Abstract
AIM: To investigate the biological function of F protein by yeast two-hybrid system.
METHODS: We constructed F protein bait plasmid by cloning the gene of F protein into pGBKT7, then recombinant plasmid DNA was transformed into yeast AH109 (a type). The transformed yeast AH109 was mated with yeast Y187 (α type) containing liver cDNA library plasmid in 2×YPDA medium. Diploid yeast was plated on synthetic dropout nutrient medium (SD/-Trp-Leu-His-Ade) containing X-α-gal for selection and screening. After extracting and sequencing plasmids from positive (blue) colonies, we underwent sequence analysis by bioinformatics.
RESULTS: Thirty-six colonies were selected and sequenced. Among them, 11 colonies were zymogen granule protein, 5 colonies were zinc finger protein, 4 colonies were zinc-α-2-glycoprotein, 1 colony was sialyltransferase, 1 colony was complement control protein factor I, 1 colony was vitronectin, and 2 colonies were new genes with unknown function.
CONCLUSION: The yeast two-hybrid system is an effective method for identifying hepatocyte proteins interacting with F protein of hepatitis C virus. F protein may bind to different proteins.
Keywords: Hepatitis C virus, F protein, Yeast two-hybrid system
INTRODUCTION
Hepatitis C virus (HCV) is the major etiologic agent of parenterally transmitted non-A, non-B hepatitis worldwide[1-3]. Most infected individuals are unable to eliminate the virus, resulting in a persistent infection in about 80% of cases. Chronically infected patients often develop progressive liver disease, cirrhosis, hepatic failure, and hepatocellular carcinoma (HCC)[4,5]. Because development of a robust cell culture system for HCV infection has remained elusive, little is known about HCV-host cell interactions and how they influence cell physiology or viral replication. There is no vaccine against HCV.
HCV, discovered by cDNA cloning in 1989, is a member of the Hepacivirus genus within the Flaviviridae family, and a 9.6-kb positive single-stranded RNA virus. The HCV genome carries a single open reading frame (ORF) flanked by nontranslated regions and encodes a single polyprotein of about 3 010 -3 033 amino acids. HCV polyprotein is cleaved by both host cell and viral proteases into at least 10 distinct structural and nonstructural protein products. The major structural proteins are a core (C) protein and two envelope proteins called E1, E2 and a short hydrophobic peptide p7. Four major nonstructural proteins called NS2, NS3, NS4, and NS5 are also generated, two of which, NS4 and NS5 are further processed into smaller polypeptides called NS4A, NS4B, NS5A, and NS5B. Most nonstructural proteins have enzymatic activities critical for viral replication. An additional HCV protein, F (for “frameshift protein”) or ARFP (for “alternate reading frame protein”), encoded by the ORF that overlaps the core gene in the +1 frame (core+1 ORF), is synthesized in vitro from the initiator codon of the polyprotein sequence followed by a +1 ribosomal frameshift operating in the region of core codons 8-14[6-9]. Antibodies to F protein have been detected in HCV patients, indicating the production of F protein during natural HCV infection[10,11]. The function of F protein in the life cycle of HCV remains unknown. To investigate the biological importance of F protein, we screened and identified the proteins interacting with HCV F protein by yeast two-hybrid system. One of the positive interacting proteins, named HCV F-interacting protein (HCV FBP2), was without known function.
MATERIALS AND METHODS
Bacterial yeast strains and plasmids
All yeast strains and plasmids for yeast two-hybrid experiments were obtained from Clontech Co. (Palo Alto, CA, USA) as components of the MATCHMAKER two-hybrid system 3. Yeast strain AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4△, gal80△, LYS2:GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3: MEL1UAS-MEL1TATA-LacZ) containing pGBKT7-53, coding for DNA-BD/mouse p53 fusing protein was used for cloning of bait plasmid. Yeast strain Y187 (MATa ura3-52, his3-200, Ade2-101, trp1-901, leu2-3, 112, gal4△ , gal80△ , met-, URA3 :GAL1UAS-GAL1TATA-lacZ MEL1) containing pTD1-1 in which pACT2 coding for AD/SV40 large T antigen fusing protein was used for cloning of library plasmids. Pretransformed human cDNA liver cell library Y187 and bacterial strain DH5a were used for cloning of each shuttle plasmid. Yeast-Escherichia coli shuttle plasmids pGBKT7 DNA-BD, pGADT7 AD, pGBKT7-53, pGADT7, pGBKT7 -Lam, pCL1 were from Clontech Co. (K1612-1). pGEM T vector was from Promega Co., USA.
Chemical agents and culture media
Taq DNA polymerase was purchased from MBI Co. T4 DNA ligase, EcoRI, and BamHI restriction endonuclease were from Takara Co. c- myc mAb secreted by 1-9E10.2 hybridoma (ATCC), goat anti-mouse IgG conjugated with horseradish peroxidase were from Zhongshan Co., China. Lithium acetate, semi- sulfate adenine, acrylamide and N, N’-bis-acrylamide were from Sigma Co., TEMED was from Boehringer Mannheim Co. Tryptone and yeast extracts were from OXOID Co. X-α-gal and cultural media YPDA, SD/ -Trp SD/-Leu, SD/-Trp/-Leu, SD/-Trp/-Leu/ -His, SD/-Trp/-Leu/-His/-Ade were from Clontech Co. Protein-G agarose was from Roche Co. pGEM-T vector was from Promega Co.
RT-PCR kit and TNT® coupled reticulocyte lysate systems were from Promega Co.[35] S-methionine (1 000 Ci/mmoL, 10 mCi/mL) was from Isotope Company of China. Amplification fluorographic reagent (#NAMP100) was from Amersham Life Sciences Co. Others were from Sigma Co.
Construction of “bait” plasmid and expression of HCV F protein
HCV- F sequences were generated by PCR amplification of HCV plasmid (HCV strain 1b) containing coding sequences for all the structural and nonstructural proteins. The sequences of the primers, containing the EcoRI and BamHI restriction enzyme sites, are sense primer: 5’-GAA TTC ATG GCA CGA ATC CTA AAC C-3’, and antisense primer: 5’-GGA TCC CTA CGC CGT CTT CCA GAA C- 3’. The PCR conditions were at 94°C for 30 s, at 60°C for 30 s, at 72°C for 30 s. Ten nanograms of PCR product was cloned with pGEM-T vector. The primary structure of insert was confirmed by direct sequencing. The fragment of encoding F was released from pGEM -T-F by digestion with EcoRI and BamHI, and ligated to pGBKT7. Vector pGBKT7 expressing proteins were fused with amino acids 1-147 of the GAL4 DNA binding domain (DNA-BD), pGADT7 expressing proteins were fused with amino acids 768-881 of the GAL4 activation domain (AD). Plasmid pGBKT7-F (Figure 1) containing full-length HCV F gene could directly express DNA binding domain, c-myc and F fusion protein. The plasmid was transformed into yeast strain AH109 by lithium acetate method[12]. Western blotting was performed to confirm the expression of the fusion protein using c-myc monoclonal antibody. Transformed AH109 (bait) was cultured on quadruple dropout media to exclude the auto-activation activity.
Figure 1.
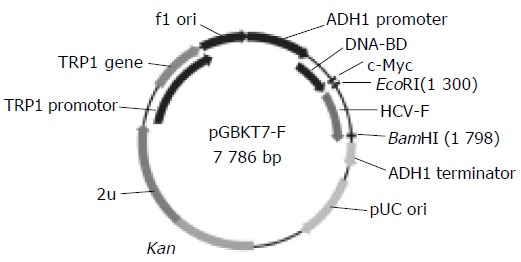
Map of “bait” plasmid pGBKT7-F.
Yeast two-hybrid library screening using yeast mating
One large (2-3 mm) fresh (<2 mo old) colony of AH109 [bait] was inoculated into 50 mL of SD/-Trp and incubated at 30°C overnight (16-24 h) with shaking at 250-270 r/min. Then the cells were spun down by centrifuging the entire 50 mL culture at 1 000 r/min for 5 min. After the supernatant was decanted, the cell pellet was resuspended in the residual liquid by vortexing. A human liver cDNA library was cloned into pACT2 and yeast reporter strain Y187 (Clontech Co., USA). The entire AH109 [bait] culture and 1 mL human liver cDNA library (1×106 cfu/mL) were combined and cultured in a 2-L sterile flask and 45 mL of 2×YPDA/Kan was added and swirled gently. After 20 h of mating, the cells were spun down and resuspended, and then spread on 50 large (150-mm) plates containing 100 mL of SD/- Ade/ -His/-Leu/-Trp (QDO). After growth for 6-15 d, the yeast colonies were transferred onto the plates containing X-α-gal to check for expression of the MEL1 reporter gene (blue colonies).
Plasmid isolation from yeast and transformation of E.coli with yeast plasmid
Approximately 1×106 colonies were screened and positive clones were identified. Yeast plasmid was isolated from positive yeast colonies by the lyticase method (Clontech Co.), and transformed into super-competence E.coli DH5α by chemical method. Transformants were plated on ampicillin SOB selection media and grown under selection. Subsequently, pACT2-cDNA constructs were re-isolated, and analyzed by restriction digests and sequencing.
Bioinformatic analysis
After the positive colonies were sequenced, the sequences were blasted with GenBank to analyze the function of the genes (http://www.ncbi.nlm.nih.gov.blast).
Cell culture and new gene cloning
The hepatoblastoma cell line HepG2 was propagated in DMEM supplemented with 10% FBS, 200 µmol/L L-glutamine, penicillin, and streptomycin. HepG2 cells were plated at a density of 1×106/well in 35-mm dishes. Total cellular RNA was isolated using TRIzol (Invitrogen Co., USA) according to the manufacturer’s instructions. cDNAs were reverse-transcribed from total RNA.
On the basis of liver cDNA library of genes of proteins interacting with HCV-F protein, the coding sequence of a new gene with unknown function, named HCV FBP2, was obtained by bioinformatics methods. The sequence of HCV FBP2 was deposited into GenBank, and the accession number was AY553876. The standard PCR cloning technique was used to amplify HCV FBP2 gene. Cytoplasmic RNA was isolated from HepG2 cells. RNA was used for RT-PCR. The primers were sense: 5’-GGA TTC ATG CCA TAT ATT CCT CTCA-3’, and antisense: 5’-GGA TCC CTA ATA CAA ATG TAT GTG G-3’ (HepG2 cDNA). The PCR conditions were at 94°C for 60 s, at 58°C for 60 s, at 72°C for 60 s, for 30 cycles. The PCR product was cloned with pGEM-T vector (Promega Co.). The primary structure of insert was confirmed by direct sequencing. The gene was ligated into yeast plasmids pGBKT7 and pGADT7.
Confirmation of true interaction in yeast
To confirm the true protein-protein interaction and exclude false positives, the plasmids of positive colonies were transformed into yeast strain Y187, then mating experiments were carried out by mating with yeast strain AH109 containing pGBKT7-F or pGBKT7-Lam. After mating, the diploid yeast was plated on SD/-Ade-His-Leu-Trp (QDO) covered by X-α-gal to test the specificity of interactions.
In vitro translation
Twenty-five microliters of mixture of TNT® reticulocytes, 2 µ L TNT ® reaction buffer, 1 µ L T7 TNT® RNA polymerase, 1 µL amino acid mixture (minus methionine, 1 mmol/L), 2 µL [35 S] -methionine, 1 µL RNasin RNase inhibitor (40 U/mL), 2 µL DNA template (pGBKT7 -F or pGADT7-library gene) (0.5 µg/mL), 16 µL ddH2O, were incubated at 30°C for 90 min.
Co-immunoprecipitation
Following reactants were combined in a 1.5-mL microcentrifuge tube on ice: 5 µL in vitro translated bait protein, 5 µL in vitro translated library protein. Only 10 µL pGBKT7-F plasmid was added into the control. The mixtures were incubated at 30°C for 1 h. Then, the following reagents were added into the reaction tubes: 470 µL co-immunoprecipitation buffer (20 mmol/L Tris-HCl [pH 7.5], 150 mmol/L NaCl, 1 mmol/L DTT, 5 µg/mL aprotinin, 0.5 mmol/L PMSF, 0.1% Tween 20), 10 µL protein-G agarose beads, 10 µL c-myc mAb, and incubated at 4°C for 2 h with continuous shaking. The tubes were centrifuged at 14 000 g for 1-2 min. The supernatants were removed, 0.5 mL TBST was added to the tubes. Rinse was repeated thrice, 15 µL SDS-loading buffer was added. The samples were heated at 80°C for 5 min. The tubes were placed on ice, and briefly centrifuged, and 10 µL was loaded onto an SDS-PAGE mini-gel for electrophoresis. After electrophoresis, the gel was transferred onto a tray containing gel fixation solution, and placed on a rotary shaker for 10 min at room temperature. The gel was rinsed with H2O, then amplification fluorographic reagent was added and shook for 20 min at room temperature, then dried at 80°C under constant vacuum. The gel was exposed to X-ray film overnight at room temperature. The film was developed by standard techniques.
RESULTS
Identification of recombinant plasmid
The full-length sequences of HCV F were generated by PCR amplification of HCV plasmid (HCV strain 1b), and a 840-bp fragment of HCV FBP2 was amplified by RT-PCR after total RNA was isolated from HepG2 cells, sequenced and analyzed by comparing to Vector NTI 6 and BLAST database homology search (http://www.ncbi.nlm.nih.gov/ blast) . After being cut by EcoRI/BamHI, the fragments were in-frame ligated respectively into pGBKT7 and pGADT7 EcoRI/BamHI sites. Restriction enzyme analysis of pGBKT7 -F, pGBKT7- FBP2, and pGADT7-FBP2 plasmids with EcoRI/BamHI yielded two bands respectively: 7 300 bp empty pGBKT7 and 486 bp HCV F, 7 300 bp empty pGBKT7 and 840 bp HCV FBP2, 7 900 bp empty pGADT7 and 840 bp HCV FBP2. The products of plasmid were amplified by PCR. Analysis of the PCR products by agarose gel electrophoresis showed the clear bands with the expected size (486 bp of F, 840 bp of FBP2). Sequences of the PCR products were correct (Figures 2A-D).
Figure 2.
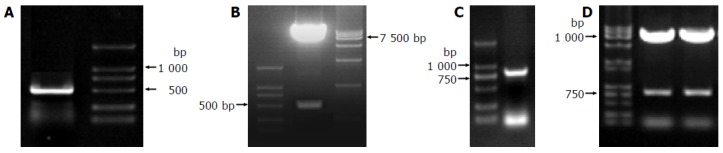
Sequences of PCR products A: a 486-bp fragment-F amplified by amplified by RT-PCR; D1: pGBKT7-FBP2; D2: pGADT7-FBP2 cut by EcoRI/RT-PCR; B: pGBKT7-F cut by EcoRI/BamHI; C: a 840-bp fragment-FBP2 BamHI.
Expression of “bait” fusion protein
Yeast strain AH109 transformed with pGBKT7 -F and pGBKT7-FBP2 stably expressed the fusion protein at high level (Figure 3) and only grew on SD/-Trp medium but not on QDO medium. Thus, the transformed yeast could be used for yeast hybrid analysis.
Figure 3.
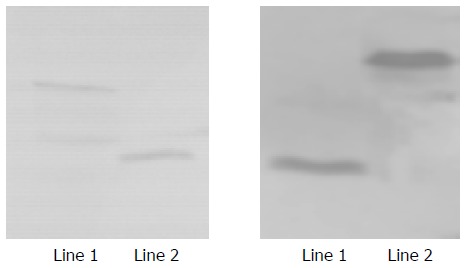
Expression of HCV F and HCV FBP2 proteins in yeast. Lane 1: HCV F protein; lane 2: positive control; lane 3: HCV FBP2 protein.
Screening of liver cell cDNA library
We isolated plasmids from the blue colonies containing only pGBKT7-F and one library plasmid other than other plasmids. Because plasmid pACT2- cDNA contains two restriction endonuclease sites of BglII, the gene fragments of the liver cell cDNA library (pACT2 - cDNA) were released by Bgl II digestion (Figure 4). The gene fragments with different lengths proved that these screened clones were positive colonies but not false positive colonies growing on SD/-Trp/-Leu/-His/-Ade culture medium after mating.
Figure 4.
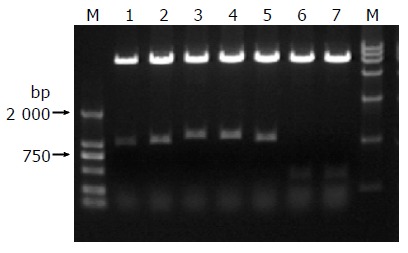
Identification of different colonies by BglII digestion.
Analysis of cDNA sequence and homology
A total of 36 positive colonies were grown on the selective SD/-trp-leu-his-ade/X-α-gal medium. These colonies were prescreened by BglII digesting to make sure that only colonies with different inserts were subjected to sequencing. Thirty -six colonies from cDNA library were sequenced. Using the BLAST program from the National Center for Biotechnology Information, two of the sequences were unknown genes and one of them was named as HCV FBP2. The full -length sequences accepted by GenBank were obtained by bioinformatics method. The other 34 sequences had a high similarity to known genes. The data are presented in Table 1.
Table 1.
Comparison between positive clones and similar sequences in GenBank
| High similarity to known genes | Similar | Homolog |
| number | (%) | |
| Homo sapiens zymogen granule protein 16 | 11 | 99 |
| Homo sapiens zinc-alpha-2-glycoprotein | 4 | 99 |
| Highly similar to Homo sapiens SL15 protein | 5 | 100 |
| Homo sapiens RAB14, member RAS oncogene family | 1 | 98 |
| Human mRNA for complement control protein factor I | 1 | 98 |
| Homo sapiens angiotensinogen (serine or cysteine) | 1 | 97–99 |
| proteinase inhibitor, clade A (alpha-1 antiproteinase, | ||
| antitrypsin), member 8) | ||
| Homo sapiens sialyltransferase 1 | 1 | 97 |
| Similar to Homo sapiens zinc finger protein 83 | 5 | 100 |
| Homo sapiens mRNA for peroxisomal ion protease | 2 | 100 |
| Homo sapiens cathepsin B mRNA | 1 | 99 |
| Homo sapiens vitronectin (serum spreading factor, | 1 | 100 |
| somatomedin B, complement S-protein) | ||
| Homo sapiens serine (or cysteine) proteinase inhibitor, | 1 | 99 |
| clade G (C1 inhibitor), member1 | ||
| Homo sapiens genes of unknown functions | 2 |
Back-across testing
We confirmed the protein-protein interaction between HCV F and a new gene named as HCV FBP2 by back-across in yeast. Expressive vector pGADT7 -HCV FBP2 was constructed and transformed into yeast Y187. This Y187 containing pGADT7-HCV FBP2 was mated with the yeast AH109 containing pGBKT7-F. The mated yeast was plated on the dropout medium. The positive colonies emerged and became blue.
In vitro co-immunoprecipitation
HCV F protein containing 161 aa, Mr = 17 710, being smaller than FBP2 containing 280 aa, Mr = 30 800 is shown in Figure 5.
Figure 5.
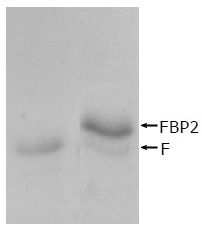
Interaction between HCV F protein and FBP2 protein identified by co-immunoprecipitation. Lane 1: HCV F protein; lane 2: interaction between HCV F and FBP2 proteins.
DISCUSSION
The core protein of HCV is a multifunctional protein[13-15]. There is evidence that this protein can directly modulate intracellular signal transduction, gene transcription, cell proliferation, and cell death, interfere with lipid metabolism and suppress host immune responses[16-19]. HCV F protein is a newly discovered HCV gene product encoded by the ORF that overlaps the core gene in the +1 frame[6-8]. F protein might have previously escaped attention because it possesses the immunodominant epitopes located in the N-terminus, in common with the core protein. A F protein analog synthesized in vitro reacts with the sera of HCV patients but not with the sera of hepatitis B patients, indicating that F protein is expressed during natural HCV infection. F protein is a very unstable protein with a half-life of <10 min in Huh7 hepatoma cells and in vitro, and is mediated by the proteasome pathway[20,21]. Immunofluorescence staining and subcellular fractionation experiments indicate that the subcellular localization of F protein is in the endoplasmic reticulum, and is similar to those of HCV core and NS5A proteins, suggesting that F protein may participate in HCV morphogenesis or replication. F protein shows more sequence diversity than the core protein[22-25]. Mutations of the core protein help virus to escape host immunologic reaction and may trigger cellular malignancy conversion. F protein might regulate cellular functions that are important for the viral life cycle or might play a role in viral morphogenesis or viral entry.
Because proteins often assemble into large complexes to perform different activities, characterization of the interaction pattern of a protein could contribute to the elucidation of the functions of protein[26]. Interaction between viral and hepatocellular proteins plays an important role in the pathogenesis of the virus and may mediate virus to enter hepatocytes. Their network interaction can change normal biological functions of proteins, influence self-replication of virus, prolong infection and develop diseases. Yeast two-hybrid 3 is an effective gene analysis method which can analyze interactions between protein and protein, protein and DNA, protein and RNA in eukaryotic cells, as well as a new genetics technique for studying interactions of proteins in physiologic condition in vivo.
Yeast two-hybrid system 3 based on the system originally designed by Fields and Song[27] takes the advantage of the properties of GAL4 protein of the yeast Saccharomyces cerevisiae. GAL4-yeast two-hybrid assay uses two expression vectors, one uses GAL4-DNA-binding domain (DBD) and the other uses GAL4-activation domain (AD) . The GAL4-DBD is fused to protein ‘X’ and GAL4-AD is fused to protein ‘Y’ to form the bait and the target of the interaction trap, respectively. A selection of host cells with different reporter genes and different growth selection markers can detect and confirm protein-protein interactions and has fewer false positives[27-30].
In this study, the “bait” plasmid pGBKT7-F was transformed into yeast strain AH109. HCV F gene was expressed in yeast cells. After the “bait” plasmid pGBKT7-F yeast strain AH109 was mated with liver cDNA library yeast strain Y187, the diploid yeast cells were plated on QDO media containing X-α-gal, 36 true positives were obtained. By sequencing analysis of isolated library plasmids, we got the sequences of 34 genes with known functions and the sequences of two genes with unknown function, one of them was named as HCV FBP2. In order to further confirm the interaction between expressed protein and HCV F protein, we performed the experiment of co-immunoprecipitation of both proteins. A strong interaction between HCV F protein and HCV FBP2 protein in vitro was observed.
We screened Homo sapiens zymogen granule (ZG) protein 16 (ZG16p) interacting with F protein from liver cDNA library. ZG protein is highly expressed in the pancreas cells and regulates ATP-sensitive K(+) conductance of ZGs. ZG16p is one of the linker proteins with a submembranous protein matrix on the inner surface of ZG. The submembranous matrix composed of sulfated proteoglycans, glycoproteins and lectin ZG16p is essential for the binding and sorting of aggregated zymogens during granule formation. ZG16p is associated with lipid microdomains (‘rafts’) and affects the protein synthesis and intracellular transport of secretory proteins and maturation of zymogens in ZGs[31-34]. HCV infection and chronic hepatopathy caused by HCV are significantly associated with diabetes, and reduction of glucose tolerance is related to the severity of chronic hepatitis C[35,36]. The result of our experiment showed that interaction occurred between HCV F and ZG16p, thus providing a new clue for revealing the function of HCV F protein and relationship between pathogenesis of HCV and diabetes mellitus.
Another important protein interacting with F protein from liver cDNA library is the Homo sapiens zinc- alpha 2-glycoprotein (Zn-α 2-GP). It is present at a high concentration in the seminal plasma and in other human body fluids. Hepatocytes are a source of protein in blood plasma and epithelia. Zn-α2-GP is present at carcinoma of prostate, breast cancer and hydroadenoma. Zn-α2-GP production is associated with tumor differentiation, decreased or absent Zn-α2-GP production is found in poorly differentiated (high Gleason grade) tumors. Zn-α2-GP production in tumors can elevate the systemic ZAG concentration and induce lipolysis leading to cachexia. Zn-α2-GP is a potential serum marker of prostate cancer that may elevate early in tumor development. Whether tumors can induce secretion of endogenous Zn-α2-GP by normal prostatic or other secretory ZAG expression can predict the risk of a progressive disease needs further study[37].
Vitronectin, also known as a serum spreading factor, is a glycoprotein present in both human plasma and serum. It is also present in different loose connective tissues, often in co-localization of elastic fibrils. Vitronectin binds to cells through interaction of the Arg-Gly-Asp (RGD) sequence in its cell-binding domain with vitronectin-specific cell surface receptors and may modulate blood coagulation and complement-induced cytolysis. Matrix-bound vitronectin plays a role in regulating pericellular proteolysis by binding to plasminogen activator inhibitor type I. Vitronectin may promote cell attachment, spreading, proliferation, and differentiation of normal and neoplastic cells[38,39].
C1 inhibitor (C1 INH) is a highly glucosylated protein. The content of glucose is up to 35-49%. It has 30% homology of amino acids of serine proteinase inhibitor superfamily members including α1 anti-trypsin, α1 anti-chymotrypsin and anti-thrombin III. C1 INH combines activated C1r or C1s to form a stable complex, leading to inactivation of serine proteinase C1. C1 INH may prevent auto-activation of C1. Because C1 INH still inhibits factors XIIa, XIa, kallikrein and fibrinolysin, it plays an important role in regulation of blood coagulation, kinin, and fibrinolytic system[40,41].
Sialyltransferase is a member of glycosyltransferase family and presents at Golgi apparatus. Sialyltransferase possesses the functions of catalyzing cellular adhesive molecule-ganglioside polysialic acidification. Ganglioside, a kind of glycosphingolipid containing sialic acid, is an important factor for cellular adhesion and malignancy cell growth in vitro culture.
HCV F protein also interacts with complement control protein factor I, peroxisomal ion protease, cathepsin B, RAS oncogene. These interacting proteins are closely correlated with carbohydrate metabolism, immunoregulation, occurrence and development of tumor. How interactions between HCV F protein and interacting proteins affect the occurrence and development of chronic hepatitis C, hepatic fibrosis and hepatocarcinoma, needs to be further elucidated.
Footnotes
Science Editor Wang XL Language Editor Elsevier HK
Supported by the National Natural Science Foundation of China, Nos. C03011402 and C30070690; and the Research and Technique Foundation of PLA during the 9th-Five year plan period, No. 98D063; and the Launching Foundation for Student Studying Abroad of PLA, No. 98H038; and the Youth Research and Technique Foundation of PLA during the 10th-Five Year Plan Period, No. 01Q138; and the Research and Technique Foundation of PLA during the 10th-Five Year Plan Period, No. 01MB135
References
- 1.Yokoyama Y, Kuramitsu Y, Takashima M, Iizuka N, Toda T, Terai S, Sakaida I, Oka M, Nakamura K, Okita K. Proteomic profiling of proteins decreased in hepatocellular carcinoma from patients infected with hepatitis C virus. Proteomics. 2004;4:2111–2116. doi: 10.1002/pmic.200300712. [DOI] [PubMed] [Google Scholar]
- 2.Miller RH, Purcell RH. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci USA. 1990;87:2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmonds P, Smith DB, McOmish F, Yap PL, Kolberg J, Urdea MS, Holmes EC. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J Gen Virol. 1994;75(Pt 5):1053–1061. doi: 10.1099/0022-1317-75-5-1053. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien TR, Kirk G, Zhang M. Hepatocellular carcinoma: paradigm of preventive oncology. Cancer J. 2004;10:67–73. doi: 10.1097/00130404-200403000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka J, Yoshizawa H. A national project for the management of viral hepatitis toward prevention of hepatocellular carcinoma in Japan. Gan To Kagaku Ryoho. 2004;31:864–870. [PubMed] [Google Scholar]
- 6.Varaklioti A, Vassilaki N, Georgopoulou U, Mavromara P. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J Biol Chem. 2002;277:17713–17721. doi: 10.1074/jbc.M201722200. [DOI] [PubMed] [Google Scholar]
- 7.Walewski JL, Keller TR, Stump DD, Branch AD. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA. 2001;7:710–721. doi: 10.1017/s1355838201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Z, Choi J, Yen TS, Lu W, Strohecker A, Govindarajan S, Chien D, Selby MJ, Ou J. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 2001;20:3840–3848. doi: 10.1093/emboj/20.14.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulant S, Becchi M, Penin F, Lavergne JP. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J Biol Chem. 2003;278:45785–45792. doi: 10.1074/jbc.M307174200. [DOI] [PubMed] [Google Scholar]
- 10.Roussel J, Pillez A, Montpellier C, Duverlie G, Cahour A, Dubuisson J, Wychowski C. Characterization of the expression of the hepatitis C virus F protein. J Gen Virol. 2003;84:1751–1759. doi: 10.1099/vir.0.19065-0. [DOI] [PubMed] [Google Scholar]
- 11.Choi J, Xu Z, Ou JH. Triple decoding of hepatitis C virus RNA by programmed translational frameshifting. Mol Cell Biol. 2003;23:1489–1497. doi: 10.1128/MCB.23.5.1489-1497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto M, Hsieh TY, Zhu N, VanArsdale T, Hwang SB, Jeng KS, Gorbalenya AE, Lo SY, Ou JH, Ware CF, et al. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-beta receptor. J Virol. 1997;71:1301–1309. doi: 10.1128/jvi.71.2.1301-1309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M, Liu Y, Cheng J, Zhang SL, Wang L, Shao Q, Zhang J, Yang Q. Transactivating effect of hepatitis C virus core protein: a suppression subtractive hybridization study. World J Gastroenterol. 2004;10:1746–1749. doi: 10.3748/wjg.v10.i12.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamoto K, Moriishi K, Miyamura T, Matsuura Y. Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein. J Virol. 2004;78:6370–6380. doi: 10.1128/JVI.78.12.6370-6380.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majeau N, Gagné V, Boivin A, Bolduc M, Majeau JA, Ouellet D, Leclerc D. The N-terminal half of the core protein of hepatitis C virus is sufficient for nucleocapsid formation. J Gen Virol. 2004;85:971–981. doi: 10.1099/vir.0.79775-0. [DOI] [PubMed] [Google Scholar]
- 16.Carlos MP, Yamamura Y, Vu Q, Conzen K, Anderson DE, Torres JV. Humoral immunity to immunodominant epitopes of Hepatitis C virus in individuals infected with genotypes 1a or 1b. Clin Immunol. 2004;111:22–27. doi: 10.1016/j.clim.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel M, Watowich SJ. Biophysical characterization of hepatitis C virus core protein: implications for interactions within the virus and host. FEBS Lett. 2004;557:174–180. doi: 10.1016/s0014-5793(03)01486-8. [DOI] [PubMed] [Google Scholar]
- 18.Hahn CS, Cho YG, Kang BS, Lester IM, Hahn YS. The HCV core protein acts as a positive regulator of fas-mediated apoptosis in a human lymphoblastoid T cell line. Virology. 2000;276:127–137. doi: 10.1006/viro.2000.0541. [DOI] [PubMed] [Google Scholar]
- 19.Sabile A, Perlemuter G, Bono F, Kohara K, Demaugre F, Kohara M, Matsuura Y, Miyamura T, Bréchot C, Barba G. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology. 1999;30:1064–1076. doi: 10.1002/hep.510300429. [DOI] [PubMed] [Google Scholar]
- 20.Ogata S, Nagano-Fujii M, Ku Y, Yoon S, Hotta H. Comparative sequence analysis of the core protein and its frameshift product, the F protein, of hepatitis C virus subtype 1b strains obtained from patients with and without hepatocellular carcinoma. J Clin Microbiol. 2002;40:3625–3630. doi: 10.1128/JCM.40.10.3625-3630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z, Choi J, Lu W, Ou JH. Hepatitis C virus f protein is a short-lived protein associated with the endoplasmic reticulum. J Virol. 2003;77:1578–1583. doi: 10.1128/JVI.77.2.1578-1583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brass V, Bieck E, Montserret R, Wölk B, Hellings JA, Blum HE, Penin F, Moradpour D. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J Biol Chem. 2002;277:8130–8139. doi: 10.1074/jbc.M111289200. [DOI] [PubMed] [Google Scholar]
- 23.Lanford RE, Notvall L, Chavez D, White R, Frenzel G, Simonsen C, Kim J. Analysis of hepatitis C virus capsid, E1, and E2/NS1 proteins expressed in insect cells. Virology. 1993;197:225–235. doi: 10.1006/viro.1993.1583. [DOI] [PubMed] [Google Scholar]
- 24.McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002;21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi ST, Polyak SJ, Tu H, Taylor DR, Gretch DR, Lai MM. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology. 2002;292:198–210. doi: 10.1006/viro.2001.1225. [DOI] [PubMed] [Google Scholar]
- 26.Emmert-Buck MR, Gillespie JW, Paweletz CP, Ornstein DK, Basrur V, Appella E, Wang QH, Huang J, Hu N, Taylor P, et al. An approach to proteomic analysis of human tumors. Mol Carcinog. 2000;27:158–165. [PubMed] [Google Scholar]
- 27.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 28.Osman A. Yeast two-hybrid assay for studying protein-protein interactions. Methods Mol Biol. 2004;270:403–422. doi: 10.1385/1-59259-793-9:403. [DOI] [PubMed] [Google Scholar]
- 29.Gietz RD, Woods RA. Screening for protein-protein interactions in the yeast two-hybrid system. Methods Mol Biol. 2002;185:471–486. doi: 10.1385/1-59259-241-4:471. [DOI] [PubMed] [Google Scholar]
- 30.Zhen Z. Progress in proteomics. Sheng Wu Gong Cheng Xue Bao. 2001;17:491–493. [PubMed] [Google Scholar]
- 31.Kalus I, Hodel A, Koch A, Kleene R, Edwardson JM, Schrader M. Interaction of syncollin with GP-2, the major membrane protein of pancreatic zymogen granules, and association with lipid microdomains. Biochem J. 2002;362:433–442. doi: 10.1042/0264-6021:3620433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodel A, An SJ, Hansen NJ, Lawrence J, Wäsle B, Schrader M, Edwardson JM. Cholesterol-dependent interaction of syncollin with the membrane of the pancreatic zymogen granule. Biochem J. 2001;356:843–850. doi: 10.1042/0264-6021:3560843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt K, Schrader M, Kern HF, Kleene R. Regulated apical secretion of zymogens in rat pancreas. Involvement of the glycosylphosphatidylinositol-anchored glycoprotein GP-2, the lectin ZG16p, and cholesterol-glycosphingolipid-enriched microdomains. J Biol Chem. 2001;276:14315–14323. doi: 10.1074/jbc.M006221200. [DOI] [PubMed] [Google Scholar]
- 34.Kleene R, Dartsch H, Kern HF. The secretory lectin ZG16p mediates sorting of enzyme proteins to the zymogen granule membrane in pancreatic acinar cells. Eur J Cell Biol. 1999;78:79–90. doi: 10.1016/S0171-9335(99)80009-0. [DOI] [PubMed] [Google Scholar]
- 35.Arao M, Murase K, Kusakabe A, Yoshioka K, Fukuzawa Y, Ishikawa T, Tagaya T, Yamanouchi K, Ichimiya H, Sameshima Y, et al. Prevalence of diabetes mellitus in Japanese patients infected chronically with hepatitis C virus. J Gastroenterol. 2003;38:355–360. doi: 10.1007/s005350300063. [DOI] [PubMed] [Google Scholar]
- 36.Paraschiv C, Graur M, Butnariu G, Mihai B, Constantinescu D. [The pathological mechanisms of glycoregulation disturbances in chronic hepatitis B and C] Rev Med Chir Soc Med Nat Iasi. 2002;107:294–297. [PubMed] [Google Scholar]
- 37.Hale LP, Price DT, Sanchez LM, Demark-Wahnefried W, Madden JF. Zinc alpha-2-glycoprotein is expressed by malignant prostatic epithelium and may serve as a potential serum marker for prostate cancer. Clin Cancer Res. 2001;7:846–853. [PubMed] [Google Scholar]
- 38.Stawowy P, Kallisch H, Veinot JP, Kilimnik A, Prichett W, Goetze S, Seidah NG, Chrétien M, Fleck E, Graf K. Endoproteolytic activation of alpha(v) integrin by proprotein convertase PC5 is required for vascular smooth muscle cell adhesion to vitronectin and integrin-dependent signaling. Circulation. 2004;109:770–776. doi: 10.1161/01.CIR.0000112583.50762.DE. [DOI] [PubMed] [Google Scholar]
- 39.Faucheux N, Schweiss R, Lützow K, Werner C, Groth T. Self-assembled monolayers with different terminating groups as model substrates for cell adhesion studies. Biomaterials. 2004;25:2721–2730. doi: 10.1016/j.biomaterials.2003.09.069. [DOI] [PubMed] [Google Scholar]
- 40.Sim RB, Tsiftsoglou SA. Proteases of the complement system. Biochem Soc Trans. 2004;32:21–27. doi: 10.1042/bst0320021. [DOI] [PubMed] [Google Scholar]
- 41.Toomayan GA, Chen LE, Jiang HX, Qi WN, Seaber AV, Frank MM, Urbaniak JR. C1-esterase inhibitor and a novel peptide inhibitor improve contractile function in reperfused skeletal muscle. Microsurgery. 2003;23:561–567. doi: 10.1002/micr.10210. [DOI] [PubMed] [Google Scholar]


