Abstract
AIM: To study the changes of human telomerase reverse transcriptase (hTERT) mRNA expression in human hepatocarcinoma cell lines (HepG2) and cholangiocarcinoma cell lines (QBC939) after HBx gene transfection and to illustrate the significance of transcriptional regulation of hTERT gene by HBx gene in the carcinogenesis.
METHODS: HepG2 and QBC939 cell lines were cultured and co-transfected with eukaryotic expression vector containing the HBx coding region and cloning vector containing enhanced green fluorescent protein (EGFP) coding sequence using lipid-mediated gene transduction technique. Thirty-six hours after transfection, EGFP expression in cells was used as the indicator of successful transfection. Flow cytometry was performed to determine the transfection efficiency. Cells were harvested and total RNA was extracted using TRIzol® reagent. The expression of hTERT mRNA in HepG2 and QBC939 cell lines was assayed by reverse transcription-polymerase chain reaction. The expression of HBx protein in both cell lines was detected by immunocytochemical staining and Western blotting.
RESULTS: Flow cytometry showed that the transfection efficiency was 46.4% in HepG2 cells and 29.6% in QBC939 cells for both HBx gene expression vector and blank vector. The expression of hTERT mRNA was meaningfully increased in HepG2 and QBC939 cell lines when transfected with HBx gene expression vector compared to those transfected with OPTI-MEM medium and blank vector. Immunocytochemical staining and Western blotting revealed HBx protein expression in HepG2 and QBC939 cells only when transfected with HBx gene.
CONCLUSION: HBx gene transfection can upregulate the transcriptional expression of hTERT mRNA. The transactiv-ation of hTERT gene by HBx gene is a newfound mechanism for pathogenesis of hepatocarcinomas and cholangioca-rcinomas after HBV infection.
Keywords: Hepatocholangiocarcinoma, Human telomerase reverse transcriptase, Gene expression, Hepatitis B virus, X protein
INTRODUCTION
As an oncogenic DNA virus, HBV has been ascertained as the major causative factor for hepatocarcinogenesis[1]. Our recent study has also shown that HBV infection may be associated with biliary tract carcinogenesis[2]. The X gene of HBV (HBx gene) encodes a viral protein (HBx protein), which performs an essential function in the development of human hepatocarcinoma[3] and cholangiocarcinoma[4]. The finding that HBx gene can upregulate telomerase activation, when transferred into hepatocarcinoma cells reveals a new mechanism for HBx gene in carcinogenesis[5]. Human telomerase reverse transcriptase (hTERT) is the rate-limiting determinant for telomerase activity and its expression parallels frequently to telomerase activity[6,7]. Our previous findings that HBx gene can upregulate hTERT mRNA expression, when transferred into normal human cholangiocytes and cholangiocarcinoma cells[8], ascertain the regulation of hTERT gene by HBx gene. To learn more about the mechanism in carcinogenesis, we transferred the eukaryotic expression vector encompassing the entire HBx coding region into both hepatocarcinoma cell line (HepG2) and cholangiocarcinoma cell line (QBC939). By assaying the expression of hTERT mRNA, we attempted to elucidate the significance of transcriptional regulation of hTERT gene by HBx gene in the carcinogenesis.
MATERIALS AND METHODS
Cell and culture
HepG2 cell line was purchased from Foundation Institute, Chinese Academy of Medical Sciences. QBC939 cell lines were obtained from Dr. Shuguang Wang (Southwest Hospital, the Third Military Medical University, Chongqing). Cells were maintained in Dulbecco’s modified Eagle’s medium containing 100 mL/L fetal bovine serum (FBS), 100 U/mL penicillin and 100 µg/mL streptomycin, and propagated at 37°C in a humidified atmosphere of 50 mL/L CO2 in air. Cells in monolayer culture passaged approximately thrice a week by incubation with 2.5 g/L trypsin/1 mmol/L versene at 37°C for approximately 5 min until the cells were shrunken. Trypsin activity was quenched by the addition of fresh medium containing FBS and cells were seeded at 1/4 split ratio.
Plasmids and transfection
The empty vector pcDNA3 is eukaryotic expression vector used as a negative control. pCMV-X is constructed by inserting the entire HBx coding region (HBV nucleotides 1 372-1 833, 465 bp) into the EcoRI/EcoRV sites of the pcDNA3 vector. Cloning vector pEGFP carries an enhanced green fluorescent protein (EGFP) gene, which was cloned between the two MCS of the pPD16.43. EGFP encoded by pEGFP can emit bright green fluorescence in eukaryotic cells. All the plasmids contain ampicillin resistance genes for propagation and selection in E. coli.
Transient transfection of plasmids into both HepG2 and QBC939 was performed using LipofectAMINE PLUS™ reagent (Gibco BRL ) according to the manufacturer’s instructions. The day before transfection, cells were trypsinized and seeded in six-well plates. At the day of transfection, cells reached over 80% confluence, and then 2.9 µg pcDNA3 or pCMV-X DNA and 0.1 µg pEGFP DNA were diluted into OPTI-MEM medium (Invitrogen) and mixed with Lipofect-AMINE PLUS™ reagent. After the cells were washed once by OPTI-MEM medium, a total of 1 mL transfection medium was added to the cells. The cells were incubated at 37°C for 3 h in a humidified atmosphere containing 50 mL/ L CO2. After 3 h of incubation, the transfection medium was replaced with fresh complete medium containing serum and the cells were further incubated for 36 h. Then the cells were harvested, and the expression of hTERT mRNA was assayed. The cells transfected with OPTI -MEM medium were used as blank control (group A) and that transfected with pcDNA3 vector were used as negative control (group C), while that transfected with pCMV-X were defined as experimental group (group B). Co-transfection of pEGFP was used to evaluate the successful transfection.
Detection of transfection efficiency by flow cytometry
Cells were trypsinized, washed with PBS for twice, followed by dilution at 1×106 cells/mL. Transfection efficiency was detected using flow cytometry by following the manufacturer’s instructions.
Extraction of total cellular RNA and RT-PCR analysis for hTERT mRNA
Total cellular RNAs were extracted from cells of different groups by TRIzol reagent (Gibco BRL ) according to the manufacturer’s instructions. Two micrograms of extracted RNA was reverse transcribed into cDNA first-strand using 200 U of Moloney Murine Leukemia Virus Reverse Transcriptase (Promega, Madison, USA) and 1 µg of oligo (dT)15 primer (Promega) in a final volume of 25 µL of enzyme buffer (Promega) for 60 min at 42°C. hTERT cDNA analysis was performed by PCR amplification of a fragment of 145 bp using primer pairs: 5’-CGGAAGAGTGTCTGG-AGCAA-3’ (sense) and 5’-GGATGAAGCGGAGTCTGGA-3’ (antisense). A 320-bp fragment of GAPDH gene was amplified as an internal control. The primers for GAPDH are 5’-GGAAGCTTGTCATCAATGG-3’ (sense) and 5’-CTGTGGTCATGAGTCCTTC- 3’ (antisense). PCR was performed with 5 µL of cDNA first- strand in a 50 µL reaction volume containing 2 mmol/L MgCl2, 1 mmol/L dNTPs, 0.4 µmol/L of each primer, and 2.5 U of Taq DNA polymerase (Promega). The reaction mixture was heated at 94°C for 5 min, followed by amplification through 33 cycles. Each cycle included denaturation at 94°C for 40 s, annealing at 60°C for 40 s and extension at 72°C for 90 s. Then 10 µL of the PCR product was electrophoresed on a 15 g/L agarose gel containing ethidium bromide (0.5 µg/mL), and visualized under ultraviolet light. The quantitative difference of bands between groups was analyzed by NIH Image software. The reverse transcription-polymerase chain reaction (RT-PCR) was repeated in triplicate and quantitative data were expressed as mean±SD.
Detection of HBx protein by Western blotting
Total cellular protein was extracted by cell lysis (20 mmol/L Tris -HCl [pH 7.5], 1 mmol/L EDTA, 1 mmol/L EGTA, 150 mmol/L NaCl, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β -glycerolphosphate, 1 µg/mL leupeptin, 1 mol/L PMSF) and quantitated by BCATM protein assay kit (Pierce Biotech). An equal amount of protein sample was separated on SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membrane. The membrane was incubated in a blocking buffer (1 g/L Tween-20, 50 g/L non-fat dry milk in TBS) for 3 h. Rabbit anti-human HBx polyclonal antibody was diluted by primary antibody dilution buffer (1 g/L Tween -20, 50 g/L BSA in TBS) at 1: 800. The membrane was then incubated with HBx antibody overnight at 4°C. The expression of HBx protein was detected using Phototope -HRP Western Blot Detection Kit (Cell Signaling Technology).
Detection of HBx protein in transferred cells by immunocy-tochemistry
The cells in different groups were cultured on cover slips and fixed with fixative (acetone and methanol). Detection of HBx protein expression in the transfected cells was performed by UltraSensitive™ Immunochemistry kit according to the manufacturer’s instructions. A rabbit anti-human HBx polyclonal antibody was used at dilution of 1:800.
Statistical analysis
Comparison between means was tested by Student’s t-test and P value lesser than 0.05 was considered statistically significant.
RESULTS
Transfection efficiency
Under fluorescence microscope, EGFP expression could only be observed in the cells of groups B and C (Figure 1). There was no such green fluorescence could be seen in group A. Transfection efficiency was 46.4% for HepG2 and 29.6% for QBC939 when detected by flow cytometry (Figure 2).
Figure 1.
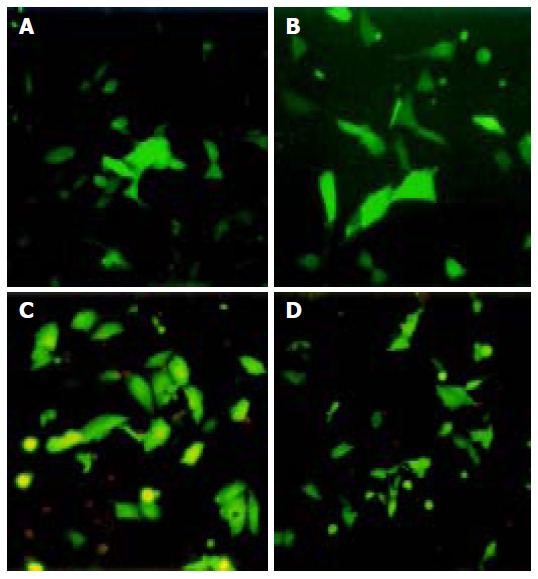
EGFP expressions in HepG2 (A and B) and QBC939 (C and D) cell lines under fluorescence microscope. After 36 h of transfection, EGFP-expressing cells were visible in group B (A and C) and group C (B and D), but not in group A.
Figure 2.
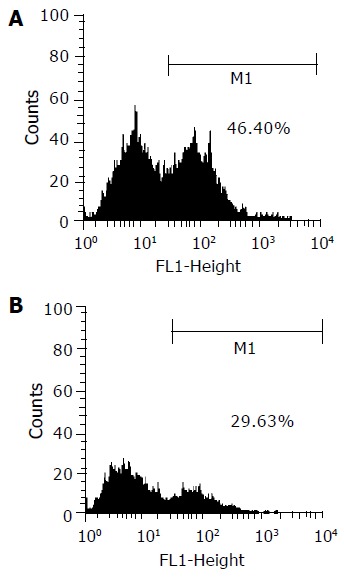
Transfection efficiency evaluated by flow cytometry. The transfection efficiencies were about 46.4% for HepG2 (A) and 29.6% for QBC939 (B).
hTERT mRNA expression
To examine the effects of HBx gene on hTERT transcription, pCMV-X expression vector was co-transfected with pEGFP into HepG2 and QBC939 cell lines. Thirty-six hours after transfection, the total cellular RNA was extracted and hTERT transcript was assayed using RT-PCR. The result showed that hTERT transcript was expressed in all groups (Figure 3). Expression of the human GAPDH gene was monitored for quantitation of the amount of RNA in all of the samples as an internal control. Relative expression of hTERT mRNA was determined by measuring band intensities of both hTERT transcript and GAPDH transcript and calculating the ratio of hTERT to GAPDH. After being analyzed by NIH Image software, hTERT mRNA expression was more distinct in group B than that in other two groups (Figure 4).
Figure 3.
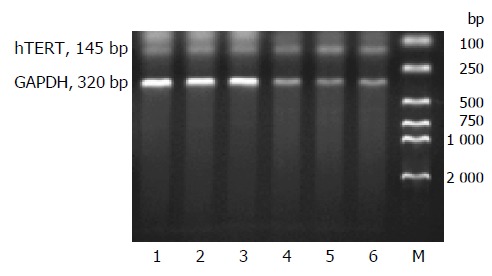
Analysis of hTERT mRNA expression by RT-PCR. RT-PCR was performed on total RNA extracted from both HepG2 (lanes 1-3) and QBC939 (lanes 4- 6) cell lines transfected with OPTI-MEM medium (lanes 1 and 4), pCMV-X (lanes 2 and 5) and pcDNA3 (lanes 3 and 6) respectively. M: DL 2000 marker.
Figure 4.
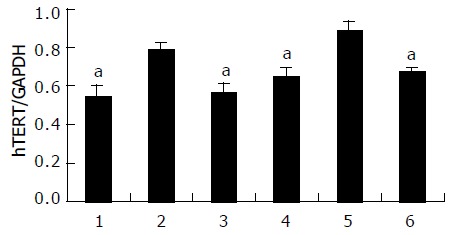
Relative quantitative changes of hTERT mRNA in HepG2 (lanes 1-3) and QBC939 (lanes 4-6) cell lines. A significant higher expression of hTERT mRNA was observed in both HepG2 and QBC939 cells after being transfected with pCMV-X vector (lanes 2 and 5) compared to those transfected with OPTI-MEM medium (lanes 1 and 4) and pcDNA3 (lanes 3 and 6). aP<0.05 vs others.
Western blotting analysis
The result of Western blot analysis showed that the expression of HBx protein could only be detected in HepG2 and QBC939 cells when transfected with HBx gene, but undetected in those when transfected with OPTI-MEM medium and blank vector (Figure 5).
Figure 5.
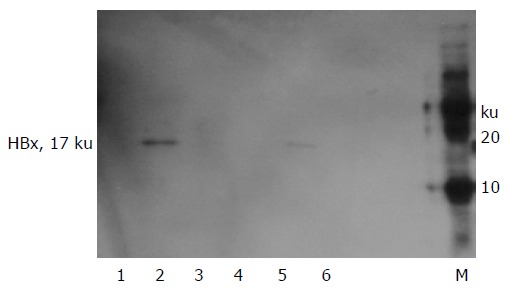
Western blot analysis of HBx protein expression in transfected HepG2 (lanes 1-3) and QBC939 (lanes 4 -6) cell lines. A marked expression of HBx protein was observed in HepG2 and QBC939 cells when transfected with pCMV -X vector (lanes 2 and 5), but there were no such protein expression in those when transfected with OPTI-MEM medium (lanes 1 and 4) and blank vector (lanes 3 and 6). M: Protein marker.
Immunocytochemical staining
A sensitive immunocytochemical staining was performed to identify whether there was HBx protein expression in transfected HepG2 and QBC939 cell lines. The results showed positive signals observed in cytoplasm of HepG2 (Figure 6) and QBC939 cells (Figure 7) in group B, while no such positive signals could be observed in HepG2 and QBC939 cells in groups A and C.
Figure 6.
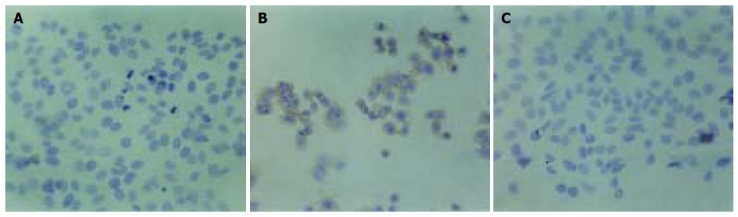
HBx protein expression in transfected HepG2 cells as assayed by UltraSensitive™ immunocytochemistry (SP×200). The cells transfected with OPTI-MEM medium (A) and blank vector (C) showed no expression of HBx protein, while some cells transfected with pCMV-X vector (B) showed visible brown positive signals.
Figure 7.
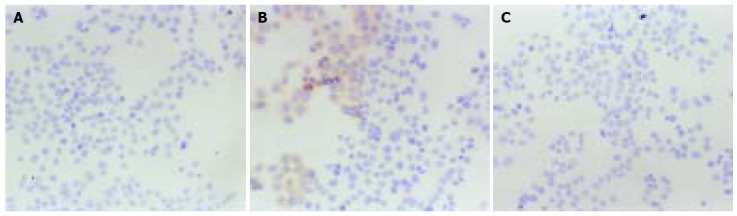
HBx protein expression in transfected QBC939 cells as detected by UltraSensitive™ immunocytochemistry (SP×200). The cells transfected with OPTI-MEM medium (A) and blank vector (C) showed no expression of HBx protein, but some cells transfected with pCMV-X vector (B) showed brown positive signals.
DISCUSSION
Telomerase/hTERT mechanisms have long been investigated for their potential role in cancer. Activation of telomerase is considered as the major episode for carcinogenesis[9] and over 85% of tumorigenic tissues and malignant cells express telomerase activity. hTERT is the rate-limiting determinant for regulating telomerase activity and also highly expressed in neoplastic tissues and tumor cells[10]. Most studies have shown that hTERT expression is usually correlated with telomerase activity and upregulation of hTERT expression is considered as the major step in carcinogenesis[11]. Multiple transcriptional factor binding sites are well identified in hTERT core promoter region and these regulatory factors make up a complex system of hTERT regulation[12]. The findings that HBV genome integration into upstream of hTERT core promoter region[13] and the cis -activation of hTERT gene transcription by HBV enhancer first elucidate a new mechanism for HBV in oncogenesis are the possible transcriptional regulation of hTERT gene[14].
HBV has been illustrated as the major causative factor for hepatocarcinogenesis[15]. Recent researches have also revealed that HBV infection might be associated with the development of cholangiocarcinoma. HBx protein, which is encoded by HBx gene, is considered as an essential oncogenic factor in oncogenesis of both human hepatocarcinomas and cholangiocarcinomas. HBx protein is a multifunctional viral oncoprotein and functions mainly as a transcriptional co- activator involving in multiple gene regulation and signaling pathways[16]. The integration of HBV genome into the hTERT promoter region[17] and upregulation of telomerase expression by HBx gene transfection[5] imply the possibility that HBx gene and protein may regulate hTERT gene transcription. In order to validate this surmise, we cultured HepG2 and QBC939 cells in vitro and transferred transiently the HBx gene into both the cell lines through lipid-mediated gene transduction technique. By observing the changes of hTERT mRNA expression, we attempted to elucidate the transcriptional regulation of hTERT gene by HBx gene. We observed that both HepG2 and QBC939 cell lines, when transfected with OPTI-MEM medium, could express hTERT mRNA quantitatively. Though there was no increase for hTERT mRNA expression in both cells when transfected with blank vector, a dramatic higher expression of hTERT mRNA was observed in both HepG2 and QBC939 cells when transfected with HBx gene. The results showed that HBx gene transfection could upregulate transcriptional expression of hTERT gene in both hepatocarcinomas and cholangiocarcinoma cells.
It is well known that some viruses contribute to carcinogenesis in two ways. One way is to transcriptionally activate proto-oncogenes of host cells by the integration of viral genome into the cellular genome and activation of the regulatory sequences (i.e., promoter or enhancer) of the viral genome[18]. This viral cis-activation mechanism can target the hTERT gene transcription[19]. Previous studies have found that HBV genome integrated in the hTERT promoter region and HBV enhancer could cis-activate the transcription of hTERT gene. Taken together with our present results, it is revealed that activation of telomerase by integration of HBV genome into telomerase gene is a new mechanism for oncogenic virus, HBV, in carcinogenesis. Another way is that some virus-encoded proteins contribute to cell transformation and carcinogenesis by modulating cellular signaling pathways that regulate cell-cycle progression, cell proliferation and differentiation, and cell apoptosis[20]. Recent evidence suggests that the activation of telomerase through transcriptional activation of the hTERT gene is another way in which human tumor-associated viruses can work[21]. Human papilloma viruses, especially its oncoproteins E6 and E7[22], which can abrogate the tumor suppressive function of p53 and Rb signaling pathways[23] and upregulate the hTERT promoter activity[24,25], play an important role in the development of cancers. The E-box elements within the hTERT promoter seem to be the target motifs responsible for the E6 activation of the hTERT transcription[26]. It is most likely that the aforementioned two ways are responsible for the upregulation of hTERT gene expression after HBx gene transfection into HepG2 and QBC939 cells. Although no direct evidence shows the existence of the integrated sites for HBx gene and the binding sites for HBx protein in hTERT promoter region, further investigations will identify these functional sites and validate the regulatory mechanisms eventually. HBx protein is a potential oncogenic factor and functions mainly as a transcriptional co-activator[16] involved in multiple gene regulation[27] and signaling pathway[28]. We suppose that upregulation of hTERT mRNA expression by HBx gene transfection is the result of the regulatory functions of binding motifs within the hTERT promoter for HBx protein or HBx-containing transcriptional complexes. But the exact mechanisms of these regulatory functions need to be elucidated through the study of transcription regulation for hTERT gene. Using immunocytochemical staining and Western blotting, we observed HBx protein expression only in HepG2 and QBC939 cells transfected with HBx gene. The expression of HBx protein in transfected cells indicates the achievement of transcription and duplication for HBx gene in some of these cells and meanwhile, demonstrates a key factor in regulating hTERT gene expression.
In conclusion, activation of telomerase and transcriptional regulation of hTERT gene involve in the development of carcinomas and act as the major targets for some tumor-associated viruses[19]. The transactivation of hTERT mRNA expression by HBx gene is a newfound mechanism for HBV in oncogenesis of hepatocarcinomas and cholangiocarcinomas. The elucidation of this mechanism may contribute to the deep understanding of the pathogenesis of HBV infection and subsequent formation of cancers.
ACKNOWLEDGMENTS
We thank Dr. Shu-Guang Wang for kindly providing us cholangiocarcinoma cell lines (QBC939) and Professor Xiao-Dong Zhang for providing us plasmids and technical assistance.
Footnotes
Science Editor Kumar M and Guo SY Language Editor Elsevier HK
References
- 1.Arbuthnot P, Kew M. Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol. 2001;82:77–100. doi: 10.1111/j.1365-2613.2001.iep0082-0077-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qu ZL, Zou SQ, Wei GH, Sun ZC, Wu XZ. In situ nucleic acid detection of HBV X gene in extrahepatic biliary tract carcinomas and its clinicopathological significance. Zhonghua WaiKe ZaZhi. 2004;42:88–91. [PubMed] [Google Scholar]
- 3.Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093–5107. doi: 10.1038/sj.onc.1206557. [DOI] [PubMed] [Google Scholar]
- 4.Qu ZL, Zou SQ, Zhen SL, Wu XZ, Cui NQ. The expression and significance of hepatitis B virus X protein in extrahepatic bile duct carcinomas and the surrounding non-cancerous tissues. Zhonghua Shiyan Waike Zazhi. 2002;19:401–402. [Google Scholar]
- 5.Zhou W, Shen Q, Gu B, Ren H, Zhang D. [Effects of hepatitis B virus X gene on apoptosis and the activity of telomerase in HepG(2) cells] Zhonghua GanZangBing ZaZhi. 2000;8:212–214. [PubMed] [Google Scholar]
- 6.Kyo S, Kanaya T, Takakura M, Tanaka M, Inoue M. Human telomerase reverse transcriptase as a critical determinant of telomerase activity in normal and malignant endometrial tissues. Int J Cancer. 1999;80:60–63. doi: 10.1002/(sici)1097-0215(19990105)80:1<60::aid-ijc12>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 7.Nagao K, Tomimatsu M, Endo H, Hisatomi H, Hikiji K. Telomerase reverse transcriptase mRNA expression and telomerase activity in hepatocellular carcinoma. J Gastroenterol. 1999;34:83–87. doi: 10.1007/s005350050220. [DOI] [PubMed] [Google Scholar]
- 8.Qu ZL, Zou SQ, Wang X. The effect of hepatitis B virus X gene transfection on expression of human telomerase reverse transcriptase mRNA in human bile duct carcinoma cell lines. Zhonghua WaiKe ZaZhi. 2004;42:1254–1257. [PubMed] [Google Scholar]
- 9.Guilleret I, Yan P, Guillou L, Braunschweig R, Coindre JM, Benhattar J. The human telomerase RNA gene (hTERC) is regulated during carcinogenesis but is not dependent on DNA methylation. Carcinogenesis. 2002;23:2025–2030. doi: 10.1093/carcin/23.12.2025. [DOI] [PubMed] [Google Scholar]
- 10.Dhaene K, Van Marck E, Parwaresch R. Telomeres, telomerase and cancer: an up-date. Virchows Arch. 2000;437:1–16. doi: 10.1007/s004280000189. [DOI] [PubMed] [Google Scholar]
- 11.Yan P, Saraga EP, Bouzourene H, Bosman FT, Benhattar J. Expression of telomerase genes correlates with telomerase activity in human colorectal carcinogenesis. J Pathol. 2001;193:21–26. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH728>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 12.Poole JC, Andrews LG, Tollefsbol TO. Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT) Gene. 2001;269:1–12. doi: 10.1016/s0378-1119(01)00440-1. [DOI] [PubMed] [Google Scholar]
- 13.Ferber MJ, Montoya DP, Yu C, Aderca I, McGee A, Thorland EC, Nagorney DM, Gostout BS, Burgart LJ, Boix L, et al. Integrations of the hepatitis B virus (HBV) and human papillomavirus (HPV) into the human telomerase reverse transcriptase (hTERT) gene in liver and cervical cancers. Oncogene. 2003;22:3813–3820. doi: 10.1038/sj.onc.1206528. [DOI] [PubMed] [Google Scholar]
- 14.Horikawa I, Barrett JC. cis-Activation of the human telomerase gene (hTERT) by the hepatitis B virus genome. J Natl Cancer Inst. 2001;93:1171–1173. doi: 10.1093/jnci/93.15.1171. [DOI] [PubMed] [Google Scholar]
- 15.Anzola M. Hepatocellular carcinoma: role of hepatitis B and hepatitis C viruses proteins in hepatocarcinogenesis. J Viral Hepat. 2004;11:383–393. doi: 10.1111/j.1365-2893.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- 16.Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol. 2001;36:651–660. doi: 10.1007/s005350170027. [DOI] [PubMed] [Google Scholar]
- 17.Horikawa I, Cable PL, Afshari C, Barrett JC. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 1999;59:826–830. [PubMed] [Google Scholar]
- 18.Van Tine BA, Kappes JC, Banerjee NS, Knops J, Lai L, Steenbergen RD, Meijer CL, Snijders PJ, Chatis P, Broker TR, et al. Clonal selection for transcriptionally active viral oncogenes during progression to cancer. J Virol. 2004;78:11172–11186. doi: 10.1128/JVI.78.20.11172-11186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horikawa I, Barrett JC. Transcriptional regulation of the telomerase hTERT gene as a target for cellular and viral oncogenic mechanisms. Carcinogenesis. 2003;24:1167–1176. doi: 10.1093/carcin/bgg085. [DOI] [PubMed] [Google Scholar]
- 20.Cheng Y. Human DNA oncogenic viruses and their transforming protein interactions with cell cycle control proteins. Chin Med J (Engl) 1999;112:291–296. [PubMed] [Google Scholar]
- 21.Veldman T, Horikawa I, Barrett JC, Schlegel R. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J Virol. 2001;75:4467–4472. doi: 10.1128/JVI.75.9.4467-4472.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al Moustafa AE, Foulkes WD, Benlimame N, Wong A, Yen L, Bergeron J, Batist G, Alpert L, Alaoui-Jamali MA. E6/E7 proteins of HPV type 16 and ErbB-2 cooperate to induce neoplastic transformation of primary normal oral epithelial cells. Oncogene. 2004;23:350–358. doi: 10.1038/sj.onc.1207148. [DOI] [PubMed] [Google Scholar]
- 23.Psyrri A, DeFilippis RA, Edwards AP, Yates KE, Manuelidis L, DiMaio D. Role of the retinoblastoma pathway in senescence triggered by repression of the human papillomavirus E7 protein in cervical carcinoma cells. Cancer Res. 2004;64:3079–3086. doi: 10.1158/0008-5472.can-03-3739. [DOI] [PubMed] [Google Scholar]
- 24.Jeong Seo E, Jung Kim H, Jae Lee C, Tae Kang H, Seong Hwang E. The role of HPV oncoproteins and cellular factors in maintenance of hTERT expression in cervical carcinoma cells. Gynecol Oncol. 2004;94:40–47. doi: 10.1016/j.ygyno.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 25.Veldman T, Horikawa I, Barrett JC, Schlegel R. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J Virol. 2001;75:4467–4472. doi: 10.1128/JVI.75.9.4467-4472.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gewin L, Galloway DA. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J Virol. 2001;75:7198–7201. doi: 10.1128/JVI.75.15.7198-7201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kekulé AS, Lauer U, Weiss L, Luber B, Hofschneider PH. Hepatitis B virus transactivator HBx uses a tumour promoter signalling pathway. Nature. 1993;361:742–745. doi: 10.1038/361742a0. [DOI] [PubMed] [Google Scholar]
- 28.Yun C, Cho H, Kim SJ, Lee JH, Park SY, Chan GK, Cho H. Mitotic aberration coupled with centrosome amplification is induced by hepatitis B virus X oncoprotein via the Ras-mitogen-activated protein/extracellular signal-regulated kinase-mitogen-activated protein pathway. Mol Cancer Res. 2004;2:159–169. [PubMed] [Google Scholar]


