Abstract
The primary antibody repertoire is generated by mechanisms involving the assembly of the exons that encode the antigen-binding variable regions of immunoglobulin heavy (IgH) and light (IgL) chains during the early development of B lymphocytes. After antigen-dependent activation, mature B lymphocytes can further alter their IgH and IgL variable region exons by the process of somatic hypermutation (SHM), which allows the selection of B cells in which SHMs resulted in the production of antibodies with increased antigen affinity. In addition, during antigen-dependent activation, B cells can also change the constant region of their IgH chain through a DNA double-strand-break (DSB) dependent process referred to as IgH class switch recombination (CSR), which generates B cell progeny that produce antibodies with different IgH constant region effector functions that are best suited for a elimination of a particular pathogen or in a particular setting. Both the mutations that underlie SHM and the DSBs that underlie CSR are initiated in target genes by activation-induced cytidine deaminase (AID). This review describes in depth the processes of SHM and CSR with a focus on mechanisms that direct AID cytidine deamination in activated B cells and mechanisms that promote the differential outcomes of such cytidine deamination.
OVERVIEW AND INTRODUCTION
Immunoglobulin genes, B cell receptors and antibodies
The B cell receptor (BCR) is expressed on the B lymphocyte cell surface where it serves as a receptor for foreign antigens (1). The BCR is comprised of two immunoglobulin (Ig) heavy (IgH) chains encoded by the IgH heavy chain locus and two Ig light (IgL) chains encoded by, for a given BCR, either the Igκ or Igλ (collectively referred to as IgL) light chain loci (Fig. 1). These three Ig loci lie on different chromosomes in both humans and mice. While there are certain differences in organization, the overall strategies for Ig gene diversification in mice and humans are very much the same (2, 3), so this review will focus mainly on the mouse. The amino-terminal portions of the IgH and IgL chains have a highly variable amino acid sequence from species to species of antibody and are called variable (V) regions. The IgH and IgL variable regions interact to generate the antigen-binding portion of the BCR/antibody. The carboxy-terminal end of IgH and IgL chains have only a few variations in their sequences and thus are called constant (C) regions.
FIGURE 1.
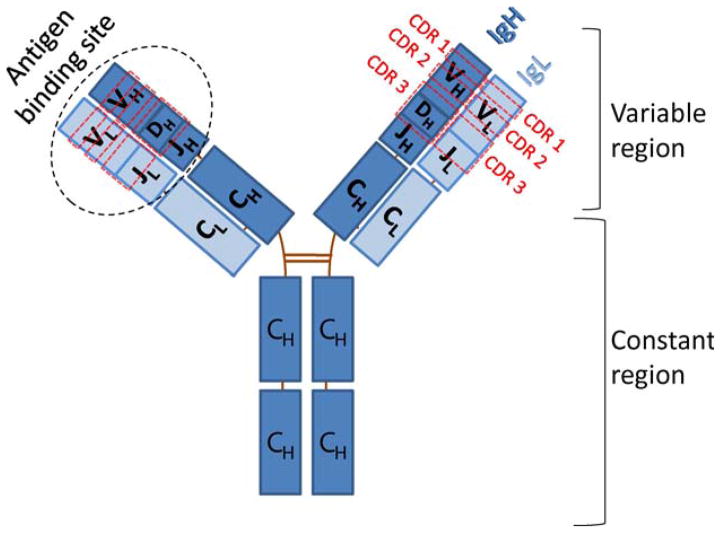
Antibody structure. The BCR is comprised of two immunoglobulin (Ig) heavy (IgH) chains encoded by the IgH heavy chain locus and two Ig light (IgL) chains. The rectangles represent Ig domains that constitute the structural units of the immunoglobulin heavy and light chains. The variable regions are assembled through V(D)J recombination of VH, DH, and JH gene segments on the heavy chain and VL and JL gene segments on the light chain. Complementarity-determining regions (CDRs) are indicated as regions in dashed red boxes: CDR 1 and 2 are encoded in the VH or VL gene segments, and CDR 3 is encoded by the VH DH JH junctional region or VL and JL junctional region. The heavy and light chain variable regions form the antigen-binding site. The constant region determines the class and effector function of the antibody molecule. doi:10.1128/microbiolspec.MDNA3-0037-2014.f1
The antigen-independent generation of an extremely large population of B cells in which individual cells express BCRs with unique antigen-binding specificity is of fundamental importance for vertebrates to generate effective humoral adaptive immune responses, as it enables B cells to recognize and respond to an enormous variety of foreign antigens. In this context, IgH and IgL variable region exons are not encoded in the germline, but rather are assembled during early B cell development prior to antigen exposure in the fetal liver and bone marrow by the V(D)J recombination process (2). V(D)J recombination generates an IgH VH(D)JH variable region exon by assembling different combinations of the numerous IgH variable (VH) segments, diversity (D) segments, and joining (JH) segments that lie within a 1 to 3 Mb region at the 5′ end of the IgH locus. V(D)J recombination assembles an IgL VLJL variable region exon from Igκ or Igλ V segments and J segments (2).
V(D)J recombination is initiated by the lymphocyte-specific RAG1 and RAG2 (“RAG”) endonuclease that recognizes conserved recombination signal sequences (RSS) that flank the V, D, and J segments (4). RAG cleaves between the RSSs and the coding sequences of a pair of involved segments, generating a pair of blunt RSS double strand break (DSB) ends that are later joined to each other and a pair of hair-pinned coding DSB ends that are processed and joined to each other (4) by the general cellular classical nonhomologous end-joining (C-NHEJ) DSB repair pathway (5, 6). Coding ends are often further diversified before they are joined, including the de novo additions of N nucleotides by the terminal deoxynucleotidyl transferase (Tdt), another lymphocyte-specific factor involved in V(D)J recombination (7). The combinatorial diversity arising from the numerous V, D, and J segments, as well as the junctional diversity that arises from junctional diversification during joining the segments, generates an enormous repertoire of primary variable region exons (8). Within the IgH and IgL variable regions there are three regions that show “hypervariability” separated by much less variable “framework” regions (FWR). As they are involved in antigen contact, these three hypervariable regions are termed complementarity-determining regions (CDRs) (9). CDR1 and CDR2 are encoded in the different germline VH and VL gene segments. The most diverse portion of the primary variable region exon is CDR3, which is generated through combinatorial assortment of V, D, and J sequences and from junctional diversification mechanisms (10).
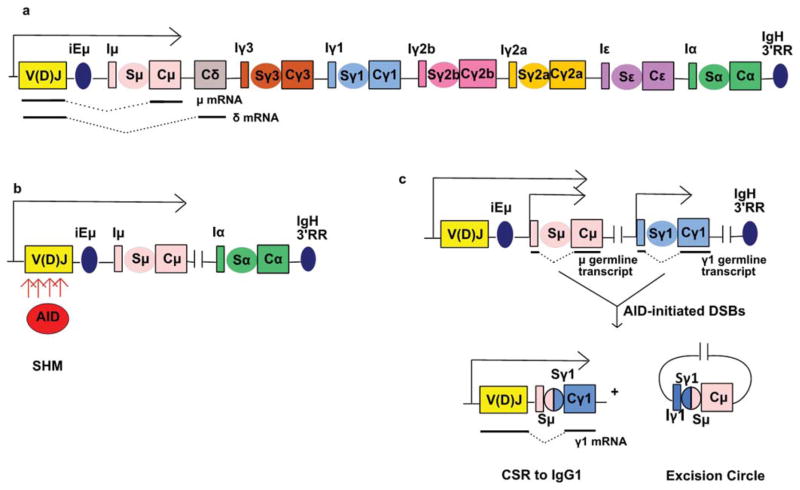
Transcription of fully assembled IgH and IgL chain genes is initiated from the promoter of the V segment used in the V(D)J exon and continues through downstream exons that encode the C regions of IgH and IgL chains (11). The mouse IgH locus contains 8 sets of exons that encode different CH regions (sometimes termed “CH genes”) within the approximately 200 kb region downstream of the JH segment and lying in the order 5′-VDJ-Cμ-Cδ-Cγ3-Cγ1-Cγ2b-Cγ2a-Cε-Cα 3′ (12) (Fig. 2a). The set of CH exons expressed with the V region exon determines the class of the BCR/antibody (e.g. IgM, IgG, IgE, IgA). Within the IgH locus, there are several B cell specific enhancers, for example iEμ, which lies between the JH segments and the Cμ exons and a 30 kb IgH 3′ regulatory region (“IgH 3′RR”), which is downstream of the Cα exons (11, 13) (Fig. 2a). Initially, the IgH variable region exon is transcribed in association with the immediately downstream Cμ exons, and in some cells, Cδ exons. Alternative RNA splicing of these primary IgH transcripts leads to differential expression of Cμ and Cδ and also to differential expression of CH sequences that specify whether the IgH chain is expressed as membrane-bound BCR or is secreted as an antibody (14). Thus, prior to antigenic stimulation, resting B cells express IgM (or IgD).
FIGURE 2.
Genomic alterations of the IgH locus. a. Organization of the IgH constant (C) region. Each C region is preceded by a switch (S) region and a noncoding “I” exon. Blue oval between V(D)J exon and Iμ represents IgH intronic enhancer (iEμ). Blue oval downstream of Cα represents IgH 3′ regulatory region (IgH 3′RR). μ and δ mRNAs are shown below the corresponding genes. Dashed line represents spliced transcript. b. AID generates point mutations and/or DNA double strand breaks (DSBs) at the V(D)J exon during somatic hypermutation (SHM). c. AID-initiated DSBs in Sμ and Sγ1 result in CSR to IgG1. μ and γ1 germline transcripts are initiated from promoters upstream of the corresponding I exons. doi:10.1128/microbiolspec.MDNA3-0037-2014.f2
During B cell development, V(D)J recombination generally occurs first at the IgH locus in progenitor (pro) B cells (2, 6). In this regard, developing B cells generate D to JH rearrangements on both IgH alleles and then append VH segments to pre-existing DJH rearrangements (15). If the first VH to DJH is in frame (productive), the resulting IgH μ heavy chain protein generates a signal that feeds back to prevent VH to DJH joining on the other DJH rearranged allele and to promote development to the precursor (pre) B cell stage. The resulting pre B cells will have a productive V(D)J IgH allele and a “frozen” DJH intermediate allele. If, due to junctional diversification, the first VH to DJH rearrangement is out of frame, the cell can move on and append a VH segment to the second DJH allele which, if productive, will again promote development to the pre B stage with the resulting pre B cells having an in-frame productive V(D)J rearrangement and out-of-frame nonproductive V(D)J rearrangement. Because about two thirds of V to DJH rearrangements are nonproductive, about 40% of normal B cells have two V(D)J rearrangements (one productive and one nonproductive). This “feedback” mechanism for the control of VH to DJH rearrangement is thought to have evolved to ensure mono-specificity of B cell clones in the context of the phenomenon of “allelic exclusion” (see reference 16 for details). Precursor B cells rearrange IgL genes and if they form a productive IgL rearrangement leading to an IgL chain that pairs with μ heavy chain, they then express the complete IgM molecule on their surface as the BCR (2, 6). These newly generated IgM+ B cells then migrate into the periphery and survey the secondary lymphoid organs, including spleen, lymph nodes and Peyer’s patches, for cognate antigen that binds their BCR.
Overview of SHM, CSR and the role of AID
An encounter with cognate antigen in the secondary lymphoid organs, usually in the context of a T-dependent immune response, can activate mature B cells. Activation can lead to the generation of B cells that secrete their BCR as a secreted antibody. Antigen-dependent B cell activation can also lead to the two somatic processes of genomic rearrangement that enhance the efficacy of the antibody response against specific antigen: SHM further diversifies the variable region exon and alters the affinity of the BCR for antigen (9) (Fig. 2b), while CSR switches the CH region exon used and alters the antibody’s antigen elimination function (17) (Fig. 2c). SHM occurs in the germinal centers (GCs) (18), specialized compartments of secondary lymphoid organs, while CSR can occur inside or outside of the GCs (19, 20). Both SHM and CSR are initiated by activation-induced cytidine deaminase (AID) (21, 22). AID is a small (24 kDa) protein that deaminates cytidine residues on single-stranded DNA (ssDNA), usually in the context of preferred sequence substrate motifs (9). Both SHM and CSR require transcription, both to promote specific AID targeting and also to contribute to formation of requisite ssDNA substrates. Both processes also co-opt activities of normal cellular base excision repair (BER) and mismatch repair (MMR) to convert AID cytidine deamination lesions to mutational and/or DSB outcomes. Each of these AID-associated processes will be discussed in depth in following sections.
During SHM, AID deaminates cytosine residues in IgH and IgL V(D)J exons and the deamination products are processed through specific repair pathways into predominantly point mutations, as well as a low frequency of small insertions and deletions (9, 23, 24). SHM produces nucleotide substitutions at all four bases, with a bias towards transitions over transversions such that approximately two thirds of nucleotide substitutions are transitions (25). The mutation frequency over the rearranged variable region exon as a whole is approximately 10−3 mutations per base pair per generation, with the highest levels found within complementarity-determining regions (CDRs) (9, 26, 27). In the context of the GC reaction, B cells with SHMs that increase antigen-binding affinity of their BCR are positively selected and those with SHMs that decrease affinity or inactivate the receptor are negatively selected via rounds of SHM, clonal expansion and affinity-based selection; in this manner SHM leads to affinity maturation of the antibody response (18).
For CSR, AID deaminates cytosine residues in long (1 to 10 kb), repetitive, noncoding, switch (S) regions that lie just upstream of each set of CH exons (except Cδ exons, which do not undergo traditional CSR) (17, 28). Deamination products at donor and acceptor S regions are processed to DNA DSBs, as well as point mutations (17, 28). CSR is completed when AID-initiated DSBs generated in two participating S regions are fused to delete intervening DNA including the Cμ exons (12, 28). A switch in expression from IgM to different IgH classes such as IgG, IgE, and IgA occurs when Cμ exons are replaced with one of the sets of downstream CH exons (e.g. Cγ, Cε, or Cα exons). Each antibody class is specialized for certain pathogen-elimination functions. For example, IgG promotes phagocytosis of antibody-coated particles, IgE triggers mast cell degranulation, and IgA defends against pathogens at mucosal surfaces (29, 30, 31). Thus, CSR alters an antibody’s effector function to one that may be better suited for a given pathogen-elimination response, while maintaining the same variable region exon and thus antibody-binding specificity.
AID FUNCTIONS THROUGH CYTIDINE DEAMINATION OF TARGET DNA
AID was discovered by a subtractive hybridization approach that employed a mouse B cell line stimulated to undergo CSR from IgM to IgA (32). AID knock-out mice were found to be specifically defective for SHM and CSR (21). Likewise, contemporaneous studies of human patients with an autosomal recessive form of hyper-IgM syndrome, characterized by high levels of IgM in the serum and profound defects in IgH CSR and SHM, showed that they had AID mutations (22). These two types of studies showed that AID is required for both SHM and for IgH CSR and, thus, can be considered a master regulator of peripheral antibody diversification. Subsequent studies further showed that AID also is required for the variable region exon DSBs that initiate the gene conversion process that diversifies chicken antibody repertoires (33).
AID target sequences for SHM
AID deaminates cytidines to uridines in ssDNA (34, 35, 36, 37) preferentially deaminating cytidines in the context of “hotspots” described by the consensus motif DGYW (WRCH on the complementary strand, D = A/G/T, Y = C/T, W = A/T, H = T/C/A, R = A/G) (38). DGYW motifs are very abundant in the tandem repeat units of S regions, with a high density of DGYW motifs a conserved feature among the S regions of species from frogs to mammals (39). V region exons contain a lower density of DGYW motifs than S regions, but their DGYW frequency is still mildly enriched compared to bulk genomic DNA (39). As genomic DNA usually is in duplex form, transcription-based mechanisms have evolved to generate the requisite ssDNA substrates for AID (this is discussed in more detail in subsequent sections).
Mammalian and frog S regions are highly enriched for AGCT motifs, a palindromic variant of the canonical DGYW, which provide AID hotspot motifs on both DNA strands and contribute to DSB generation (see below). DGYW motifs are considered favored mutational hotspots, but not perfect predictors of mutability, since identical DGYW motifs within a given sequence undergo different levels of mutation (40). In addition, DGYW motifs are ubiquitous throughout the genome, but only undergo AID-initiated mutations in a subset of genes and, in most cases, at frequencies orders of magnitude less than at Ig gene targets (38, 41, 42). Thus, additional targeting mechanisms are important, including substrate sequence context beyond the DGYW motif, and higher level mechanisms including transcription (described in later sections). In addition, there is evidence that differential repair of AID cytidine deamination lesions also can influence final mutation and DSB outcome (9, 42).
AID-initiated lesions are processed by normal repair pathways to yield mutations and DSBs
The point mutations and DSBs that occur during SHM and CSR are generated in two steps (9). In the first step, AID deaminates cytidines to uridines (U) in V(D)J exons during SHM, or in S regions during CSR to produce uracil:guanine (U/G) mismatches. The second step involves error-prone resolution of the U/G lesion by co-opted BER and/or MMR pathway activities (Fig. 3). Normally the BER and MMR pathways repair such lesions in an error-free manner. How activities of the BER and MMR pathways that evolved to maintain genome fidelity are coerced into contributing to generating mutations and DSBs downstream of AID lesions is understood only in part. Below we will describe current knowledge of enzymatic processes involved in the two steps leading from AID-generated U/G mismatches to mutations or DSBs, starting with a description of the normal BER and MMR pathways.
FIGURE 3.
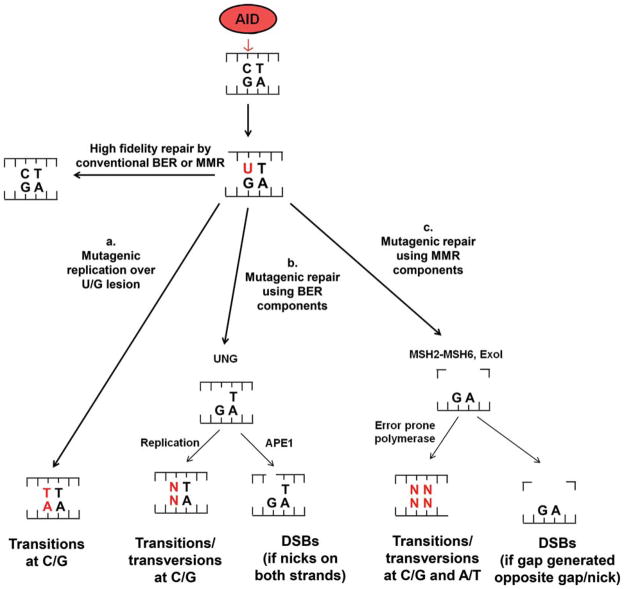
Mechanisms of AID cytidine deamination in SHM and CSR. AID deaminates cytidine (C) to uridines (U). The U/G lesion may be repaired with high fidelity (i.e. to C/G) by conventional base excision repair (BER) or mismatch repair (MMR). Mutagenic outcomes during SHM and CSR are generated by the following processes. a. Replication over the U/G lesion produces transition mutations at C/G base pairs. b. Uracil-DNA-Glycosylase (UNG) of the BER pathway excises the U creating an abasic site. Replication over the abasic site generates transition and transversion mutations at C/G base pairs. N indicates any nucleotide A,G,C, or T. AP endonuclease 1 (APE1) may create a nick at the abasic site. Nicks on both DNA strands may lead to DSBs. c. MSH2-MSH6 of the mismatch repair pathway recognize the U/G mismatch. Exo1 excises the patch of DNA containing the mismatch. Error-prone polymerase resynthesizes the patch leading to spreading of mutations to A/T base pairs. Overlapping gaps may lead to DSBs. doi:10.1128/microbiolspec.MDNA3-0037-2014.f3
BER repairs damaged bases by the following general steps: (1) recognition and excision of a damaged base (e.g. uracil) from the DNA backbone by an initiating DNA glycosylase (e.g. uracil-DNA-glycosylase) to create an abasic site; (2) cleavage of the DNA backbone at the abasic site by an apurinic/apyridimic (AP) endonuclease, generating a ssDNA nick adjacent to the abasic site; (3) processing of the nick to a single-nucleotide gap; (4) filling in of the gap by DNA polymerase β; and finally (5) sealing of the nick by DNA ligase 1 or DNA ligase 3 (43). An alternative to this one-nucleotide short-patch form of BER is long-patch BER. In long-patch BER, after nicking of the DNA by AP endonuclease, DNA polymerase β/δ/ε displaces and polymerizes an approximately 2 to 10 bp long tract of DNA (43, 44). The displaced strand is removed by activity catalyzed by the flap structure-specific endonuclease 1 (FEN1), and a remaining nick sealed by DNA ligase 1 (43, 44). MMR functions primarily in repair of base–base mismatches by a process involving (1) recognition of the mismatch by the MSH2-MSH6 heterodimer; (2) recruitment of a complex of MLH1 and PMS2 (MutLα); (3) excision of the patch of DNA surrounding the mismatch by exonuclease-1 (Exo1) to generate a gap; (4) gap-filling by DNA polymerase δ bound to PCNA; and (5) ligation by DNA ligase 1 to seal the nick (45, 46).
During SHM, replication over the initiating U/G lesion can produce transition (purine > purine or pyrimidine > pyrimidine) mutations at C/G base pairs (Fig. 3a) (9). In addition, the uracil can also be excised by UNG of the BER pathway which leads to an abasic site. Replication over the abasic site can lead to both transversions (purine > pyrimidine or pyrimidine > purine) as well as transitions at initiating C/G base pairs (Fig. 3b) (9). Thus, UNG deficiency produces mainly transition mutations at C/G base pairs (9, 25). As AID only deaminates C’s, the mutagenic BER processes described above cannot account for SHM at A and T residues. Rather, SHMs at A’s and T’s depends largely on components of the MMR pathway (47, 48). The MSH2-MSH6 heterodimer recognizes the U/G mismatch (49, 50), Exo1 then excises the patch of DNA containing the mismatch (51), and an error-prone polymerase such as polymerase η resynthesizes the patch (52, 53). Thus, mutations are “spread” from the C/G sites of deamination to nearby A/T sites.
The generation of DSBs during CSR also employs the activities of the BER and MMR pathways (Fig. 3b and c) (9, 20). Following the excision of uracil by UNG, AP endonuclease 1 (APE1) may create a nick at the abasic site (20, 44) (Fig. 3b). Adjacent nicks on opposite strands, for example in the context of AGCT motifs, may be sufficient to generate a DSB, particularly if target motifs are very dense as they are in S regions (54). Overlapping gaps generated during MMR can also lead to DSBs during CSR (55, 56) (Fig. 3c). Accordingly, while BER-deficiency or MMR-deficiency alone reduces CSR, combined BER and MMR deficiencies (e.g. UNG and MSH2 deficiency) abrogate CSR (9, 55). AID-initiated SHMs also accompany the DSBs that are generated in CSR-activated B cells, but in this case only mutations at C/G residues are found with little or no spreading to A/T residues. Such differential targeting/outcomes of AID activity during SHM and CSR will be discussed in more depth below.
UNG and MMR double deficiency, in addition to ablating CSR, also eliminates both C/G transversion mutations and spreading of mutations in the context of V region SHM, leaving only C/G transition mutations and also eliminates C/G transversion mutations in S regions during CSR (50, 55, 57). Thus in the absence of BER and MMR, V and S regions exhibit only the footprint of AID deamination, strongly supporting the two-step model in which AID deamination is followed by processing of the resulting lesions by BER and MMR to DSBs, transversion mutations at C and G residues, and transition and transversion mutations at A and T residues.
SOMATIC HYPERMUTATION
SHM occurs during germinal center responses
The process of somatic hypermutation (SHM) introduces point mutations in the assembled V regions of IgH and IgL genes of mature activated B cells (9). SHM takes place in the GC, a specialized structure found in B cell follicles of peripheral lymphoid organs (e.g lymph nodes and spleen) where rapidly proliferating B cells accumulate after primary immunization (18). Consistent with the role of AID in SHM, GC B cells express high levels of AID (32). In the GC, antigen-activated B cells, usually with the help of T cells, undergo multiple rounds of SHM and Darwinian-like selection for clones with high-affinity antigen binding followed by clonal expansion, leading to the evolution of B cells that express BCRs with increased affinity for the antigen. This process of affinity maturation is fundamental to the production of high-affinity antibodies to particular pathogens (18, 58).
SHM targets in the endogenous IgH and IgL loci
Both productive and nonproductive IgH and IgL variable region exon alleles of B cells undergoing a GC response are subject to SHM (59, 60); however, only mutations in productive alleles affect the BCR and influence the fate of GC B cells that bind the antigen (58). GC B cells with SHMs that decrease BCR affinity for antigen or lead to auto-reactivity are not selected. In addition, B cells in which SHMs alter residues necessary for normal BCR functions (e.g. certain framework residues necessary for proper folding) are lost as BCR tonic (e.g. ligand-independent) signaling is required for survival (61). Given the strict selection for and against SHMs on the productive V(D)J alleles, SHMs on the nonproductive allele, which are not selected for or against, are considered a better indicator of intrinsic mutational patterns that are not biased by antigen selection (62, 63).
SHMs concentrate prominently within the CDRs of the V region exon (9). This accumulation does not appear to be merely due to selection since nonproductive V exons and passenger V transgenes (that provide a transcribed V(D)J exon that is not involved in BCR expression) also show preferential accumulation of SHMs in CDRs (40, 63). Preferential targeting of SHM to CDRs may reflect evolutionary pressure to direct SHM to parts of the V region that bind antigen and away from the intervening FWR, which are important for the Ig’s structural integrity (64). However, the mechanism by which SHMs preferentially accumulate in the CDRs is not known. Differential AID targeting is a likely possibility, with CDRs of VH exons containing somewhat more AID hotspots than other regions of the V exon (40). In this regard, although various codons can encode a given amino acid, CDRs preferentially use those that contain AID hotspot motifs (65, 66). However, it is notable that in nonproductively rearranged V exons, identical AID hotspot motifs (e.g. AGCT) mutate more when located in the CDRs as compared to when located in FWRs (40, 67). Thus, the underlying sequence of CDRs or their flanking regions, beyond AID target motifs, also may have a role in recruiting AID activity for SHM. In addition, the location of the CDRs in terms of their distance from the transcriptional start site (68) and/or other aspects of the overall structure of the V region exon could play a role in directing AID activity. Finally, the possibility of differential repair of AID deamination lesions might also contribute (42). Clearly, there is much to be learned about how AID is targeted within V exons during SHM.
The question of whether or how specific features of V exon sequences contribute to AID targeting must take into account findings that non-Ig sequences, including β-globin, chloramphenicol acetyltransferase and Ig Cκ sequences, are apparently robust substrates when inserted in place of the V exon in transgenic passenger alleles (69, 70, 71). However, the degree of SHM of these transgenes, while sometimes approaching that of the bona fide V exon, is quite dependent on integration site and copy-numbers (69). Thus, it is possible that features of the integration site or chromatin structural alterations associated with tandem transgene arrays could influence AID targeting in unknown ways. In this regard, targeting of such non-Ig sequences as single copies in place of an endogenous V exon will be required to conclusively address the degree to which these sequences undergo SHM relative to V exon sequences in a physiological endogenous setting. If non-Ig sequences mutate as well as V sequences in the endogenous V location, it would imply that there is nothing absolutely specific to the V exon sequence that targets high levels of SHM. In this case, the question would be why and how SHMs are focused on the CDRs, which may imply evolution of V exon to suppress SHM at FWRs versus CDRs. If non-Ig sequences do not mutate as well as V sequences when expressed in single copies in the normal physiological location, it would support the notion that CDRs evolved to specifically support AID-initiated SHMs.
Mutational versus deletional outcomes during SHM
SHM, in contrast to CSR, is generally considered to involve predominantly point mutations and much less frequently DSBs (9). However, deletions have been found at relatively high frequency in some studies of nonproductive V exons (24) and passenger V exons (72). Such deletions generally would result from DSBs, which lead to deletions either through resection or by joining to another DSB in the same V exon (73). Such internal deletions are frequent in S regions in accord with their high DSB frequency (see below). DSBs can also lead to insertions, which have also been found in V exons in association with SHM (24). Together, deletions and insertions are often generically referred to as “indels” (74). DSBs and associated indels must occur at some frequency during SHM but are likely mostly selected against in productive V exon alleles, since they could disrupt reading frame or overall V region structure. The wide variability of the levels of indels found in V exons that have undergone SHM in different experiments (24, 63, 75, 76) could reflect most samples coming from productive alleles, the possibility that different V exon sequences have different propensities to undergo DSBs, limitations of sample size, or other factors. High-throughput sequencing of the Ig variable region exons from HIV-1 infected patients that produce rare broadly neutralizing antibodies have revealed that certain of these broadly neutralizing antibodies are extensively mutated and harbor very frequent indels (77, 78). How these anti-HIV broadly neutralizing antibodies accumulate such high levels of SHM and indels during affinity maturation is still speculative (78). An important question is whether AID-induced SHMs in some unmutated or affinity matured V exons can generate new sequences that further promote or direct DSBs and SHMs.
Mechanisms that target SHM to specific variable region exon targets
The mechanisms by which AID is targeted to its substrates is of great interest given the potentially deleterious consequences of AID’s mutagenic activity. Off-target AID activities can activate oncogenes via mutations or translocations and, thereby, contribute to cancer (79, 80). In this regard, transcription has been shown to be a key factor for targeting AID to V exons (9, 26, 81, 82) as well as to S regions (see below). Correspondingly, deleting the V promoter eliminates SHM (83). In addition, non-Ig promoters can support SHM at least to some degree (83, 84, 85), suggesting that the V promoter per se may not direct AID targeting; but rather that such targeting is provided by transcription in general. Consistent with a key role for transcription, the spatial distribution of SHMs in a V exon is influenced by distance from the transcription start site (TSS), with the TSS defining the 5′ boundary of SHM (9, 76) and SHM frequency decreasing with distance from the TSS (68).
Ig enhancers, which are known transcriptional regulators (13, 82), promote SHM within transgenic V(D)J substrates (84, 86, 87). However, such enhancers, including the IgH intronic enhancer (iEμ), intronic Igκ enhancer (iEκ) and 3′ Igκ enhancer were deleted in mice and found to not be required for SHM (87, 88, 89). The difference between the transgene and endogenous findings may reflect redundancy of tested enhancer elements with other enhancers or other types of elements in the endogenous setting (82). The 30 kb IgH 3′RR contains a number of different enhancers and deletion of several of them in the endogenous locus can abolish germline CH transcription and CSR to most CHs (91; see below), without affecting V(D)J transcription or SHM (92). However, recent studies showed that complete deletion of the 30 kb IgH 3′RR in the endogenous locus completely abolishes germline CH transcription and CSR (93) and also severely impairs SHM (94). However, the impairment of SHM in the absence of the 30 kb IgH 3′ RR was accompanied by only marginal reduction in transcription (94). These studies imply that the full IgH 3′RR contains elements that may impact AID targeting during SHM via mechanisms beyond transcription (94), as has also been suggested by mutation targeting studies in chicken Igκ ((95), see below). Another possibility is that the type of transcription (e.g. sense versus anti-sense) is important.
AID must be directed to its intended Ig gene substrate sequences versus other sequences to maintain specificity and reduce its potential off-target mutational activity. In addition, once at a target sequence, AID must gain access to a requisite ssDNA substrate. Transcription and transcription-related mechanisms have been implicated in facilitating both of these steps. In this context, a number of AID-associated proteins have been described that may contribute to these activities (96, 97). Transcriptional stalling has been implicated in directing AID to its targets (71, 81). The transcription associated factor Suppressor of Ty5 homolog (Spt5) associates with both AID (98) and RNA pol II (Pol II) (99, 100). Co-localization of Pol II, AID and Spt5 on genomic sites is predictive of AID-induced mutated sites, suggesting that Spt5 may target AID to genomic loci by recruiting AID to sites of stalled Pol II (98). The factors that lead to Pol II stalling in AID targets are unknown.
How AID gains access to the ssDNA substrate following targeting
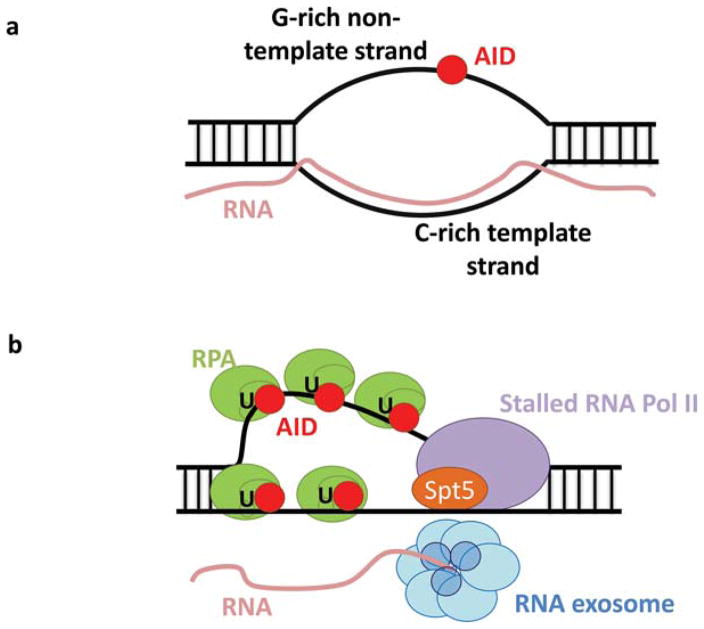
Following recruitment to targets, AID must gain access to the ssDNA template. Purified AID deaminates the nontemplate strand of mammalian S regions transcribed in vitro because mammalian S regions loop out the ssDNA nontemplate strand in the form of R loops upon transcription (35; see below). Purified AID does not deaminate T7-transcribed V exon substrates, which do not form R loops (101). However, serine 38 (S38)-phosphorylated AID in association with replication protein A (RPA) deaminates V exon substrates in vitro but only on the nontemplate strand, suggesting that RPA may assist in stabilizing ssDNA template for AID deamination and/or enhance further AID recruitment (101). Consistent with this model, disrupting the S38 phosphorylation of AID dramatically reduces SHM in GCB cells and CSR in activated B cells in vivo (102, 103). Both strands of DNA are targeted for SHM. Thus, while transcriptional stalling may target AID, and co-factors such as RPA may stabilize ssDNA substrates, a mechanism must exist to provide AID with access to the template strand, which may be masked by nascent RNA transcripts. The RNA exosome has been implicated in this role as it allows AID to robustly deaminate both strands of T7 transcribed substrates in vitro and is required for normal levels of CSR in vivo (104). The RNA exosome is an evolutionarily conserved exonuclease that processes nuclear RNA precursors and degrades RNA in the nucleus and cytoplasm (105). A working model suggests that once AID is brought to a target via stalled Pol II and Spt5, the RNA exosome displaces or degrades the nascent RNA, thus making the template strand available for deamination, which may in vivo be further augmented by RPA association (104) (Fig. 4b). Negative supercoiling is an additional mechanism that has been proposed to make both DNA strands available as ssDNA substrates (106, 107).
FIGURE 4.
Transcriptional targeting of AID. a. R-loop structure. An R loop forms from G-rich RNA transcribed from the C-rich template strand forming a stable RNA-DNA hybrid with the C-rich template strand and looping out the G-rich nontemplate strand as ssDNA. b. A working model suggests that once AID is brought to a target via stalled Pol II and Spt5, the RNA exosome displaces or degrades the nascent RNA, thus making the template strand available for deamination, which may in vivo be further augmented by RPA association. Figure adapted from reference 104. doi:10.1128/microbiolspec.MDNA3-0037-2014.f4
Factors that promote AID activity at “on-” and “off-target” genes
As transcription occurs in a large number of genes in activated B cells, transcription alone cannot explain occurrence of SHM (and CSR) specifically within Ig loci and also within just a limited number of “off-target” genes, including potential B cell oncogenes, that undergo SHM at lower levels (42, 108, 109, 110). Thus many transcribed genes in B cells do not appear to undergo SHM. In this regard, unknown aspects of sequence context of AID targeting motifs in V(D)J exon, type of transcription (see above), aspects of specific chromatin structure, or cis elements beyond those involved in transcription may contribute to focusing SHM to Ig substrates and also contribute to AID mutation to particular off-targets. Recent studies of the DT40 chicken B cell line identified within the chicken Igκ gene a diversification activator (DIVAC) that targets SHM to the V exon (95, 111, 112). The DIVAC contains multiple, redundant transcription factor binding motifs yet enhances SHM without stimulating a major increase in transcription (at least as measured by steady-state transcript levels), suggesting that it may target SHM by a transcription-independent mechanism (95). Notably, various mouse and human Ig enhancers also served as very strong DIVACs in this system, suggesting possible functional redundancy between enhancers and DIVACs in promoting SHM in the chicken Igκ locus, by mechanisms that do not involve influences on transcription per se (95). If so, however, then there still must be additional elements that function in this context in mammalian cells since mouse iEμ and mouse iEκ functioned as DIVACs in the DT40 assay (95), but their deletion does not markedly impair SHM in mouse GC B cells (88, 90).
IgH CLASS SWITCH RECOMBINATION
Overview
During class switch recombination (CSR), AID introduces cytidine deamination C to U mismatch lesions into large repetitive S regions that flank the upstream portion of CH genes. These AID-initiated lesions are then converted into DSBs by co-opted BER and MMR activities (Fig. 3b and c). Deletional end-joining, usually via C-NHEJ of the upstream end of a DSB in Sμ to the downstream end of a DSB in a downstream S region, juxtaposes the V(D)J exon and downstream CH gene to effect CSR. Whether there are mechanisms that promote such deletional joining versus the inversional joining alternative is unknown (6). The primary IgH class switching event involves switching from IgM to the various IgGs, IgE, and IgA, in which DSBs in the “donor” Sμ is joined to “acceptor” Sγ, Sε, or Sα. CSR also can in some cases occur successively. For example, an initial CSR event from Sμ to Sγ1 can generate a hybrid Sμ/Sγ1 donor S region that can subsequently successively undergo CSR with Sε to generate an IgE-producing B cell (30). Downstream “CSR“ events between Sγ and Sε can also occur in cells in which Sμ has been truncated (113); in theory such a pathway could also contribute to successive CSR but the physiological significance of such downstream CSR events in normal B cells remains to be determined. The targeting of AID to S-region sequences shares at least some common mechanistic aspects with AID targeting during SHM. A primary mechanistic feature of AID targeting for both SHM and CSR is the requirement for transcription, which again both contributes to directing AID to its target S region sequence and which contributes to providing AID with access to ssDNA template.
Targeting of specific IgH CSR events via differential activation of transcription in various CH genes
Each set of CH exons is part of a “germline” transcription unit referred to as a CH gene, even though its transcribed RNA does not encode a known protein product (17) (Fig. 2). In these germline CH genes, transcription is initiated from a cytokine activation-specific promoter upstream of a noncoding “I” (for “intervening”) exon, continues through the associated S region, and terminates downstream of the CH exons (17, 114). Different cytokines secreted by T cells and other immune cells can stimulate transcription from different I region promoters and, thereby, transcriptionally direct CSR to the CH region most appropriate for a given type of pathogen or setting of pathogen infection (17, 20). As an example of such regulation, B cells activated through the CD40/CD40L pathway of B and T cell interaction that are also exposed to interleukin-4 (IL-4) (a cytokine secreted by T helper cells) will activate germline transcription from the IL-4 inducible Iγ1 and Iε promoters, and thereby direct CSR to IgG1 and IgE (114, 115, 116).
Induction of such germline CH transcription occurs on both productive and nonproductive alleles (117), frequently leading to CSR to the same CH gene on both alleles (118, 119), showing that CSR is a directed recombinational process. Deletion of the I exon abolishes germline CH transcription from the associated I promoter and, thus, eliminates CSR to the corresponding S regions (120, 121). Moreover, replacement of I promoters with heterologous promoters ectopically targets CSR under B cell activation conditions in which it would not normally occur, implicating a direct role for transcription in directing CSR (122, 123, 124, 125). In addition to the cytokine and/or activation of specific I region promoters, differential regulation of transcription from these promoters is modulated by the IgH 3′RR located just downstream of the IgH locus (126).
The 30 kb region of IgH 3′RR contains multiple DNase I hypersensitive sites that correspond to enhancers (13). Combined deletion of one subset of these enhancers severely impairs germline CH transcription from all I region promoters, except the Iγ1 promoter, and, correspondingly, severely impairs CSR to the transcriptionally inhibited CH genes (91, 126). Deletion of the entire 30 kb region deletes additional enhancers and abrogates CSR to all CH genes, including Cγ1, confirming that the IgH 3′RR is a master regulator of CSR, and implicating differential activity of elements within it in controlling CSR (93) and, as described earlier SHM (94). The IgH 3′RR has been implicated in differentially regulating CSR to different CH genes by a “promoter competition” mechanism, by which certain activated I region promoters compete with and exclude interaction of other I promoters with the IgH 3′RR (126, 127). For example, LPS treatment of B cells induces germline transcription of Cγ2b and Cγ3 and CSR to these CH genes. However, including IL-4 along with LPS in the treatment activates the Iγ1 and Iε promoters which are proposed to inhibit germline transcription from Iγ2b and Iγ3 promoters via competition for the IgH 3′RR, resulting in inhibition of CSR to Cγ2b and Cγ3 and activation of CSR to Cγ1 and Cε (127). Such regulation is consistent with the finding that the IgH 3′RR forms chromosomal loops with activated I region promoters (128, 129).
Targeting AID activity within S regions
Once AID is targeted to S regions via transcriptionally related mechanisms, other features of targeting may overlap at least in part with those discussed for SHM, but there also are notable differences that are discussed below. Mammalian S regions are 1 to 10 kb long sequences composed primarily of tandem repetitive units (39). The deletion of S regions and their replacement with a sequence that lacks S-region features severely impairs CSR to the associated CH exon, demonstrating that the S region plays a specific role in CSR (117, 130, 131). Mouse and human Sμ, Sα, and Sε are comprised of pentameric repeats, while the Sγ regions are comprised of 49 to 52 bp long repeats, that are enriched in smaller repeats including the AGCT and other DGYW targeting motifs (3, 39). In addition, mammalian S regions are C/G-rich and G-rich on the nontemplate strand (39). In contrast to mammalian S regions, the Sμ of Xenopus (frog) is A/T-rich and contains a high density of DGYW sequences, in particular AGCT motifs (39, 132). As mentioned earlier, the palindromic nature of the AGCT motif may make it an optimal substrate for generating DSB breaks by providing AID hotspot motifs on both DNA strands (130, 131), with the high density of such motifs in S regions thereby promoting DSBs.
Transcription through mammalian S regions generates stable R loops in vivo (133). Such R loops result from G-rich RNA transcribed from the C-rich nontemplate S-region strand forming a stable RNA-DNA hybrid with the C-rich DNA template strand and looping out the G-rich nontemplate strand as ssDNA (134, 135, 136, 137, 138) (Fig. 4a). In biochemical experiments, AID robustly deaminates the nontemplate strand of a T7 transcribed S-region substrate that forms an R loop but not a C-rich substrate that does not form an R loop (35). R-loop formation during CSR in B cells is also abolished by inversion of the S-region sequence, which decreases, but does not eliminate, CSR (117). Based on such findings, the formation of R loops in mammalian S regions has been suggested to have evolved to enhance AID access to S regions (35, 138). However, as mentioned, inversion experiments show that R-loop formation is not absolutely required for S-region function in CSR (117). Likewise, Xenopus Sμ regions, which are A/T-rich and do not form R loops support substantial CSR when substituted for mouse S regions in activated B cells (131). We note, though, that it remains possible that R-loop formation in mammalian S regions evolved to enhance some other CSR function, for example by playing a role in S-region synapsis (see below).
As for SHM, AID targeting in CSR also appears to involve Pol II stalling, as revealed by accumulation of Pol ll in transcribed S regions (139, 140). R loops generated in the transcribed S region may enhance stalling (139, 140). Spt5 also has been implicated in recruiting AID in the context of stalled Pol II in S regions during CSR (98). In vitro, RPA facilitates AID access to the nontemplate strand of T7 transcribed Xenopus Sμ by association with S38-phosphorylated AID (101) and also binds R loop forming S regions in a S38-phosphorylated-AID-dependent manner (101, 141, 142). Thus, the prevention of AID S38-phosphorylation by an S38A mutation severely impairs RPA association and CSR in mouse B cells without affecting AID catalytic activity, supporting the notion that RPA interaction with S38-phosphorylated AID is important for CSR in vivo (102, 103, 143). The S38-phosphorylation of AID by PKA (143) may be important for CSR in a feedback loop mechanism that involves RPA and downstream repair factors (141, 144). Within transcribed mammalian S regions, template DNA is likely shielded from AID activity by stably hybridized RNA in the form of R loops (133). In this regard, the RNA exosome could function to displace/degrade the nascent RNA, facilitating targeting to the template strand (104) (Fig. 4b). In vitro, at least, the RNA exosome facilitates AID access to both DNA strands of non-R-loop-forming transcribed substrates (104, 145). Whether it plays such a role in vivo is unknown (104). Finally, RPA may stabilize the ssDNA substrates and augment AID activity in a phosphorylation-dependent manner (101, 143).
Evolution of CSR from SHM
Evolutionarily, SHM precedes CSR, with SHM emerging in early jawed vertebrates and CSR emerging in amphibians (146, 147). It has been proposed that Xenopus Sμ evolved to employ mechanisms utilized for AID targeting during SHM of variable region exons, which are not C/G-rich and which do not form R loops when transcribed (131). In this context, the AGCT-rich Xenopus Sμ region may have evolved via duplication of AGCT motif-dense CDR regions of V exons and in that context would target AID by SHM-like mechanisms. The novel features of mammalian S regions (C/G-richness, R-loop formation, much higher AGCT content) may have evolved to further enhance AID targeting or other aspects of CSR (e.g. synapsis, see below; 148). Further understanding of how S-region structure contributes to CSR may be illuminated by studying the divergent S regions of other species which may have evolved alternative or additional solutions for optimizing S-region substrates for CSR. For example, with respect to base composition, duck Sμ is C/G-rich but G:C content is equal between the two strands, while the putative duck Sα has an almost even distribution of the four bases on both strands with only a minor enrichment of G on one strand (39, 149). Assays of the abilities of these divergent S regions to support AID targeting and/or CSR in mammalian B cells could provide new insights into the S-region elements and the types of functions they support.
Joining AID-initiated S-region DSBs to complete CSR: overview
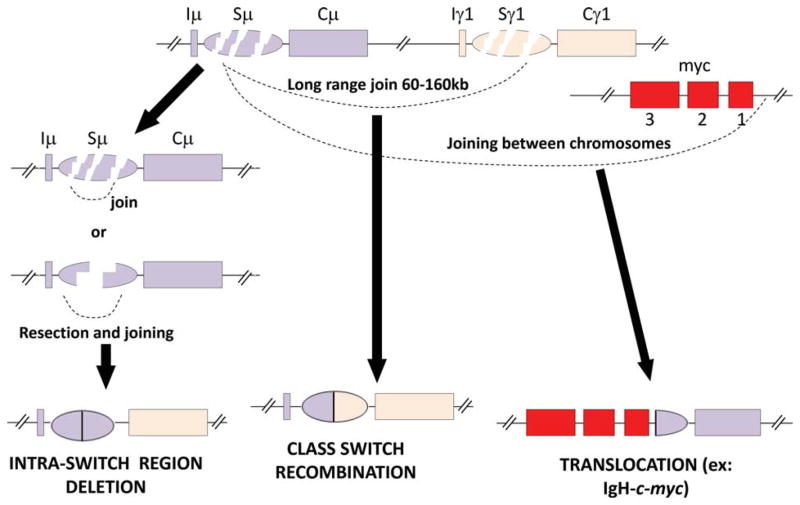
Productive CSR requires the upstream end of a DSB in Sμ to join to the downstream end of a DSB in a downstream S region that lies between 60 and 160 kb away, depending on the targeted S region. AID deaminates multiple cytosines within a given targeted S region (57); this may lead to multiple DSBs, with Sμ thought to be a particularly strong target. In addition to joining to DSBs in other S regions, S-region DSBs may be directly joined back together, be joined back together following end resection, or be joined to another DSB within the same S region (Fig. 5). The latter two outcomes result in internal S-region deletions (ISDs). In addition, DSBs generated in an S region may participate in chromosomal translocations by joining to other non-S-region DSBs on the cis chromosome or to DSBs on other chromosomes (80). Yet, in activated B cell populations in vitro, CSR joins can occur in up to 50% or more cells over a 4-day period (102), raising the question of the nature of the mechanisms that promote CSR events over substantial distances within the IgH locus.
FIGURE 5.
Outcomes of DSBs in S regions. DSBs within a S region may be directly joined back together or be joined back together following end resection, leading to intra-switch region deletions. Alternatively, a DSB generated in one S region may join to a DSB in another S region over a long-range (60 to 160 kb), which may lead to CSR. In addition, DSBs generated in an S region may participate in chromosomal translocations by joining to other non-S-region DSBs on the cis chromosome or to DSBs on other chromosomes. doi:10.1128/microbiolspec.MDNA3-0037-2014.f5
Factors that promote S-region synapsis during CSR
For two DSBs in different genomic locations to be joined, they must be simultaneously broken and physically juxtaposed (synapsed). Thus, joining of two separate DSBs will be influenced by the frequency of DSBs at each site (which reflects both frequency of generation and time of persistence) and by the frequency with which the DSB target sequences are synapsed (6, 150). During V(D)J recombination, appropriate pairs of RSSs are likely synapsed by stochastic mechanisms that are enhanced by locus contraction. Then, RAG1/2 binds to and cleaves the synapsed pairs of RSS ends and subsequently holds them in a post-cleavage complex in which the appropriate ends are joined (e.g. coding end to coding end and RSS end to RSS end) exclusively by C-NHEJ (4, 5, 6). In contrast, AID can clearly act on S regions in the absence of their synapsis, as evidenced by frequent ISDs and other experiments (151). Thus, the question arises as to how joining between DSBs in two separate S regions occurs at sufficient levels to yield physiologic levels of CSR, as opposed to just being rejoined or joined as ISDs.
Studies of recombinational IgH class switching in the absence of S regions or AID have provided insight. Specifically, B cells in which Sμ and Sγ1 are replaced with yeast I-SceI meganuclease target sites can undergo I-SceI-dependent recombinational class switching to IgG1 at levels approaching those of the wild type (WT) (152, 153). High-throughput analyses of joining frequencies between these widely separated IgH locus I-SceI-generated DSBs indicated that high level joining frequencies also occur in other cell types (e.g. T cells); in addition, similarly separated I-SceI or Cas-9/gRNA separated DSBs joined at high frequency in other chromosomal locations (e.g. around the c-myc gene) (153). Indeed, high-throughput DSB joining assays showed that DSBs within so-called megabase or submegabase topological “domains” (154, 155, 156, 157) are joined at surprisingly high frequency in several tested sites across the genome (6, 150).
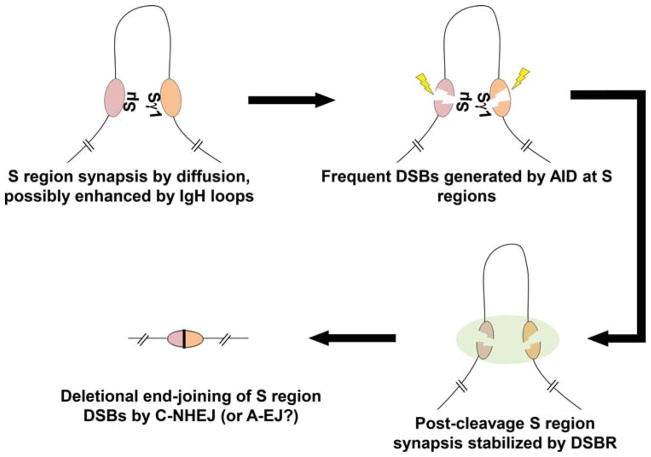
The occurrence of sequences, such as of different S regions, within such megabase domains would generate a much greater chance of them being transiently synapsed by diffusion or related mechanisms (153). Thus, if two sequences in such a domain also were frequently broken, they would have a higher probability of being joined when broken while synapsed, thereby driving frequent joining (6, 153) (Fig. 6). CSR has been speculated to have evolved to exploit this general property of gene organization in chromatin (6, 17, 153). In addition, IgH-locus-specific mechanisms may further promote S-region synapsis in activated B cells; such mechanisms might involve the organization of the locus into specific loops via interactions of activated I-region promoters with enhancers and or other types of intra-locus interactions (158). While AID has no known properties of stabilizing the synapsis of two S-region DSBs similar to RAG holding the DSBs in a post-cleavage complex, other factors including those of the DNA damage response regulated by the ataxia telangiectasia mutated kinase (ATM), as discussed below, may contribute such stabilization activities that increase the duration of S-region DSB synapsis to further promote CSR (97, 159, 160) (Fig. 6).
FIGURE 6.
Synapsis and end-joining. The roles of synapsis and tethering in promoting long-range joining are shown. We propose that S regions are synapsed by diffusion, and that synapsis is possibly enhanced by proximity of S regions resulting from chromatin organization into megabase/submegabase domains. Post-cleavage, synapsis may be maintained by general DSB response (DSBR) factors, promoting the joining of S-region DSB ends by classical nonhomologous end-joining (C-NHEJ) and possibly alternative end-joining (A-EJ). doi:10.1128/microbiolspec.MDNA3-0037-2014.f6
Contribution of the ATM-dependent DNA damage response to both synapsis and joining during CSR
DSBs, including AID-dependent S-region DSBs during CSR, activate the ATM-dependent DNA damage response. In the ATM-dependent DNA damage response, ATM phosphorylates substrates that mediate cell cycle checkpoints, such as p53, and DNA repair factors, including the proteins histone H2AX, mediator of DNA damage checkpoint protein 1 (MDC1), the nibrin (Nbs1) component of the Mre11-Rad50-Nbs1 complex (MRN), and tumor suppressor p53 binding protein 1 (53BP1) (160). ATM-dependent double-strand-break response factors (DSBR factors) form foci that extend mega bases along chromatin flanking DSB sites and provide docking sites for protein complexes that bind and tether broken DNA ends (159, 161). DSBs in two different S regions are well within the range of overlapping DSBR foci, which might contribute to stabilizing the synapsis of broken S regions (159). Thus, deficiency for ATM or for several of its substrates, including H2AX, MDC1, and 53BP1 reduce CSR while increasing AID-dependent IgH breaks and translocations (162, 163).
Of the ATM substrates implicated in CSR, 53BP1 appears to have a specialized and especially critical role in CSR. In this regard, CSR in 53BP1-deficient B cells is profoundly reduced (164, 165). In addition, while overall levels of genome instability in CSR-activated 53BP1-deficient B cells are similar to those of ATM-or H2AX-deficient B cells, most of the instability arising in the context of 53BP1 deficiency occurs at the IgH locus, in contrast to the more wide spread genomic instability found in the context of ATM or H2AX deficiency (162, 163). Proposed roles for 53BP1 include preventing resection of DSB ends (166, 167), influencing repair pathway choice between C-NHEJ and alternative end-joining (A-EJ) (166), and promoting long-range joining (168). In the latter context, one potential function of 53BP1 would be to stabilize synapsed S regions once they are broken. ATM-dependent phosphorylation of 53BP1 recruits Rif1 to DSBs where it counters resection of DNA breaks that would inhibit CSR (169, 170, 171). The ATM-dependent DSB response also contributes directly to the joining of CSR DSBs via the C-NHEJ pathway discussed next (172).
End-joining of synapsed DSB ends to complete IgH CSR
DSBs in two synapsed S regions are end-joined to complete CSR. Most CSR DSBs appear to be joined by C-NHEJ, one of the two major known DSB joining pathways. C-NHEJ DSB end-joining may generate “direct” (or “blunt”) DSB joins, which involves ligating blunt DNA ends (either generated as blunt ends or blunted by end-processing) or, alternatively, C-NHEJ can employ base pairing interactions between short (usually 1 to 4 bp) stretches of homology (“micro-homologies”, MH) present in single-strand overhangs (173, 174). Consistent with C-NHEJ, most CSR junctions are direct or use short MHs (175, 176). C-NHEJ and a number of C-NHEJ factors were originally discovered based on the exclusive role of C-NHEJ for joining during V(D)J recombination (177). During C-NHEJ, DSBs are bound by the Ku70 and Ku80 end-binding complex (Ku), which promotes end ligation carried out by the XRCC4/ligase 4 (Lig4) complex (173). These factors Ku70, Ku80, XRCC4, and Lig4 are considered the “core” C-NHEJ factors as they are required for joining of all types of broken ends during C-NHEJ (174). Ku also recruits DNA-PKcs which appears to function both in end synapsis (178) and also by recruiting other factors, most notably the Artemis endonuclease, that process certain types of ends before they can be joined by core C-NHEJ factors (173).
Although V(D)J recombination and, thus, mature B cell development is strictly dependent on C-NHEJ (177, 179, 180, 181, 182), this developmental requirement can be circumvented by providing developing B cells with preassembled IgH and IgL variable region exons in their germline (“HL mice”). Germline inactivation of Ku, XRCC4 or Lig4 in HL mice, or conditional inactivation of the latter two factors in mature B cells, demonstrated that CSR can occur at up to 40% of normal levels in the absence of core C-NHEJ factors (175, 183, 184). In such C-NHEJ deficient B cells, unjoined AID-initiated DSBs progress into AID-dependent IgH locus chromosomal breaks and translocations (73, 175). DNA-PKcs and Artemis deficiencies generally have a milder effect on CSR levels (185, 186, 187, 188), consistent at least in part with major roles for these factors in processing certain types of ends but not others for C-NHEJ. In addition, Artemis-independent functions of DNA-PKcs, for example DSB end synapsis, can be provided by functionally redundant activities of the ATM kinase (189). Despite their more modest contribution to CSR joining than core C-NHEJ factors, the roles of DNA-PKcs and Artemis in CSR are clear since in the absence of either factor, activated B cells accumulate significantly increased levels of IgH locus chromosomal breaks and translocations (190).
CSR in the absence of C-NHEJ is carried out by one or more alternative end-joining pathways (5, 174). A-EJ has been described in many ways, including being considered MH-dependent. Yet, depending on context, A-EJ in mammalian cells is not necessarily MH-dependent (174). Thus, perhaps the best working definition is any end-joining that occurs in the absence of the core C-NHEJ factors (174, 191). In the absence of XRCC4 or Lig4, nearly all CSR junctions have MHs at their junctions in contrast to only 50% of CSR junctions having MHs in WT B cells (175). Notably, a fraction of these MHs in CSR junctions from C-NHEJ deficient cells are longer than those typically associated with C-NHEJ (174, 175, 176). The use of long MH may be influenced by sequence context, with ends from more related S regions having a greater chance to produce long MHs to support joining by A-EJ (175, 192). Currently, however, the degree to which A-EJ contributes to normal CSR in the presence of intact C-NHEJ remains to be determined.
A-EJ during CSR likely represents more than one pathway. These pathways include a Ku-independent pathway, as Ku-deficient cells also have reduced yet substantial CSR that is somewhat less MH-dependent than that of XRCC4 or Lig4 deficient B cells, and a Lig4-independent pathway that uses a different ligase downstream of Ku and other C-NHEJ components, as B cells lacking both Ku and Lig4 undergo CSR similarly to B cells lacking Ku (184). Factors thus far implicated in A-EJ during CSR include components of other known DNA repair pathways, including poly (ADP-ribose) polymerase 1 (PARP-1), which may provide a DSB end recognition function, Mre11 and the C-terminal-binding protein 1-interacting protein (CtIP), which may be involved in end-processing, and X-ray repair cross-complementing protein 1 (XRCC1), ligase 1, and ligase 3, which may be involved in end-joining (174). A-EJ, including the potential role of this pathway in CSR and translocations, has been recently reviewed in depth (174).
PERSPECTIVE
Elucidating mechanisms that promote differential targeting and outcome of AID activity in SHM and CSR is a major ongoing question. While many advances have been made, the question of how AID cytidine deamination activity is specifically targeted during SHM and CSR remains in substantial part unanswered. As outlined above, transcription of target sequences is required for CSR and SHM. Yet transcription per se is not sufficient to explain the specificity of AID targeting since most transcribed genes in CSR-activated or GC B cells are not subject to detectable AID deamination. In this context, the non-Ig loci that are recurrent targets of lower level AID targeting in CSR-activated or GC B cells also show no readily apparent similarity with S regions and V(D)J exons at the sequence level (193). A longstanding mystery involves the differential targeting of AID to V exons during the GC SHM response versus to S regions during CSR. Thus, AID acts on the S regions in CSR-activated B cells, but not (detectably) on the adjacent V(D)J exons, even though they are actively transcribed (194, 195). Likewise, some GC B cells have robust SHM within their IgH V exons in the absence of CSR (196, 197, 198). How AID achieves such specificity with respect to physiological targets is unknown. Another unanswered question is how the mutational outcome of AID activity is targeted to the three CDRs within variable region exons and the relative contributions of cellular selection versus actual targeting. Clearly, in all of these contexts, there are likely, yet to be defined, sequence motifs that may couple with unique transcription features of particular genes and contribute to make them AID targets. The chicken DIVAC elements, which function to enhance SHM through unknown mechanisms, are one potential example (95, 111, 112).
Once AID cytidine deamination is targeted, various mechanisms may lead to differential outcomes of this potentially mutagenic activity during SHM and CSR. In this regard, the level of AID targeting to a particular sequence is usually estimated based on the accumulation of mutations or DSBs and rearrangements/translocations. However, it remains possible that, for at least some sequences, the level at which such genomic alterations are found at particular sequences may be influenced by their predisposition to undergo high fidelity versus mutagenic repair outcomes of the AID-generated cytidine deamination lesions (42). It also is generally considered that AID targeting can preferentially lead to point mutational versus DSB outcomes, respectively, during SHM and CSR. However, more rigorous analyses need to be performed to determine the extent to which these generalizations actually apply, since AID activity at S regions clearly can lead to point mutations and AID activity on V regions can lead to DSBs. Beyond this potential caveat, sequences likely play an important role in the DSB outcome of AID activity in S regions, with a prime example being the abundance in S regions of the palindromic AGCT motif that could promote DSBs by leading to AID targeting on both DNA strands (6, 9, 26). In addition, other aspects of target sequences, differential repair pathways, or SHM versus CSR specific co-factors that favor the generation of point mutation versus DSBs may also play a role.
Another question that is not fully answered is how AID activity leads to C/G mutations in cell lines (199, 200, 201, 202, 203) or in CSR-activated B cells (194) versus spreading of the C/G mutations to A/T base pairs in GC B cells (204, 205). Error-prone DNA polymerases during BER and MMR in GC B cells have been implicated in the spreading process that generates mutations at A/T base pairs (206, 207), raising the possibility that differential expression of these enzymes or other related factors in GC B cells could contribute to SHM spreading (208). However, since V(D)J exons are not AID targets in B cells activated in culture for CSR, this notion has not yet been tested directly.
Acknowledgments
We thank Feilong Meng, Ming Tian, and Vipul Kumar for helpful comments. This work was supported by NIH R01 AI077595 (to FA) and a fellowship from the Cancer Research Institute (to LSY). FWA is an Investigator of the Howard Hughes Medical Institute.
References
- 1.Harwood NE, Batista FD. New insights into the early molecular events underlying B cell activation. Immunity. 2008;28(5):609–619. doi: 10.1016/j.immuni.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 3.Pan-Hammarstrom Q, Zhao Y, Hammarstrom L. Class switch recombination: a comparison between mouse and human. Adv Immunol. 93:1–61. doi: 10.1016/S0065-2776(06)93001-6. [DOI] [PubMed] [Google Scholar]
- 4.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 5.Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 6.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152(3):417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alt FW, Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc Natl Acad Sci USA. 1982;79(13):4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334(6181):395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 9.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 10.Stewart AK, Schwartz RS. Immunoglobulin V regions and the B cell. Blood. 1994;83(7):1717–1730. [PubMed] [Google Scholar]
- 11.Roy AL, Sen R, Roeder RG. Enhancer-promoter communication and transcriptional regulation of Igh. Trends Immunol. 2011;32(11):532–539. doi: 10.1016/j.it.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muramatsu M, Nagaoka H, Shinkura R, Begum NA, Honjo T. Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- 13.Pinaud E, Marquet M, Fiancette R, Peron S, Vincent-Fabert C, Denizot Y, Cogne M. The IgH locus 3′ regulatory region: pulling the strings from behind. Adv Immunol. 2011;110:27–70. doi: 10.1016/B978-0-12-387663-8.00002-8. [DOI] [PubMed] [Google Scholar]
- 14.Chen K, Cerutti A. New insights into the enigma of immunoglobulin D. Immunol Rev. 2010;237(1):160–179. doi: 10.1111/j.1600-065X.2010.00929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3(6):1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mostoslavsky R, Alt RW, Rajewsky K. The lingering enigma of the allelic exclusion mechanism. Cell. 2004;118(5):539–544. doi: 10.1016/j.cell.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, Manis J, Alt FW. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 18.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 19.Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol. 2010;28:243–273. doi: 10.1146/annurev-immunol-030409-101314. [DOI] [PubMed] [Google Scholar]
- 20.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 22.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102(5):565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 23.Wilson PC, de Bouteiller O, Liu YJ, Potter K, Banchereau J, Capra JD, Pascual V. Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. J Exp Med. 1998;187(1):59–70. doi: 10.1084/jem.187.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goossens T, Klein U, Kuppers R. Frequent occurrence of deletions and duplications during somatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc Natl Acad Sci USA. 1998;95(5):2463–2468. doi: 10.1073/pnas.95.5.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12(20):1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 26.Maul RW, Gearhart PJ. AID and somatic hypermutation. Adv Immunol. 2010;105:159–191. doi: 10.1016/S0065-2776(10)05006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 28.Matthews AJ, Zheng S, DiMenna LJ, Chaudhuri J. Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair. Adv Immunol. 2014;122:1–57. doi: 10.1016/B978-0-12-800267-4.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woof JM, Kerr MA. The function of immunoglobulin A in immunity. J Pathol. 2006;208(2):270–282. doi: 10.1002/path.1877. [DOI] [PubMed] [Google Scholar]
- 30.Wu LC, Zarrin AA. The production and regulation of IgE by the immune system. Nat Rev Immunol. 2014;14(4):247–259. doi: 10.1038/nri3632. [DOI] [PubMed] [Google Scholar]
- 31.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8(1):34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 32.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274(26):18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 33.Arakawa H, Hauschild J, Buerstedde JM. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295(5558):1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 34.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci USA. 2003;100 (7):4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422(6933):726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 36.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197(10):1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424(6944):103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 38.Rogozin IB, Diaz M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J Immunol. 2004;172 (6):3382–3384. doi: 10.4049/jimmunol.172.6.3382. [DOI] [PubMed] [Google Scholar]
- 39.Hackney JA, Misaghi S, Senger K, Garris C, Sun Y, Lorenzo MN, Zarrin AA. DNA targets of AID evolutionary link between antibody somatic hypermutation and class switch recombination. Adv Immunol. 2009;101:163–189. doi: 10.1016/S0065-2776(08)01005-5. [DOI] [PubMed] [Google Scholar]
- 40.Dorner T, Brezinschek HP, Brezinschek RI, Foster SJ, Domiati-Saad R, Lipsky PE. Analysis of the frequency and pattern of somatic mutations within nonproductively rearranged human variable heavy chain genes. J Immunol. 1997;158(6):2779–2789. [PubMed] [Google Scholar]
- 41.Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim Biophys Acta. 1992;1171(1):11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 42.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451(7180):841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 43.Krokan HE, Bjoras M. Base excision repair. Cold Spring Harbor Perspect Biol. 2013;5(4):a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66(6):981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7(5):335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 46.Saribasak H, Gearhart PJ. Does DNA repair occur during somatic hypermutation? Semin Immunol. 2012;24(4):287–292. doi: 10.1016/j.smim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neuberger MS, Di Noia JM, Beale RC, Williams GT, Yang Z, Rada C. Somatic hypermutation at A.T pairs: polymerase error versus dUTP incorporation. Nat Rev Immunol. 2005;5(2):171–178. doi: 10.1038/nri1553. [DOI] [PubMed] [Google Scholar]
- 48.Neuberger MS, Rada C. Somatic hypermutation: activation-induced deaminase for C/G followed by polymerase eta for A/T. J Exp Med. 2007;204(1):7–10. doi: 10.1084/jem.20062409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiesendanger M, Kneitz B, Edelmann W, Scharff MD. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J Exp Med. 2000;191(3):579–584. doi: 10.1084/jem.191.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen HM, Tanaka A, Bozek G, Nicolae D, Storb U. Somatic hypermutation and class switch recombination in Msh6(−/−)Ung(−/−) double-knockout mice. J Immunol. 2006;177(8):5386–5392. doi: 10.4049/jimmunol.177.8.5386. [DOI] [PubMed] [Google Scholar]
- 51.Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, Sack SZ, Parris T, Edelmann W, Scharff MD. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat Immunol. 2004;5(2):224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- 52.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2(6):537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 53.Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med. 2007;204(1):17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418(6893):99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 55.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16(2):163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 56.Chahwan R, Edelmann W, Scharff MD, Roa S. AIDing antibody diversity by error-prone mismatch repair. Semin Immunol. 2012;24(4):293–300. doi: 10.1016/j.smim.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J Exp Med. 2006;203(9):2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neuberger MS. Antibody diversification by somatic mutation: from Burnet onwards. Immunol Cell Biol. 2008;86(2):124–132. doi: 10.1038/sj.icb.7100160. [DOI] [PubMed] [Google Scholar]
- 59.Pech M, Hochtl J, Schnell H, Zachau HG. Differences between germ-line and rearranged immunoglobulin V kappa coding sequences suggest a localized mutation mechanism. Nature. 1981;291(5817):668–670. doi: 10.1038/291668a0. [DOI] [PubMed] [Google Scholar]
- 60.Roes J, Huppi K, Rajewsky K, Sablitzky F. V gene rearrangement is required to fully activate the hypermutation mechanism in B cells. J Immunol. 1989;142(3):1022–1026. [PubMed] [Google Scholar]
- 61.Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90(6):1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 62.Betz AG, Neuberger MS, Milstein C. Discriminating intrinsic and antigen-selected mutational hotspots in immunoglobulin V genes. Immunol Today. 1993;14(8):405–411. doi: 10.1016/0167-5699(93)90144-a. [DOI] [PubMed] [Google Scholar]
- 63.Betz AG, Rada C, Pannell R, Milstein C, Neuberger MS. Passenger transgenes reveal intrinsic specificity of the antibody hypermutation mechanism: clustering, polarity, and specific hot spots. Proc Natl Acad Sci USA. 1993;90(6):2385–2388. doi: 10.1073/pnas.90.6.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jolly CJ, Wagner SD, Rada C, Klix N, Milstein C, Neuberger MS. The targeting of somatic hypermutation. Semin Immunol. 1996;8(3):159–168. doi: 10.1006/smim.1996.0020. [DOI] [PubMed] [Google Scholar]
- 65.Wagner SD, Milstein C, Neuberger MS. Codon bias targets mutation. Nature. 1995;376(6543):732. doi: 10.1038/376732a0. [DOI] [PubMed] [Google Scholar]
- 66.Kepler TB. Codon bias and plasticity in immunoglobulins. Mol Biol Evol. 1997;14(6):637–643. doi: 10.1093/oxfordjournals.molbev.a025803. [DOI] [PubMed] [Google Scholar]
- 67.Foster SJ, Dorner T, Lipsky PE. Somatic hypermutation of VkappaJkappa rearrangements: targeting of RGYW motifs on both DNA strands and preferential selection of mutated codons within RGYW motifs. Eur J Immunol. 1999;29(12):4011–4021. doi: 10.1002/(SICI)1521-4141(199912)29:12<4011::AID-IMMU4011>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 68.Rada C, Milstein C. The intrinsic hypermutability of antibody heavy and light chain genes decays exponentially. EMBO J. 2001;20(16):4570–4576. doi: 10.1093/emboj/20.16.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yelamos J, Klix N, Goyenechea B, Lozano F, Chui YL, Gonzalez Fernandez A, Pannell R, Neuberger MS, Milstein C. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature. 1995;376(6537):225–229. doi: 10.1038/376225a0. [DOI] [PubMed] [Google Scholar]
- 70.Azuma T, Motoyama N, Fields LE, Loh DY. Mutations of the chloramphenicol acetyl transferase transgene driven by the immunoglobulin promoter and intron enhancer. Int Immunol. 1993;5(2):121–130. doi: 10.1093/intimm/5.2.121. [DOI] [PubMed] [Google Scholar]
- 71.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4(1):57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 72.Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity. 2000;13(5):589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- 73.Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, Fazeli A, Feldman L, Nussenzweig A, Nussenzweig M, Alt FW. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci USA. 2010;107(7):3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Briney BS, Willis JR, Crowe EF., Jr Location and length distribution of somatic hypermutation-associated DNA insertions and deletions reveals regions of antibody structural plasticity. Genes Immun. 2012;13 (7):523–529. doi: 10.1038/gene.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiss U, Zoebelein R, Rajewsky K. Accumulation of somatic mutants in the B cell compartment after primary immunization with a T cell-dependent antigen. Eur J Immunol. 1992;22(2):511–517. doi: 10.1002/eji.1830220233. [DOI] [PubMed] [Google Scholar]
- 76.Lebecque SG, Gearhart PJ. Boundaries of somatic mutation in rearranged immunoglobulin genes: 5′ boundary is near the promoter, and 3′ boundary is approximately 1 kb from V(D)J gene. J Exp Med. 1990;172 (6):1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O’Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Program NCS, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev. 2013;254(1):225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20(40):5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- 80.Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- 81.Storb U. Why does somatic hypermutation by AID require transcription of its target genes? Adv Immunol. 2014;122:253–277. doi: 10.1016/B978-0-12-800267-4.00007-9. [DOI] [PubMed] [Google Scholar]
- 82.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6(8):573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 83.Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9(1):105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- 84.Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77(2):239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 85.Tumas-Brundage K, Manser T. The transcriptional promoter regulates hypermutation of the antibody heavy chain locus. J Exp Med. 1997;185(2):239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sharpe MJ, Milstein C, Jarvis JM, Neuberger MS. Somatic hypermutation of immunoglobulin kappa may depend on sequences 3′ of C kappa and occurs on passenger transgenes. EMBO J. 1991;10(8):2139–2145. doi: 10.1002/j.1460-2075.1991.tb07748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Terauchi A, Hayashi K, Kitamura D, Kozono Y, Motoyama N, Azuma T. A pivotal role for DNase I-sensitive regions 3b and/or 4 in the induction of somatic hypermutation of IgH genes. J Immunol. 2001;167 (2):811–820. doi: 10.4049/jimmunol.167.2.811. [DOI] [PubMed] [Google Scholar]
- 88.Inlay MA, Gao HH, Odegard VH, Lin T, Schatz DG, Xu Y. Roles of the Ig kappa light chain intronic and 3′ enhancers in Igk somatic hypermutation. J Immunol. 2006;177(2):1146–1151. doi: 10.4049/jimmunol.177.2.1146. [DOI] [PubMed] [Google Scholar]
- 89.van der Stoep N, Gorman JR, Alt FW. Reevaluation of 3′ Ekappa function in stage- and lineage-specific rearrangement and somatic hypermutation. Immunity. 1998;8(6):743–750. doi: 10.1016/s1074-7613(00)80579-8. [DOI] [PubMed] [Google Scholar]
- 90.Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci USA. 2005;102(40):14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M, Cogne M. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 2001;15(2):187–199. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- 92.Morvan CL, Pinaud E, Decourt C, Cuvillier A, Cogne M. The immunoglobulin heavy-chain locus hs3b and hs4 3′ enhancers are dispensable for VDJ assembly and somatic hypermutation. Blood. 2003;102 (4):1421–1427. doi: 10.1182/blood-2002-12-3827. [DOI] [PubMed] [Google Scholar]
- 93.Vincent-Fabert C, Fiancette R, Pinaud E, Truffinet V, Cogne N, Cogne M, Denizot Y. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 2010;116(11):1895–1898. doi: 10.1182/blood-2010-01-264689. [DOI] [PubMed] [Google Scholar]
- 94.Rouaud P, Vincent-Fabert C, Saintamand A, Fiancette R, Marquet M, Robert I, Reina-San-Martin B, Pinaud E, Cogne M, Denizot Y. The IgH 3′ regulatory region controls somatic hypermutation in germinal center B cells. J Exp Med. 2013;210(8):1501–1507. doi: 10.1084/jem.20130072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Buerstedde JM, Alinikula J, Arakawa H, McDonald JJ, Schatz DG. Targeting of somatic hypermutation by immunoglobulin enhancer and enhancer-like sequences. PLoS Biol. 2014;12(4):e1001831. doi: 10.1371/journal.pbio.1001831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol. 2012;12(7):517–31. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Daniel JA, Nussenzweig A. The AID-induced DNA damage response in chromatin. Mol Cell. 2013;50(3):309–321. doi: 10.1016/j.molcel.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, Reina San-Martin B, Barreto V, Nieland TJ, Root DE, Casellas R, Nussenzweig MC. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143(1):122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12(3):343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12(3):357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430(7003):992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 102.Cheng HL, Vuong BQ, Basu U, Franklin A, Schwer B, Astarita J, Phan RT, Datta A, Manis J, Alt FW, Chaudhuri J. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc Natl Acad Sci USA. 2009;106 (8):2717–2722. doi: 10.1073/pnas.0812304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McBride KM, Gazumyan A, Woo EM, Schwickert TA, Chait BT, Nussenzweig MC. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med. 2008;205(11):2585–2594. doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, Januszyk K, Gregory RI, Deng H, Lima CD, Alt FW. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144(3):353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7(7):529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 106.Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci USA. 2004;101(35):12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr Opin Immunol. 2006;18 (2):164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 108.Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti RS, Klein U, Kuppers R, Rajewsky K, Dalla-Favera R. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci USA. 1998;95 (20):11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple protooncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412(6844):341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 110.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280(5370):1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 111.Kohler KM, McDonald JJ, Duke JL, Arakawa H, Tan S, Kleinstein SH, Buerstedde JM, Schatz DG. Identification of core DNA elements that target somatic hypermutation. J Immunol. 2012;189(11):5314–5326. doi: 10.4049/jimmunol.1202082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Blagodatski A, Batrak V, Schmidl S, Schoetz U, Caldwell RB, Arakawa H, Buerstedde JM. A cis-acting diversification activator both necessary and sufficient for AID-mediated hypermutation. PLoS Genet. 2009;5(1):e1000332. doi: 10.1371/journal.pgen.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang T, Franklin A, Boboila C, McQuay A, Gallagher MP, Manis JP, Khamlichi AA, Alt FW. Downstream class switching leads to IgE antibody production by B lymphocytes lacking IgM switch regions. Proc Natl Acad Sci USA. 2010;107(7):3040–3045. doi: 10.1073/pnas.0915072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lutzker S, Rothman P, Pollock R, Coffman R, Alt FW. Mitogen- and IL-4-regulated expression of germ-line Ig gamma 2b transcripts: evidence for directed heavy chain class switching. Cell. 1988;53(2):177–184. doi: 10.1016/0092-8674(88)90379-0. [DOI] [PubMed] [Google Scholar]
- 115.Rothman P, Chen YY, Lutzker S, Li SC, Stewart V, Coffman R, Alt FW. Structure and expression of germ line immunoglobulin heavy-chain epsilon transcripts: interleukin-4 plus lipopolysaccharide-directed switching to C epsilon. Mol Cell Biol. 1990;10(4):1672–1679. doi: 10.1128/mcb.10.4.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Esser C, Radbruch A. Rapid induction of transcription of unrearranged S gamma 1 switch regions in activated murine B cells by interleukin 4. EMBO J. 1989;8(2):483–488. doi: 10.1002/j.1460-2075.1989.tb03401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shinkura R, Tian M, Smith M, Chua K, Fujiwara Y, Alt FW. The influence of transcriptional orientation on endogenous switch region function. Nat Immunol. 2003;4(5):435–441. doi: 10.1038/ni918. [DOI] [PubMed] [Google Scholar]
- 118.Radbruch A, Sablitzky F. Deletion of Cmu genes in mouse B lymphocytes upon stimulation with LPS. EMBO J. 1983;2(11):1929–1935. doi: 10.1002/j.1460-2075.1983.tb01681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Radbruch A, Muller W, Rajewsky K. Class switch recombination is IgG1 specific on active and inactive IgH loci of IgG1-secreting B-cell blasts. Proc Natl Acad Sci USA. 1986;83(11):3954–3957. doi: 10.1073/pnas.83.11.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jung S, Rajewsky K, Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259 (5097):984–987. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- 121.Zhang J, Bottaro A, Li S, Stewart V, Alt FW. A selective defect in IgG2b switching as a result of targeted mutation of the I gamma 2b promoter and exon. EMBO J. 1993;12(9):3529–3537. doi: 10.1002/j.1460-2075.1993.tb06027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bottaro A, Lansford R, Xu L, Zhang J, Rothman P, Alt FW. S region transcription per se promotes basal IgE class switch recombination but additional factors regulate the efficiency of the process. EMBO J. 1994;13(3):665–674. doi: 10.1002/j.1460-2075.1994.tb06305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Seidl KJ, Bottaro A, Vo A, Zhang J, Davidson L, Alt FW. An expressed neo(r) cassette provides required functions of the 1gamma2b exon for class switching. Int Immunol. 1998;10(11):1683–1692. doi: 10.1093/intimm/10.11.1683. [DOI] [PubMed] [Google Scholar]
- 124.Lorenz M, Jung S, Radbruch A. Switch transcripts in immunoglobulin class switching. Science. 1995;267(5205):1825–1828. doi: 10.1126/science.7892607. [DOI] [PubMed] [Google Scholar]
- 125.Qiu G, Harriman GR, Stavnezer J. Ialpha exon-replacement mice synthesize a spliced HPRT-C(alpha) transcript which may explain their ability to switch to IgA. Inhibition of switching to IgG in these mice. Int Immunol. 1999;11(1):37–46. doi: 10.1093/intimm/11.1.37. [DOI] [PubMed] [Google Scholar]
- 126.Cogne M, Lansford R, Bottaro A, Zhang J, Gorman J, Young F, Cheng HL, Alt FW. A class switch control region at the 3′ end of the immunoglobulin heavy chain locus. Cell. 1994;77(5):737–747. doi: 10.1016/0092-8674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 127.Seidl KJ, Manis JP, Bottaro A, Zhang J, Davidson L, Kisselgof A, Oettgen H, Alt FW. Position-dependent inhibition of class-switch recombination by PGK-neor cassettes inserted into the immunoglobulin heavy chain constant region locus. Proc Natl Acad Sci USA. 1999;96(6):3000–3005. doi: 10.1073/pnas.96.6.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27(5):711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yan Y, Pieretti J, Ju Z, Wei S, Christin JR, Bah F, Birshtein BK, Eckhardt LA. Homologous elements hs3a and hs3b in the 3′ regulatory region of the murine immunoglobulin heavy chain (Igh) locus are both dispensable for class-switch recombination. J Biol Chem. 2011;286 (31):27123–27131. doi: 10.1074/jbc.M111.230995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Han L, Masani S, Yu K. Overlapping activation-induced cytidine deaminase hotspot motifs in Ig class-switch recombination. Proc Natl Acad Sci USA. 2011;108(28):11584–11589. doi: 10.1073/pnas.1018726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zarrin AA, Alt FW, Chaudhuri J, Stokes N, Kaushal D, Du Pasquier L, Tian M. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat Immunol. 2004;5(12):1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- 132.Mussmann R, Courtet M, Schwager J, Du Pasquier L. Microsites for immunoglobulin switch recombination breakpoints from Xenopus to mammals. Eur J Immunol. 1997;27(10):2610–2619. doi: 10.1002/eji.1830271021. [DOI] [PubMed] [Google Scholar]
- 133.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4(5):442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 134.Reaban ME, Griffin JA. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature. 1990;348(6299):342–344. doi: 10.1038/348342a0. [DOI] [PubMed] [Google Scholar]
- 135.Daniels GA, Lieber MR. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995;23(24):5006–5011. doi: 10.1093/nar/23.24.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tian M, Alt FW. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J Biol Chem. 2000;275(31):24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- 137.Mizuta R, Iwai K, Shigeno M, Mizuta M, Uemura T, Ushiki T, Kitamura D. Molecular visualization of immunoglobulin switch region RNA/DNA complex by atomic force microscope. J Biol Chem. 2003;278 (7):4431–4434. doi: 10.1074/jbc.M209262200. [DOI] [PubMed] [Google Scholar]
- 138.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol. 2008;28(1):50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, Gearhart PJ. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206(6):1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med. 2009;206 (8):1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vuong BQ, Lee M, Kabir S, Irimia C, Macchiarulo S, McKnight GS, Chaudhuri J. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat Immunol. 2009;10(4):420–426. doi: 10.1038/ni.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12(1):62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, Li G, Schrum JP, Manis JP, Alt FW. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438(7067):508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 144.Vuong BQ, Herrick-Reynolds K, Vaidyanathan B, Pucella JN, Ucher AJ, Donghia NM, Gu X, Nicolas L, Nowak U, Rahman N, Strout MP, Mills KD, Stavnezer J, Chaudhuri J. A DNA break- and phosphorylation-dependent positive feedback loop promotes immunoglobulin class-switch recombination. Nat Immunol. 2013;14(11):1183–1189. doi: 10.1038/ni.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Keim C, Kazadi D, Rothschild G, Basu U. Regulation of AID, the B-cell genome mutator. Genes Dev. 2013;27(1):1–17. doi: 10.1101/gad.200014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cannon JP, Haire RN, Rast JP, Litman GW. The phylogenetic origins of the antigen-binding receptors and somatic diversification mechanisms. Immunol Rev. 2004;200:12–22. doi: 10.1111/j.0105-2896.2004.00166.x. [DOI] [PubMed] [Google Scholar]
- 147.Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol. 2004;16(4):257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 148.Larson ED, Duquette ML, Cummings WJ, Streiff RJ, Maizels N. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr Biol. 2005;15(5):470–474. doi: 10.1016/j.cub.2004.12.077. [DOI] [PubMed] [Google Scholar]
- 149.Lundqvist ML, Middleton DL, Hazard S, Warr GW. The immunoglobulin heavy chain locus of the duck. Genomic organization and expression of D, J, and C region genes. J Biol Chem. 2001;276(50):46729–46736. doi: 10.1074/jbc.M106221200. [DOI] [PubMed] [Google Scholar]
- 150.Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148(5):908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dudley DD, Manis JP, Zarrin AA, Kaylor L, Tian M, Alt FW. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc Natl Acad Sci USA. 2002;99(15):9984–9989. doi: 10.1073/pnas.152333499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zarrin AA, Del Vecchio C, Tseng E, Gleason M, Zarin P, Tian M, Alt FW. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315(5810):377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- 153.Gostissa M, Schwer B, Chang A, Dong J, Meyers RM, Marecki GT, Choi VW, Chiarle R, Zarrin AA, Alt FW. IgH class switching exploits a general property of two DNA breaks to be joined in cis over long chromosomal distances. Proc Natl Acad Sci USA. 2014;111(7):2644–2649. doi: 10.1073/pnas.1324176111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. Organization of the mitotic chromosome. Science. 2013;342(6161):948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Bluthgen N, Dekker J, Heard E. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kenter AL, Feldman S, Wuerffel R, Achour I, Wang L, Kumar S. Three-dimensional architecture of the IgH locus facilitates class switch recombination. Ann N Y Acad Sci. 2012;1267:86–94. doi: 10.1111/j.1749-6632.2012.06604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3(2):149–153. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]
- 160.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141(1):27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146(5):905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM, Lou Z, Bassing CH, Manis JP, Chen J, Carpenter PB, Alt FW. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21(2):201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 163.Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, Nussenzweig A, Nussenzweig MC. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440(7080):105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5(5):481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 165.Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, Nussenzweig MC, Chen J. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165(4):459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207(4):855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, Nussenzweig A, Nussenzweig MC. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42(3):319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, Sleckman BP, Nussenzweig A. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456(7221):529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49(5):858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, Nussenzweig A, Casellas R, Robbiani DF, Nussenzweig MC. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339(6120):711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339(6120):700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kumar V, Alt FW, Oksenych V. Functional overlaps between XLF and the ATM-dependent DNA double strand break response. DNA Repair. 2014;16:11–22. doi: 10.1016/j.dnarep.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 175.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449(7161):478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 176.Stavnezer J, Bjorkman A, Du L, Cagigi A, Pan-Hammarstrom W. Mapping of switch recombination junctions, a tool for studying DNA repair pathways during immunoglobulin class switching. Adv Immunol. 2010;108:45–109. doi: 10.1016/B978-0-12-380995-7.00003-3. [DOI] [PubMed] [Google Scholar]
- 177.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260(5105):207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 178.Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 179.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig MC, Li GC. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382(6591):551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 180.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396 (6707):173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 181.Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, Bronson RT, Malynn BA, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin SH, Greenberg ME, Alt FW. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95(7):891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 182.Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, Davidson L, Cheng HL, Sekiguchi JM, Frank K, Stanhope-Baker P, Schlissel MS, Roth DB, Alt FW. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7(5):653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 183.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, Charneau P, de Villartay JP. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J Exp Med. 2007;204(7):1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Boboila C, Yan C, Wesemann DR, Jankovic M, Wang JH, Manis J, Nussenzweig A, Nussenzweig M, Alt FW. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med. 2010;207(2):417–427. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Manis JP, Tian M, Alt FW. Mechanism and control of class-switch recombination. Trends Immunol. 2002;23(1):31–39. doi: 10.1016/s1471-4906(01)02111-1. [DOI] [PubMed] [Google Scholar]
- 186.Cook AJ, Oganesian L, Harumal P, Basten A, Brink R, Jolly CJ. Reduced switching in SCID B cells is associated with altered somatic mutation of recombined S regions. J Immunol. 2003;171(12):6556–6564. doi: 10.4049/jimmunol.171.12.6556. [DOI] [PubMed] [Google Scholar]
- 187.Bosma GC, Kim J, Urich T, Fath DM, Cotticelli MG, Ruetsch NR, Radic MZ, Bosma MJ. DNA-dependent protein kinase activity is not required for immunoglobulin class switching. J Exp Med. 2002;196 (11):1483–1495. doi: 10.1084/jem.20001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kiefer K, Oshinsky J, Kim J, Nakajima PB, Bosma GC, Bosma MJ. The catalytic subunit of DNA-protein kinase (DNA-PKcs) is not required for Ig class-switch recombination. Proc Natl Acad Sci USA. 2007;104 (8):2843–2848. doi: 10.1073/pnas.0611359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S, Di Virgilio M, Heidkamp G, Alt FW, Nussenzweig A, Nussenzweig M. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell. 2009;34(3):285–297. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Franco S, Murphy MM, Li G, Borjeson T, Boboila C, Alt FW. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy chain class switch recombination. J Exp Med. 2008;205(3):557–564. doi: 10.1084/jem.20080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang Y, Gostissa M, Hildebrand DG, Becker MS, Boboila C, Chiarle R, Lewis S, Alt FW. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv Immunol. 2010;106:93–133. doi: 10.1016/S0065-2776(10)06004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pan-Hammarstrom Q, Jones AM, Lahdesmaki A, Zhou W, Gatti RA, Hammarstrom L, Gennery AR, Ehrenstein MR. Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells. J Exp Med. 2005;201(2):189–194. doi: 10.1084/jem.20040772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Duke JL, Liu M, Yaari G, Khalil AM, Tomayko MM, Shlomchik MJ, Schatz DG, Kleinstein SH. Multiple transcription factor binding sites predict AID targeting in non-Ig genes. J Immunol. 2013;190(8):3878–3888. doi: 10.4049/jimmunol.1202547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Nagaoka H, Muramatsu M, Yamamura N, Kinoshita K, Honjo T. Activation-induced deaminase (AID)-directed hypermutation in the immunoglobulin Smu region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J Exp Med. 2002;195(4):529–534. doi: 10.1084/jem.20012144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30(4):173–181. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 196.Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188 (9):1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rosner K, Winter DB, Kasmer C, Skovgaard GL, Tarone RE, Bohr VA, Gearhart PJ. Impact of age on hypermutation of immunoglobulin variable genes in humans. J Clin Immunol. 2001;21(2):102–115. doi: 10.1023/a:1011003821798. [DOI] [PubMed] [Google Scholar]
- 198.van Es JH, Meyling FH, Logtenberg T. High frequency of somatically mutated IgM molecules in the human adult blood B cell repertoire. Eur J Immunol. 1992;22(10):2761–2764. doi: 10.1002/eji.1830221046. [DOI] [PubMed] [Google Scholar]
- 199.Martin A, Bardwell PD, Woo CJ, Fan M, Shulman MJ, Scharff MD. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 2002;415(6873):802–806. doi: 10.1038/nature714. [DOI] [PubMed] [Google Scholar]
- 200.Sale JE, Neuberger MS. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 1998;9(6):859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- 201.Denepoux S, Razanajaona D, Blanchard D, Meffre G, Capra JD, Banchereau J, Lebecque S. Induction of somatic mutation in a human B cell line in vitro. Immunity. 1997;6(1):35–46. doi: 10.1016/s1074-7613(00)80240-x. [DOI] [PubMed] [Google Scholar]
- 202.Bachl J, Wabl M. An immunoglobulin mutator that targets G. C base pairs. Proc Natl Acad Sci USA. 1996;93(2):851–855. doi: 10.1073/pnas.93.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sale JE, Bemark M, Williams GT, Jolly CJ, Ehrenstein MR, Rada C, Milstein C, Neuberger MS. In vivo and in vitro studies of immunoglobulin gene somatic hypermutation. Philos Trans R Soc B. 2001;356 (1405):21–28. doi: 10.1098/rstb.2000.0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Milstein C, Neuberger MS, Staden R. Both DNA strands of antibody genes are hypermutation targets. Proc Natl Acad Sci USA. 1998;95 (15):8791–8794. doi: 10.1073/pnas.95.15.8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wagner SD, Neuberger MS. Somatic hypermutation of immunoglobulin genes. Annu Rev Immunol. 1996;14:441–457. doi: 10.1146/annurev.immunol.14.1.441. [DOI] [PubMed] [Google Scholar]
- 206.Reynaud CA, Delbos F, Faili A, Gueranger Q, Aoufouchi S, Weill JC. Competitive repair pathways in immunoglobulin gene hypermutation. Philos Trans R Soc B. 2009;364(1517):613–619. doi: 10.1098/rstb.2008.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Diaz M, Lawrence C. An update on the role of translesion synthesis DNA polymerases in Ig hypermutation. Trends Immunol. 2005;26 (4):215–220. doi: 10.1016/j.it.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 208.Ouchida R, Ukai A, Mori H, Kawamura K, Dolle ME, Tagawa M, Sakamoto A, Tokuhisa T, Yokosuka T, Saito T, Yokoi M, Hanaoka F, Vijg J, Wang JY. Genetic analysis reveals an intrinsic property of the germinal center B cells to generate A:T mutations. DNA Repair. 2008;7(8):1392–1398. doi: 10.1016/j.dnarep.2008.04.014. [DOI] [PubMed] [Google Scholar]