissive temperature, plates were fixed every 2-4 days with 10% formalin and stained with 0.1% crystal violet. Crystal violet was extracted from the plates using 10% acetic acid and its absorbance at 595 nm was quantified using a Tecan microplate reader. Quantifications were corrected for plating efficiency.
Cell cycle distribution of TRF2ts MEFs with or without Mad2l2 knockdown was determined by propidium-iodide (PI) staining, as well as by BrdU incorporation, and acquired on a BD Fortessa using FACSDiva software (BD Biosciences) and analyzed with FlowJo (TreeStar, Ashland, OR) software.
Analysis of tetraploidy (8N) and aneuploidy (>4N) was based on DNA-content analysis by flow-cytometry of PI-stained TRF2ts cells grown at 32°C and for 48 h at 39°C.
Cell cycle synchronisation was done as follows. For G1 cells, RPE cells were kept in 2,5 mM Thymidine (T1895, Sigma) for 16 h, released for 6 h, treated with 10 μM RO3306 (Calbiochem) for 16 h. Synchronised cells were released for 1 h, mitotic cells were collected by mitotic shake off and replated to let them continue into G1 3.5 h prior to irradiation.
For G2 cells, RPE cells were kept in 2.5 mM Thymidine for 16 h, released for 6 h and treated with 2.5 mM Thymidine for 16 h. Synchronised cells were released for 4.5 h prior to irradiation to let cells progress to G2.
Plasmids
Cells were transduced as before11 with the following pLKO-puro shRNA lentiviruses obtained from Mission library clones (Sigma) against mouse genes: Mad2l2 sh1 (TRCN0000012843: 5′-CCAGACTCCAAGTGCTCTTAT-3′), Mad2l2 sh2 (TRCN0000012844: 5′-CCAGTGGAGAAGTTTGTCTTT-3′), Mad2l2 sh3 (TRCN0000012845: 5′-CGAGCCTTCATCCTTAAGATT-3′), Mad2l2 sh4 (TRCN0000012846: 5′-CATCTTCCAGAAGCGCAAGAA-3′), Rev1 sh pool (pool of TRCN0000120297: 5′-CAGCAGTGCTTGTGAGGTATT-3′, TRCN0000120298: 5′-GCCGAGATCAACTATGGAATA-3′, TRCN0000120299: 5′-GCTGGAATGAAGACGGTGTAA-3′, TRCN0000120300: 5′-GCCCTCCGTATTGAAATCAAA-3′, TRCN0000120301: 5′-GCACGTTGATATGGACTGCTT-3′), Rev1 sh1 (TRCN0000120298: 5′-GCCGAGATCAACTATGGAATA-3′), 53bp1 sh1 (TRCN0000081778: 5′-GCTATTGTGGAGATTGTGTTT-3′) and against human genes: MAD2L2 sh (TRCN0000006570: 5′-CCCGGAGCTGAATCAGTATAT-3′), 53BP1 sh (TRCN0000018865: 5′-GATACTTGGTCTTACTGGTTT-3′), RIF1 sh (TRCN0000155431: 5′-CGCATTCTGCTGTTGTTGATT-3′), PTIP sh1 (TRCN0000129174: 5′-CTTCAGATTCATCACCGGAAA-3′), PTIP sh2 (TRCN0000322819: 5′-GCCATGTTCACAGCATATTAT-3′), RNF8 sh: (TRCN0000003439: 5′-GAAGCCGTTATGAATGTGAAA-3′), RNF168 sh (TRCN0000034137: 5′-GCAGTCAGTTAATAGAAGAAA-3′), REV3 sh (TRCN0000244436: 5′-TGACCTGTCTGAGACTATTTA-3′), RAD51 sh (TRCN0000018879: 5′-CGGTCAGAGATCATACAGATT-3′).
For knockdown of Ctip and EXO1 TRF2ts MEFs were transduced as before11 with pRetroSuper-blas shRNA retroviruses containing the following target sequences: Ctip sh1: 5′-GCAAGGTTTACAAGTCAAAGT-3′, Ctip sh2: 5′-GCAGACCTTTCTCAGTATA-3′, Ctip sh3: 5′-GCATTAACCGGCTACGAAA-3′, Exo1 sh1: 5′-GCATTTGGCACAAGAATTA-3′. Retroviral shRNAs used for knockdown of Rnf8, Ligase4 or TRF2, have been described before11. The 53bp1 shRNA vector used in CSR assays was obtained from Origene (FI334760) and contained the following target sequence: 5′-GGAAGTTGAAACCAGTGTGATTAGTATTG-3′.
For localization and complementation experiments human MAD2L2 cDNA was Gateway-cloned (Invitrogen) into pMSCV-retroviral vectors containing coding sequences for N-terminal FLAG or GFP epitopes. MAD2L2 deletion constructs were generated through Gateway-cloning (Invitrogen). The target sequences of Mad2l2 sh4 or the human MAD2L2 shRNA were changed respectively to 5′-GATTTTTCAGAAACGCAAGAA-3′ or 5′-TCCCGAACTGAATCAGTATAT-3′ to create RNAi-resistant epitope-tagged MAD2L2RR in pMSCV. GFP-RIF1, FLAG-53BP1-DB and PTIP-GFP expression vectors were described before23, 24. The target sequence of the human RIF1 shRNA was changed to 5′-GCAGCGAAGTTGAAACTTGAA-3′ to create RNAi-resistant GFP-RIF1.
qRT-PCR
RNA was isolated using Trizol reagent (Ambion). cDNA was generated using AMV first strand cDNA synthesis kit for RT-PCR (Roche) according to the manufacturer’s instructions. Quantitative RT-PCR was performed in triplicate using Power SYBR Green PCR Master Mix (Applied Biosystems) on the StepOnePlus real-time PCR system.
The following primers were used: for mouse Mad2l2 Fw 5′-ACACTCCACTGCGTCAAACC-3′ and Rev 5′-AAAGACAAACTTCTCCACTGGGC-3′; for human MAD2L2 Fw 5′-CGAGTTCCTGGAGGTGGCTGTGCATC-3′ and Rev 5′-CTTGACGCAGTGCAGCGTGTCCTGGATA-3′; for human RNF8 Fw 5′-TGGAAGAGCTAAATCGCAGCA-3′ and Rev 5′-TCAAGGTGACAGCCTCAATGAA-3′; for human RNF168 Fw 5′-GGCGAGTTTATGCTGTCCCT-3′ and Rev 5′-GCCGCCACCTTGCTTATTTC-3′; for mouse Rev1 Fw 5′-AAGCCAGATGGTCAGTACCACC-3′ and Rev 5′-TCCATTGAACGTCCAACTCCCG-3′; for human REV3 Fw 5′-AGGACTCGAAGTCACCTATGC-3′ and Rev 5′-AGAGGTAACCCCAGGAATGC-3′; for mouse Ctip Fw 5′-GTGCTGGGTCGGAGCAG-3′ and Rev 5′-TTGACTTGTAAACCTTGCACTTCC-3′; for mouse Exo1 Fw 5′-TAAACACGTCGAGCCTGTCC-3′ and Rev 5′-CAGAGCCCAGGAACCTTGTT-3′; for human RAD51 Fw 5′-GTGGAGCTAATGGCAATGCAG-3′ and Rev 5′-TTAGCTCCTTCTTTGGCGCA-3′; for mouse Hprt Fw 5′-CTGGTGAAAGGACCTCTCG-3′ and Rev 5′-TGAAGTACTCATTATAGTCAAGGGCA-3′; for human GAPDH Fw 5′-GAAGGTGAAGGTCGGAGTC-3′ and Rev 5′-GAAGATGGTGATGGGATTTC-3′.
Immunoblotting
Whole-cell lysates were prepared by scraping cells in SDS sample buffer; protein concentration was determined by standard BCA protein assay (Pierce) and equal amounts of protein were separated on precast 4-12% Bis-Tris or 3-8% Tris-acetate gels (Invitrogen). Immunoblotting was done according to standard methods using IRDye800CW- and IRDye680-labeled secondary antibodies for detection on the Odyssey Infrared imager (LI-COR) or using HRP-conjugated secondary antibodies for detection by enhanced chemiluminescence (Supersignal, Thermo Scientific). Primary antibodies used were against MAD2L2 (sc135977, Santa Cruz, 1:500), phospho-ATM S1981 (#4526, Cell Signaling, 1:1,000), phospho-KAP1 S824 (Bethyl, 1:1,000), 53BP1 (NB100-305, Novus, 1:500), phospho-H2AX S139 (clone JBW301, Upstate, 1:1,000), CHK2 (611570, BD, 1:1,000), phospho-CHK1 S345 (#2348, Cell Signaling, 1:1,000), phospho-ATR S428 (Cell Signaling, 1:1,000); RPA34 (RPA34-20, Gene Tex, 1:1,000); phospho-RPA32 S4/S8 (A300-245A, Bethyl, 1:1,000); CTIP (sc22838, Santa Cruz, 1:500); γ-tubulin (GTU-88, Sigma, 1:5,000); β-catenin (610154, BD Biosciences, 1:2,000); GFP (a-11122, Invitrogen, 1:500); RIF1 (sc55979, Santa Cruz, 1:1,000); PTIP (ABE69, Millipore and ab2614, Abcam, 1:500); actin (CP01, Calbiochem, 1:5,000). Immunoblotting for tubulin, β-catenin or actin served as loading controls.
Immunofluorescence (IF)
For IF cells were grown on eight-well chamber slides (Lab-Tek) and in cases of MAD2L2 detection first pre-extracted for 30 s to 5 min with 0.5% Triton/PBS on ice, then fixed for 10 min in 4% paraformaldehyde in PBS. Cells were permeabilized for 5 min (MEFs) or 10 min (U2OS) in 0.5% Triton/PBS, washed twice in PBS, incubated for 1 h in blocking solution (0.02% Triton, 5% NGS, 5% FCS in PBS) and overnight at 4°C with primary antibodies in blocking solution. IF for GFP-PTIP in U2OS was done as described24. Primary antibodies used were against phospho-ATM S1981 (#4526, Cell Signaling, 1:1,000), phospho-H2AX Ser 139 (05-636, Millipore and #2577S, Cell Signaling, 1:500), 53BP1 (A300-272A, Bethyl, 1:2,000), mRIF1 (gift from R. Chapman, 1:500)22, RIF1 (A300-569A, Bethyl, 1:1,000), MAD2L2 (sc135977, Santa Cruz, 1:100), FLAG (F3165 or F1804 at 1:500, F7425 at 1:400, Sigma), GFP (a-11122, Invitrogen, 1:500). Cells were washed three times with 0.02% Triton/PBS, followed by incubation with Alexa Fluor 488 or 568 goat anti-mouse or anti-rabbit IgG secondary antibodies (Invitrogen) in blocking solution for 1 h. After 4 washes in 0.02% Triton/PBS, slides were mounted in Vectashield (Vector Laboratories) containing DAPI. Confocal fluorescence images were obtained on a Leica SP5 confocal system equipped with an Ar, Kr and HeNe laser system. Images were taken with a 63x NA 1.32 oil objective and standard LAS-AF software. Possible crosstalk between fluorochromes was avoided by careful selection of imaging conditions. A minimum of 100 cells per condition per experiment was analysed. DDR foci in MEFs were captured using the Metafer4/MetaCyte platform (MetaSystems, Germany) equipped with an AxioImager Z2 microscope (Carl Zeiss, Germany). Images of random selections of cells were acquired with an EC ‘Plan-Neofluar’ 40x/0,75 objective. Analysis was done using MetaCyte software, carefully fine-tuned for each antibody. A minimum of 500 cells was analysed per condition per experiment for p-ATM, γ-H2AX, 53BP1 and RIF1. All assessments of IR-induced DDR foci were done 3 h after irradiation with 5Gy, with the exception of Fig. 3a, which is 30 min. after 10Gy.
Metaphase chromosome analysis
Cell harvesting, preparation of metaphase spreads and telomere FISH with a FITCOO-(CCCTAA)3 peptide nucleic acid custom probe (Biosynthesis) for metaphase chromosome analysis was done as described5.
Digital images of metaphases were captured using the Metafer4/MSearch automated metaphase finder system (MetaSystems, Germany) equipped with an AxioImager Z2 microscope (Carl Zeiss, Germany). After scanning metaphase preparations at 10x magnification, high-resolution images of metaphases were acquired using a ‘Plan-Apochromat′ 63x/1,40 oil objective. Chromosome fusions were quantified from >30 metaphases.
For monitoring mitotic telomeres by PNA FISH, IMR90 (E6/E7) cells were transduced with pMX-Flag-53BP1 (1-1711) or its mutant T1608A/S1618A (TASA) and pMX-GFP-RNF8 or its mutant T198A and selected with 0.2 μg/ml puromycin, for 3 days before plating. Cells were transfected with siRNAs targeting endogenous RNF8 (ThermoFisher, D-006900-01, target sequence: 5′-AGAAUGAGCUCCAAUGUAUUU-3′) and 53BP1 (ThermoFisher, D-003548-01, target sequence: 5′-GAGAGCAGAUGAUCCUUUA-3′) and the following siRNAs: PTIP siRNA pool (ThermoFisher, D-012795-01-0010, target sequences: 5′-UUAAAGCGAGCAAGUAUUA-3′, 5′-GAGCCUGGGUUGAUUAACU-3′, 5′-GUUGAUGAGUAUAAGACUA-3′ and 5′-GCCAAUGCAGUGCUGUUUA-3′), RIF1 (ThermoFisher, D-027983-02, target sequence: 5′-AGACGGUGCUCUAUUGUUA-3′), MAD2L2 (ThermoFisher, D-003272-10: 5′-GAGAAAUUCGUCUUUGAGA) and non-targeting control siRNA (ThermoFisher, D-001210-02, target sequence: 5′-UAAGGCUAUGAAGAGAUAC-3′) using RNAiMax (Invitrogen) and synchronized with a double-thymidine block. Cells were subsequently processed as previously described16.
Pulsed-field gel electrophoresis and in-gel detection of telomeric DNA
Analysis of 3′ single-strand G-overhangs at mouse telomeres was performed as described5 by pulse-field gel electrophoresis and in-gel hybridisation of a 32P-labelled telomeric repeat (CCCTAA)4 oligonucleotide to native DNA. Briefly, Trf2−/−; p53−/−;TRF2ts MEFs were harvested at the permissive temperature as well as after growth at the non-permissive temperature. Agarose plugs were prepared containing 1 × 106 cells per plug and digested overnight with 1 mg ml−1 proteinase K at 50 °C. Before loading on a 1% agarose gel, plugs were digested overnight with 60 U of MboI enzyme per plug. After running the gel on a CHEF-DR III pulsed-field gel electrophoresis system (BioRad), the gel was dried, pre-hybridized in Church Mix containing 7% SDS and hybridized overnight with a 32P-labeled (CCCTAA)4 probe, all under non-denaturing conditions. Signal was captured on a phosphorimager and analyzed with AIDA Image Analyzer software. To confirm that the G-overhang signal detected with this method was indeed derived from 3′ single-stranded telomeric TTAGGG repeats, digestion with 3′ Exonuclease and hybridization with a 32P-labeled (TTAGGG)4 probe specific for the C-rich strand was performed on separate plugs. After capturing the hybridization signal from single-stranded telomeric DNA, the gel was denatured and re-hybridized with the same probes to obtain a total telomere signal for the purpose of quantification.
Repair and clonogenic survival assays
To quantify the repair of DSBs by NHEJ, U2OS cells transduced with shRNAs were transfected with pEJ5-GFP (NHEJ), as well as an mCherry expression vector and pCAGGS-Isce-I-Trex230. Transfections were done using Mirus TransIT LT-1 (Mirus Bio LLC, USA). The fraction of mCherry-positive cells that was GFP-positive was determined 72 h post-transfection by flow-cytometry on a BD LSRFortessa using FACSDiva software. Quantification was carried out with FlowJo software (TreeStar, Ashland, OR).
For random plasmid integration assays 500,000 U2OS cells were seeded in 6 cm dishes 24 h prior to transfection with 3.9 μg of NdeI-linearized pMSCVblas-GFP and 0.1 μg of mCherry plasmids using ViaFect (Promega). Transfected cells were trypsinized the following day and seeded in 10 cm dishes at different densities for colony formation. Selection was initiated the following day with blasticidin at 5 ug/ml. Cells were fixed with 4% paraformaldehyde (PFA) 10-14 days after plating and stained with 0.1% crystal violet. Colony counting was performed on a ColCount (Oxford Optronix) and values were normalized for plating and transfection efficiencies.
For clonogenic survival assays U2OS cells were seeded in duplicate in 6-well plates at 100, 200, and 400 cells per well for unirradiated cells, and at 400, 800, and 1600 cells per well for irradiated cells. Cells were grown for <20 h before gamma-irradiation and then fixed after 10-12 days of recovery with 4% PFA and stained with 0.1% crystal violet. Colonies with >50 cells were counted on a GelCount (Oxford Optronix) and the surviving fraction was determined.
CSR assays
Research involving animals was performed in accordance with protocols approved by the animal facility at Toronto Centre for Phenogenomics (Toronto). Primary B cell extraction was performed as described before23. Briefly, mature B-lymphocytes were isolated from spleens of 8-12 week old C57BL/6 mice by depletion of CD43+ cells using CD43 microbeads (Miltenyi Biotech) according to the manufacturer’s instructions. Purified B cells were resuspended at a concentration of 106 cells/ml in the presence of 25 μg/ml LPS (Sigma-Aldrich) to allow B cell proliferation. Both mouse CH12F3-231 and primary B cells were infected with lentiviral/retroviral particles obtained from the supernatant of HEK293T cell cultures cotransfected with the lentiviral/retroviral shRNA constructs and the vector expressing MLV Gag-Pol (for retrovirus) or the lentiviral psPAX2 packaging vector with VSV-G envelope (2:1:1 ratio). Briefly, the viral supernatant was concentrated at 25,000 g for 90 min at 25°C before resuspension in RPMI culture medium. Infection was subsequently performed as described23 and followed by selection with puromycin.
IgM to IgA switching was assayed in CH12F3-2 cells activated with 1 ng/ml TGF-β1 (Preprotech), 10 ng/ml IL-4 (Preprotech) and 1 μg/ml agonist anti-CD40 (BD). IgA expression was measured by flow-cytometry using biotin-conjugated anti-mouse IgA antibody (1:200; eBioscience) followed by an anti-biotin-APC antibody at 0, 24 and 48h post-activation. Proliferation of the different transduced CH12F3-2 cell lines was performed using CFSE (Invitrogen) following the manufacturer’s guidelines. CSR assays in both cell types were done in triplicate for every independent experiment. Ex-vivo isotype switching to IgG1 in primary B cells was assayed by flow-cytometry using an anti-IgG1-biotin (1:200; BD) followed by APC-conjugated anti-biotin (Miltenyi Biotech) and 10 μg/ml propidium iodide 5 days post-isolation.
For semi-quantitative RT-PCR analysis of AID mRNA, IgM and IgA germline transcripts total RNA was extracted with TRIzol® (Invitrogen) and cDNA was synthesized by random priming using the Superscript III reverse transcriptase kit (Invitrogen). Mouse Aicda transcripts were amplified using oligonucleotides 5′-GTGCCACCTCCTGCTCACTGG-3′ and 5′-TTCATGTAGCCCTTCCCAGGC-3′. IgM (μ) germline transcripts were amplified using oligonucleotides 5′-AAAATGTCCGCTGGGCTAAG-3′ and 5′-AGAACAGTCCAGTGTAGGCAGTAGA-3′, IgA (Iα) using 5′-CCTGGCTGTTCCCCTATGAA-3′ and 5′-GAGCTGGTGGGAGTGTCAGTG-3′ and Gapdh transcripts were amplified using oligonucleotides 5′AGCCTCGTCCCGTAGACAA-3′ and 5′-AATCTCCACTTTGCCACTGC-3′. 53BP1 depletion is known to impair CSR3 and was included as control.
TSCE analysis
Telomere sister-chromatid exchanges were detected by chromosome-orientation FISH (CO-FISH), essentially as described32 with minor modifications. In brief, for CO-FISH cells were grown in 10 μM BrdU:BrdC (3:1) for 24 h with the addition of 0.2 μg/ml demecolcine for the final 2 h. Slides were treated with RNAse A (0.5 mg/ml) for 10 min at 37 °C, stained with Hoechst 33258 (0.5 μg/ml) in 2xSSC for 15 min at room temperature and exposed to 5.4e3 J/m2 365-nm UV light (Stratalinker 2400 UV irradiator). Following digestion with Exonuclease III (10 U/μl, Promega, Madison, WI) for 10 min at room temperature, slides were dehydrated through an ethanol series (70%, 95% and 100%) and incubated sequentially with Cy3-TelG 5′-[TTAGGG]3-3′ and FITC-TelC 5′-[CCCTAA]3-3′ probes at room temperature.
Telomere mobility
Telomere mobility analysis was done as described13 with minor adjustments. TRF2ts cells expressing eGFP-TRF1 and mCherry-BP1-2 (53BP1 aa 1220-1711) were seeded on Willco wells and left to attach overnight at 32°C. Prior to imaging, cells were incubated at 37°C for 30 min in Leibovitz’s L15 medium inside the Microscope climate chamber. Cells were monitored every minute for 20 min (t=20 frames) at 37°C. 5 μm Z-stacks at 0.5 μm steps in both eGFP and mCherry channels were obtained with a 63x 1.4 oil objective using ZEN software on an Observer Z1 microscope system (Carl Zeiss, Germany). Images were obtained at 2×2 binning with 512×512 pixels. Tracking of telomeres was performed with FIJI software, cells were registered with the Rigid body option from the StackReg plugin. Particles were then tracked with the particle detector and tracker plugin from Mosaic. Telomeres to track were selected on basis of colocalization with mCherry-BP1-2, and tracked for at least 17 out of 20 frames.
Database
To address if changes in MAD2L2 mRNA expression occur in human cancers we consulted the Oncomine database at https://www.oncomine.org/resource/login.html.
Extended Data
Extended Data Figure 1.
Related to Figure 1
a, A functional genetic screen for telomere-induced genomic instability regulators (TIGIRs) identifies independent shRNAs against Mad2l2 and the previously identified regulators of NHEJ-mediated telomere fusion Atm, Nbs1 (a.k.a. Nbn), Rad50, 53bp1 (a.k.a. Trp53bp1) and Rnf8. Listed are the independent shRNAs enriched >1.5 fold in at least 2 out of 3 TIGIR-screens, with their average ratio of enrichment over all 3 screens. Ratios reflect shRNA abundance after 12 days of telomere uncapping at 39°C followed by 4 days recovery at 32°C versus shRNA abundance after growth for 4 days at 32°C. b, Additional shRNAs targeting Mad2l2 or known TIGIRs that were enriched >1.5 fold in 1 of 3 screens, with their ratio. Not shown are additional shRNAs against factors not previously implicated in control of telomere fusion, that were significantly enriched in these TIGIR-screens but await validation. c, qRT-PCR analysis of Mad2l2 expression levels in TRF2ts MEFs transduced with 4 independent shRNAs targeting Mad2l2 and used in Fig. 1b (Error bars: s.d.). d, Survival assay of TRF2ts cells infected with control or Mad2l2 sh4 shRNAs, complemented with empty control or RNAi-resistant Flag-Mad2l2RR and grown as indicated. e, Western blot showing expression of endogenous MAD2L2 and exogenous Flag-MAD2L2 in TRF2ts cells used in d and in Fig. 1c. f, Photograph of shRNA-transduced TRF2ts cells grown for 12 days at 39°C. Scale bar, 100 μm. g, None of 10 shRNAs targeting Rev1 or Rev3 were significantly enriched in any of 3 independent DDR TIGIR-screens. h, shRNA-mediated Rev1 knockdown does not increase survival upon prolonged telomere uncapping in survival assays of TRF2ts MEFs. Of note, Rev3 knockdown compromised viability and was therefore not informative in these assays. i, qRT-PCR analysis of mouse Rev1 expression levels of cells shown in h (Error bars: s.d.).
Extended Data Figure 2.
Related to Figure 2
a, Representative metaphase spreads of TRF2ts MEFs transduced as indicated, harvested after 24 h at 39°C for telomere fluorescence in situ hybridization (FISH). b, Representative telomeric single-strand G-overhang (ss TTAGGG) and total telomere (total TTAGGG) analysis in TRF2ts MEFs at 32°C and after 48 h at 37°C or 39°C. c-d, Mad2l2 knockdown does not affect cell cycle parameters in TRF2ts MEFs. Cell cycle phase analysis was based on PI staining of asynchronously growing cells (c), as well as on 1 h incubation with BrdU, followed by detection of BrdU incorporation and PI staining for DNA content (d) (n=3, ± s.d.). e, MAD2L2 is required for sister-telomere fusion upon activation of DNA repair in mitosis. Sister-telomere fusions were quantified in IMR90 cells expressing exogenous WT 53BP1 and RNF8, or 53BP1-T1608A/S1618A (TASA) and RNF8-T198A (TA) mutant alleles, and depleted for endogenous RNF8 and 53BP1, as well as depleted for MAD2L2, RIF1 or PTIP (n=4). f, Examples of DNA content profiles of PI stained TRF2ts cells transduced with the indicated shRNAs and grown at 32°C or for 48 h at 39°C. Analysis of the fraction of cells with 8N (tetraploid) or > 4N (aneuploid) DNA content was done on corresponding dotplots of which the results are shown in Fig. 2c.
Extended Data Figure 3.
Related to Figure 2
a, qRT-PCR analysis of MAD2L2 expression levels in U2OS cells infected with control or MAD2L2 shRNAs, and used in the repair assays shown in Fig. 2d (Error bars: s.d.). b, c, qRT-PCR analysis of RAD51 (b) and MAD2L2 (c) expression levels in RAD51-depleted, E6E7-expressing U2OS cells used in the assays shown in Fig. 2e (Error bars: s.d.). d, Clonogenic survival assays of U2OS cells transduced with non-targeting control or 53BP1, RIF1 or MAD2L2 shRNAs and treated with the indicated doses of IR (n=3-4, ± s.e.m.). e, Western blot analysis of 53BP1, MAD2L2 and RIF1 in U2OS cells transduced with the indicated shRNAs. f, CSR in shRNA-transduced primary B cells (n=2, ± s.d.). g, Western blot analysis of MAD2L2 and 53BP1 in CH12F3-2 B cells and mouse primary B cells transduced with the indicated shRNAs. h, MAD2L2 depletion does not affect cellular proliferation in murine B cells. CH12F3-2 cells transduced with control, 53bp1 or Mad2l2 shRNAs were loaded with CFSE and analysed at 0, 24 and 48 h post-stimulation by flow-cytometry. Profiles from all time points are plotted in the same histogram. i, MAD2L2 depletion does not affect the transcription of critical genes implicated in CSR. RT-PCR analysis of AID (Aicda) mRNA, IgM (GLT IgM) and IgA (GLT IgA) germline transcript levels using 2-fold serial dilutions of cDNA made from activated CH12F3-2 B cells transduced with the indicated shRNAs. Gapdh was used as a control for transcript expression.
Extended Data Figure 4.
Related to Figure 3
a, Representative images of IF detection of p-ATM(S1981), γ-H2AX, 53BP1 and RIF1 in TRF2ts MEFs transduced with control or Mad2l2 shRNAs and grown at 32°C or for 12 h at 39°C to induce telomere uncapping. DNA was stained with DAPI. b, Quantification of the number of p-ATM, γ-H2AX, 53BP1 and RIF1 foci per cell in TRF2ts MEFs transduced with control or Mad2l2 shRNAs and grown at 32°C or for 3 and 12 h at 39°C (n=2, ± s.d.). c, Quantifications of 53BP1, RIF1 and PTIP foci in U2OS cells transduced with control or MAD2L2 shRNAs, 3 h after 5Gy (n=2, ± s.d.).
Extended Data Figure 5.
Related to Figure 3
a, Telomeric single-strand G-overhang assay of TRF2ts MEFs transduced with control or Mad2l2 sh4 shRNAs, showing that the increase in overhang signal upon Mad2l2 knockdown is due to 3′ terminal sequences because the signal is removed by treatment with Escherichia coli 3′ exonuclease EXO1. b, Mad2l2 knockdown causes increased ss telomeric G-overhang signals in TRF2ts;Lig4−/−;TRF2ts MEFs. c, Quantification of relative telomeric G-overhang signals in TRF2ts;Lig4−/− MEFs transduced with control or Mad2l2 shRNAs and grown at 32°C or for 12 or 24 h at 39°C (n=2, ± s.e.m.). d, qRT-PCR analysis of Mad2l2 expression levels in TRF2ts;Lig4−/− MEFs infected with control or Mad2l2 shRNA lentivirus (Error bars: s.d.). e, Survival assays of TRF2ts MEFs transduced with control or Mad2l2 shRNAs and subsequently with control, Ctip or Exo1 shRNAs. f, Quantification of the survival assays shown in e. g, qRT-PCR analysis of Ctip and EXO1 expression levels of cells shown in e, f, h and in Fig. 3f (Error bars: s.d.). h, Growth curves at 39°C of TRF2ts MEFs transduced with non-targeting control or Mad2l2 shRNAs and subsequently with control, Ctip or Exo1 shRNAs (Error bars: s.e.m.).
Extended Data Figure 6.
a, TSCE analysis in shRNA-transduced TRF2ts MEFs grown at 32°C or for 12 h at 39°C to uncap telomeres (n=2, ± s.d., counting >1,000 chromosomes per condition, per experiment). TSCE frequency in control cells is set at 1 (corresponding to an average of 6.9% of chromosomes with a TSCE event). Shown on the right are examples of chromosomes without and with TSCE in cells quantified on the left. b, qRT-PCR analysis of Mad2l2 expression levels in 53bp1−/−, Rif1−/−, Ptip−/− and Ptip+/+ MEFs used in the chromosome fusion analysis shown in Fig. 4a and in c (Error bars: s.d.). c, Percentage of chromosomes fused upon TRF2 inhibition in the Ptip+/+ MEFs matching with the Ptip−/− MEFs shown in Fig. 4a (n=2, ± s.e.m.). d, Analysis of different types of telomere fusions in Rif1−/− MEFs. Depletion of MAD2L2 in Rif1−/− MEFs does not reduce inter-chromosomal telomere fusions induced by TRF2 inhibition, indicating epistasis. However, irrespective of TRF2 inhibition, MAD2L2 depletion in Rif1−/− MEFs induces association between sister-telomeres, causing an increase in total fusions scored for MAD2L2-depleted Rif1−/− MEFs, as also visible in Fig. 4a (n=2, ± s.e.m., >1,300-2,000 chromosomes were analysed per condition, per independent experiment). e, Explanation of scoring different types of telomere fusions shown in d.
Extended Data Figure 7.
Related to Figure 4
a, Schematic overview of C-terminal and N-terminal deletion mutants of MAD2L2. b, c, Expression analysis in U2OS cells of GFP-tagged WT MAD2L2 and C- and N-terminal MAD2L2 deletion mutants (b) and of FLAG-tagged WT MAD2L2 and MAD2L2-C70R and −L186A (c). d, Analysis of DDR foci formation of GFP-tagged WT MAD2L2 and C- and N-terminal MAD2L2 deletion mutants by IF detection of GFP and γH2AX in U2OS at 3 h after IR (n=2 for N-terminals, n=3 for C-terminals, ± s.e.m.). e, Analysis of WT, C70R and L186A MAD2L2 accumulation into DDR foci by IF detection of MAD2L2 and 53BP1 in U2OS at 3 h post IR (n=2, ± s.e.m.). f, g, Expression analysis in TRF2ts MEFs of GFP-tagged MAD2L2 and C- and N-terminal MAD2L2 deletion mutants (f) and of FLAG-tagged WT MAD2L2 and MAD2L2-C70R, −L186A, −T103A and −T103D (g). h, Quantification of chromosome fusions after 24 h of telomere deprotection at 39°C in TRF2ts MEFs transduced with control or Mad2l2 shRNAs and complemented with empty vector control or RNAi-resistant GFP-tagged WT MAD2L2 and C- or N-terminal MAD2L2 deletion mutants (Error bars: s.e.m.). i, Quantification of survival assays of TRF2ts MEFs transduced with control or Mad2l2 shRNAs and subsequently with empty vector control, WT MAD2L2, MAD2L2 C70R, L186A, T103A or T103D retroviruses (n=2, ± s.e.m.).
Extended Data Figure 8.
Related to Figure 4
a, Representative images of IF detection of endogenous MAD2L2 and γH2AX or 53BP1 in U2OS cells transduced with control, 53BP1, RNF8, RNF168, RIF1, PTIP or REV3 shRNA lentiviruses, irradiated with 5Gy and processed for IF after 3h (quantifications are shown in Fig. 4e). b, Western blot or qRT-PCR analysis of PTIP, RNF8, RNF168 and REV3 levels in shRNA-transduced U2OS cells (Error bars: s.d.). c, Quantification and representative images of IF detection of γH2AX and GFPMAD2L2 in Ptip+/+ or Ptip−/− MEFs, 3h after 5Gy (n=2, ± s.d.). d, Representative images of IF for 53BP1 and exogenous Flag-MAD2L2 WT, C70R or L186A in U2OS cells depleted for endogenous MAD2L2 with lentiviral shRNA, processed for IF 3h after 5Gy (quantifications are shown in Extended Data Fig. 7e). e, Quantification and representative IF images of GFP-MAD2L2 localisation to uncapped telomeres in TRF2ts MEFs transduced with control or 53BP1 shRNAs (n=2, ± s.d.).
Extended Data Figure 9.
Related to Figure 4
a, Schematic representation of the 53BP1 alleles with wild-type and substituted S/TQ sites used to address MAD2L2 IRIF localisation dependence on 53BP1. Representative IF images are displayed that show colocalisation of endogenous MAD2L2 with 53BP1-DB-WT and −8A, but not with 53BP1-DB-7A or −28A. Quantifications are provided in Fig. 4f. b, Top: Representative IF images showing colocalisation of endogenous RIF1 with 53BP1-DB-WT and −8A, but not 53BP1-DB-7A or −28A. Quantifications are presented in Fig. 4f. Bottom: Representative IF images showing colocalisation of GFP-PTIP with 53BP1-DB-WT and −7A, but impaired colocalisation of GFP-PTIP with 53BP1-DB-8A or −28A. c, Schematic representation of GFP-tagged wild-type RIF1 (WT) and GFP-tagged RIF1 lacking the N-terminal HEAT repeats (ΔHEAT), used to address MAD2L2 IRIF localisation dependence on the HEAT repeats of RIF1. Representative IF images are shown. Quantifications are presented in Fig. 4g. d, Cell cycle phase distributions of the RPE cells used in Fig. 4h to address cell cycle dependence of endogenous RIF1 and MAD2L2 localisation to IRIFs (n=3, ± s.d.).
Extended Data Figure 10.
Related to Figure 4
a, Frequency distribution (left) and scatter plot (right) of total distances travelled by uncapped telomeres in control or Mad2l2 knockdown cells. b, qRT-PCR analysis of Mad2l2 expression levels and Western blot analysis of 53BP1 proteins levels in TRF2ts cells used in the experiments shown in a and c (Error bars: ± s.d.). c, Distance travelled by 10 representative uncapped telomeres for each condition. While multiple uncapped telomeres in 53BP1-depleted cells have reduced mobility, this is not seen for uncapped telomeres in MAD2L2-depleted cells. d, Model of the role of MAD2L2 in promoting NHEJ.
Acknowledgements
We thank A. Konishi, G. Celli and T. de Lange for TRF2-I468A, Trf2flox/-;p53−/−MEFs and Trf2flox/-;p53−/−;Lig4−/− MEFs, P. Bouwman, M. Pieterse, T. Halazonetis, O. Kallioniemi, B. Gerritsen, B. Morris and P. Halonen for composing and providing the DDR TRC shRNA sub-library, members of the NKI Genomics Core Facility for technical support, A. van Kessel for MAD2L2 cDNA, E. Hendrickson for repair plasmids, R. Chapman and S. Boulton for mRIF1 antibody, 53bp1−/−, and Rif1−/−MEFs, E. Callen and A. Nussenzweig for 53BP1 and PTIP expression vectors and for Ptip−/− MEFs, N. de Wind for Rev1−/− and Rev3−/− MEFs, B. van den Broek for help with telomere mobility analysis, I. de Krijger, Z. Yalcin and M. Simonetta from the Jacobs group for contributing to end-resection and interaction analysis and members of the R. Medema lab for discussion. This work was supported by grants from the European Research Council (ERCStG 311565) and Dutch Cancer Society (KWFNKI2007-3907) to J.J.L.J, a grant from the Canadian Institutes for Health Research (CIHR MOP89754) to D.D., a La Caixa fellowship to S.S.B and a CIHR postdoctoral fellowship to A.O.. D.D. is a Canada Research Chair (Tier 1) in the Molecular Mechanisms of Genome Integrity and J.J.L.J is an EMBO Young Investigator.
Footnotes
Online Content Methods, along with any additional Extended Data display items, are available in the online version of the paper; references unique to these sections appear only in the online paper.
The authors declare no competing financial interests.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014;24:108–17. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waters LS, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–54. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 2008;22:1221–30. doi: 10.1101/gad.1634008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–81. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 8.Murnane JP. Telomere dysfunction and chromosome instability. Mutat Res. 2012;730:28–36. doi: 10.1016/j.mrfmmm.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA Ligase IV-Dependent NHEJ of Deprotected Mammalian Telomeres in G1 and G2. Curr Biol. 2002;12:1635. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 10.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–8. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 11.Peuscher MH, Jacobs JJ. DNA-damage response and repair activities at uncapped telomeres depend on RNF8. Nat Cell Biol. 2011;13:1139–45. doi: 10.1038/ncb2326. [DOI] [PubMed] [Google Scholar]
- 12.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–71. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 13.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–8. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng Y, Guo X, Ferguson DO, Chang S. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460:914–8. doi: 10.1038/nature08196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dimitrova N, de Lange T. Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of nonhomologous end joining (NHEJ) in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol. 2009;29:5552–63. doi: 10.1128/MCB.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orthwein A, et al. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014;344:189–93. doi: 10.1126/science.1248024. [DOI] [PubMed] [Google Scholar]
- 17.Daniel JA, Nussenzweig A. The AID-induced DNA damage response in chromatin. Mol Cell. 2013;50:309–21. doi: 10.1016/j.molcel.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panier S, Durocher D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol. 2013;14:661–72. doi: 10.1038/nrm3659. [DOI] [PubMed] [Google Scholar]
- 19.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–7. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–4. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Virgilio M, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–5. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman JR, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49:858–71. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escribano-Diaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–83. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Callen E, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–80. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara K, et al. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase zeta and REV1. J Biol Chem. 2010;285:12299–307. doi: 10.1074/jbc.M109.092403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khalaj M, et al. A missense mutation in Rev7 disrupts formation of Polzeta, impairing mouse development and repair of genotoxic agent-induced DNA lesions. J Biol Chem. 2014;289:3811–24. doi: 10.1074/jbc.M113.514752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–60. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 28.Sharma S, et al. REV1 and polymerase zeta facilitate homologous recombination repair. Nucleic Acids Res. 2012;40:682–91. doi: 10.1093/nar/gkr769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rimkus C, et al. Expression of the mitotic checkpoint gene MAD2L2 has prognostic significance in colon cancer. Int J Cancer. 2007;120:207–11. doi: 10.1002/ijc.22155. [DOI] [PubMed] [Google Scholar]
Additional references to Full Methods
- 30.Bennardo N, Gunn A, Cheng A, Hasty P, Stark JM. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 2009;5:e1000683. doi: 10.1371/journal.pgen.1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato H, et al. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–41. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 32.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–90. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]