Abstract
Limb girdle muscular dystrophy 2H is caused by mutations in the gene encoding the E3 ubiquitin ligase, TRIM32. Previously, we generated and characterized a Trim32 knockout mouse (T32KO) that displays both neurogenic and myopathic features. The myopathy in these mice is attributable to impaired muscle growth, associated with satellite cell senescence and premature sarcopenia. This satellite cell senescence is due to accumulation of the SUMO ligase PIASy, a substrate of TRIM32. The goal of this investigation was to identify additional substrates of TRIM32 using 2D fluorescence difference gel electrophoresis (2D-DIGE) in order to further explore its role in skeletal muscle. Because TRIM32 is an E3 ubiquitin ligase, we reasoned that TRIM32's substrates would accumulate in its absence. 2D-DIGE identified 19 proteins that accumulate in muscles from the T32KO mouse. We focused on two of these proteins, NDRG2 and TRIM72, due to their putative roles in myoblast proliferation and myogenesis. Follow-up analysis confirmed that both proteins were ubiquitinated by TRIM32 in vitro; however, only NDRG2 accumulated in skeletal muscle and myoblasts in the absence of TRIM32. NDRG2 overexpression in myoblasts led to reduced cell proliferation and delayed cell cycle withdrawal during differentiation. Thus, we identified NDRG2 as a novel target for TRIM32; these findings further corroborate the hypothesis that TRIM32 is involved in control of myogenic cells proliferation and differentiation.
Introduction
Limb girdle muscular dystrophies (LGMDs) are hereditary muscle-wasting disorders involving muscles of the pelvis and shoulder girdle. LGMD type 2H (LGMD2H) is a mild autosomal-recessive muscular dystrophy with highly variable phenotypes that range from asymptomatic to wheelchair bound. Facial weakness, scapular winging, hypertrophied calves and Achilles tendon contractions are commonly observed in LGMD2H patients (1). The age of onset ranges between the first and fourth decades. LGMD2H was originally mapped to a specific mutation (D489N) in the TRIM32 gene although other mutations in the C terminal region have now been identified (2). TRIM32 is an E3 ubiquitin ligase, containing the tripartite motif (RING finger, B-box, coiled-coil), common to all TRIM family members (3). Currently, seven mutations in the TRIM32 gene have been linked to LGMD2H, i.e. two missense mutations [c.1459G>A (p.D487N), c.1180G>A (p.R394H) (4)], one codon deletion [c.1761–1763delGAT (p.D588 del) (4)], three frameshift mutations [c.1559delC (p.T520TfsX13) (4), c.1753-1766dup (p.I590LfsX38) (5), c.1560delC (p.C521VfsX13) (6)] and one intragenic deletion that removes the entire open reading frame [del 30 586 bp + insert 2 bp (6)]. The first-described LGMD2H missense mutation (p.D487N) (1) also causes sarcotubular myopathy (STM), an allelic disorder that is characterized by a more severe muscular dystrophy phenotype than LGMD2H (7). Interestingly, six of the LGMD2H mutations are clustered in the conserved C-terminal NHL domain of TRIM32 (NHL domain was named after proteins NCL-1, HT2A and Lin-41 that contain repeats folded into a six-bladed β propeller). Using molecular modeling, it has been predicted that at least some of these mutations in the C-terminus might cause conformational changes that could impact protein–protein interactions and homodimerization (4) and therefore impair normal biological functions of TRIM32.
Besides muscular dystrophy, some mutations in TRIM32 cause an autosomal-recessive multisystemic disorder called Bardet-Biedl syndrome (BBS). BBS is due to a missense mutation P130S in the BBOX zinc-finger domain of TRIM32 (8). This disorder is associated with obesity, retinopathy, diabetes, polydactyly, renal abnormalities, learning disability and hypogenitalism. Intriguingly, no muscle abnormalities are associated with BBS, suggesting that BBS and LGMD2H mutations disrupt different biological functions of TRIM32.
Previously, we created genetically modified mice lacking TRIM32 (Trim32 knockout mouse, T32KO) and observed that they display both myopathic and neurogenic phenotypes (9). The muscles of these mice demonstrated a mild muscular dystrophy and displayed dystrophic features similar to those in patients with the muscular disorders LGMD2H and STM. T32KO mice showed a decrease in the concentration of neurofilament proteins in the brain and a reduced motor axon diameter. These axonal changes led to a shift toward a slower motor unit type and concomitant reduction in fast myosin in T32KO soleus muscle (9). In accordance with our findings, neurogenic features were also evident in LGMD2H patients, where a slight dominance of type I muscle fibers, decreased motor and sensory nerve conduction velocities and myopathic and neurogenic electromyography abnormalities in the leg muscles were observed (4,6).
Although TRIM32 appears to be expressed in numerous tissues, it is unclear why C-terminal TRIM32 mutations lead to a muscle phenotype, while those in the BBOX lead to BBS. Initial investigations focused on a role for TRIM32 as a regulator of muscle atrophy, because another TRIM family member (MURF1) induces muscle atrophy through turnover of myofibrillar proteins, following denervation or fasting. By using the siRNA approach, it was demonstrated that TRIM32 is involved in muscle wasting specifically by controlling turnover of thin filaments (10). We also explored a role for TRIM32 in muscle atrophy using T32KO mice and found that TRIM32 is dispensable for muscle atrophy (11). Furthermore, we showed that increased expression of TRIM32 mainly occurs during muscle growth, as opposed to atrophy, suggesting that it is rather a positive regulator of muscle growth. In mature adult muscle, TRIM32 expression is very low and appears to be primarily restricted to satellite cells (11,12). We found that TRIM32 plays a role in regeneration by affecting satellite cell cycle progression via modulation of the SUMO ligase PIASy (PIAS4). Impaired degradation of PIASy leads to increased SUMOylation of cellular proteins and premature senescence of T32KO myoblasts (11).
In the current investigation, we used 2D fluorescence difference gel electrophoresis (2D-DIGE) followed by LC–MS-MS to identify novel substrates of TRIM32 in skeletal muscle. We searched for proteins in muscle extracts that accumulated in the absence of TRIM32, reasoning that loss of ubiquitination would lead to accumulation of TRIM32's substrates. Although 19 proteins were identified, we focused on two of them, NDRG2 (N-myc downstream-regulated gene) and TRIM72 (an E3 ubiquitin ligase aka mitsugumin 53, MG53), because of their previously identified roles in myoblast proliferation, myogenesis and membrane repair (13–15). We show that NDRG2 is a bona fide substrate of TRIM32 in vivo. Accumulation of NDRG2 due to loss of TRIM32 in T32KO muscle is associated with decreased myoblast proliferation and delayed cell cycle withdrawal during myogenic differentiation. Thus, NDRG2 is a novel substrate of TRIM32, accumulation of which is deleterious to muscle satellite cell function.
Results
2D-DIGE and LC–MS-MS revealed 37 proteins with altered abundance in T32KO muscles
In this study, we examined proteins from the cytosolic, membrane and myofibrillar fractions of gastrocnemius muscles isolated from T32KO and WT mice using high-resolution 2D fluorescence difference gel electrophoresis (2D-DIGE) and LC–MS-MS. The reproducibility of 2-DIGE and homogeneity between the samples were ensured by comparing samples from five T32KO and five WT animals. All samples were prepared and run under identical conditions.
Comparison between T32KO and WT muscle extracts revealed 36 proteins with altered abundance in T32KO muscle. Furthermore, 19 of the identified proteins demonstrated increased abundance and 17 proteins demonstrated decreased abundance. All proteins identified in this study are listed in the Supplementary Material, Table S1.
Of the identified proteins, many were associated with energy metabolism, transcription, translation, signaling and the cellular stress response. Mitochondrial aconitate hydratase, cytoplasmic carbonic anhydrase 3, ferritin light chain, myoglobin, probable C->U editing enzyme APOBEC-2 and N-myc down-regulated protein 2 (NDRG2) were identified among the significantly increased proteins in T32KO muscles (1.5×–2×). Accumulation of annexin A4, serotransferrin, heat shock cognate 71 kDa protein and TRIM72 (≥1.35×) were also found in T32KO muscles. Phosphoglucomutase-1, l-lactate dehydrogenase, 78 kDa glucose-regulated protein (AKA heat shock 70 kDa protein 5), mitochondrial 60 kDa heat shock protein and mitochondrial aspartate aminotransferase were significantly reduced in T32KO gastrocnemius (1.37×–2.96×). Among the cytoskeletal proteins identified, troponin T, fast skeletal muscle isoform, was shown to decrease in T32KO muscles (1.69×).
Of these candidates, two proteins were selected for further validation and study, NDRG2 and TRIM72, both of which accumulated in the absence of TRIM32. Given the association of these proteins with processes of myoblast proliferation, myogenesis and cell membrane repair (13–15), and the muscle specific phenotypes associated with TRIM32 mutations, we carried out follow-up analyses on NDRG2 and TRIM72.
Validation of NDRG2 and TRIM72 accumulation in T32KO muscle
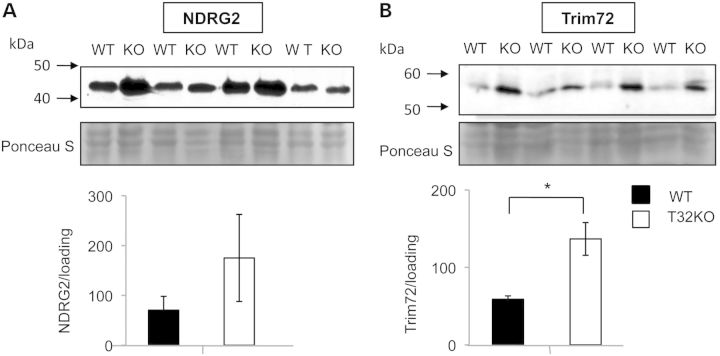
Independent verification of the differential expression of NDRG2 and TRIM72 in T32KO versus WT gastrocnemius muscles was carried out by western blot analyses with specific antibodies. In agreement with the proteomic data, both NDRG2 and TRIM72 proteins were detected at higher levels in T32KO muscle compared with WT muscles (Fig. 1A). These increases in NDRG2 and TRIM72 in T32KO gastrocnemius muscles occurred at the protein level, because no significant differences were found by qRT PCR analysis of the cDNA samples (not shown). These results suggest that both NDRG2 and TRIM72 could be endogenous substrates for TRIM32.
Figure 1.
Levels of NDRG2 and Trim72 proteins are elevated in total muscle lysates of WT and T32KO mice. Western blot analysis of NDRG2 and Trim72 proteins expressed in gastrocnemius from WT and T32KO muscles. Gastrocnemius western blot was stained for NDRG2 with NDRG2 Ab (A) or for Trim72 with Trim72 Ab (B); 40 mkg of total protein was loaded per lane. Samples from four animals per group were analyzed and representative blots are shown. The Ponceau S stained blots are shown as a loading control. Densitometry data normalized to loading, data are shown as mean ± SEM (*P < 0.05). Statistical analysis by t-test.
TRIM32 ubiquitinates NDRG2 and TRIM72 in vitro
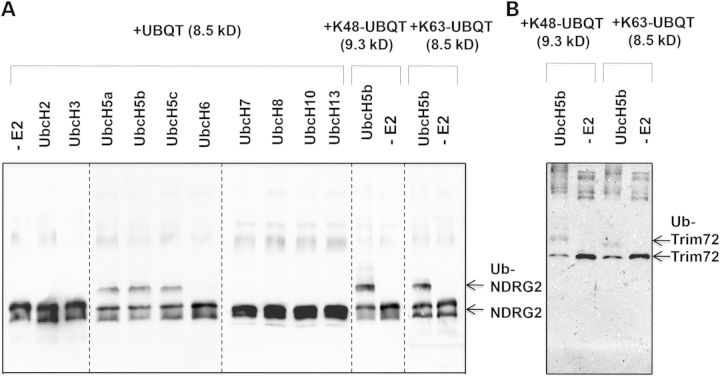
To determine whether NDRG2 and TRIM72 are substrates of TRIM32, we carried out in vitro ubiquitination assays. We found that TRIM32 ubiquitinates NDRG2 in conjunction with the E2 ligases UbcH5a, UbcH5b and UbcH5c in the presence of His-tagged ubiquitin (Fig. 2A and B) or GST-tagged ubiquitin (data not shown). The majority of the modified NDRG2 appeared to be present in the monoubiquitinated form. Di-ubiquitinated or tri-ubiquitinated species were hardly detected, as judged by the presence of bands differing by 10 kDa increments on western blot. TRIM32 was able to attach ubiquitin to NDRG2 via both lysines 48 and 63, as detected using mutated ubiquitins in which all lysines were mutated to arginine except for a single lysine at either position 48 or 63, respectively (Fig. 2A). TRIM32 also ubiquitinates TRIM72, but only in the presence of the E2 ligase UbcH5b, using WT (not shown) and K48 and K63 mutated ubiquitins. The majority of ubiquitinated TRIM72 was in the monoubiquitinated form (Fig. 2B). These results demonstrate that TRIM32 is able to ubiquitinate NDRG2 and TRIM72 in vitro.
Figure 2.
Trim32 is able to ubiquitinate recombinant NDRG2 and Trim72 proteins in vitro. (A) Trim32 ubiquitinated NDRG2 in conjunction with UbcH5a, UbcH5b and UbcH5C. Recombinant Trim32 was incubated in the presence of ATP, ubiquitin, E1 and several different E2 s as specified in the figure. Lane (–E2) indicates no E2 was added. The panel shows blot stained with anti-NDRG2 Ab. (B) Trim32 ubiquitinated Trim72 in conjunction with UbcH5b E2. The panel shows blot stained with anti-Trim72 Ab. Trim32 specifically ubiquitinated NDRG2 and TRim72 as evidenced by ∼10 kDa increment shifts in the molecular mass of the target proteins.
NDRG2 is reduced in polyubiquitinated protein fractions from T32KO muscles
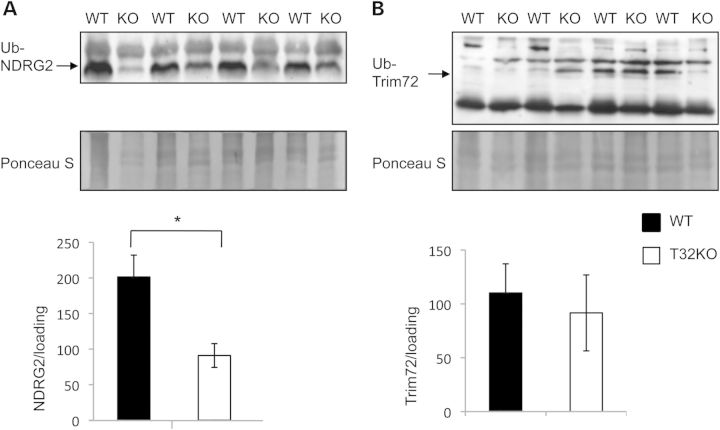
We examined the polyubiquitinated fractions of T32KO and WT muscle extracts to assess NDRG2 and TRIM72 ubiquitination levels in vivo. For this assessment, we isolated polyubiquitinylated proteins from total gastrocnemius lysates using a ubiquitin-affinity resin (anti-ubiquitin monoclonal antibody covalently coupled to agarose gel, ‘MBL’). Ubiquitin-enriched fractions were analyzed by western blotting using NDRG2- and TRIM72-specific antibodies. This analysis revealed that NDRG2 is reduced in polyubiquitinated fractions from T32KO muscles (Fig. 3A and B). No difference was observed in the levels of TRIM72 that were detected in ubiquitinated fractions. These results, together with the data on excessive accumulation of NDRG2 in T32KO muscles, strongly suggest that TRIM32 regulates turnover of NDRG2 via ubiquitination, leading to subsequent proteasomal degradation.
Figure 3.
Levels of NDRG2 protein are significantly reduced in ubiquitinated fractions from T32KO muscles. (A and B) Western blot analysis of levels of NDRG2 and Trim 72 proteins in ubiquitinated fractions eluted from ubiquitin-affinity resin. Total muscle lysates from T32KO and WT muscles were incubated with ubiquitin-affinity resin (anti-ubiquitin monoclonal antibody (clone FK2) covalently coupled to agarose gel, ‘MBL’) for 2 h at RT, extensively washed, and bound proteins were eluted by reducing sample buffer. Panel A shows blot stained with anti-NDRG2 Ab, and panel B shows blot stained with anti-Trim72 Ab. Samples from four animals per group were analyzed and representative blots are shown. The Ponceau S stained blots are shown as a loading control. Densitometry data normalized to loading, data are shown as mean ± SEM (*P < 0.05). Statistical analysis by t-test.
TRIM32 mediates ubiquitination and degradation of NDRG2 in primary myoblasts
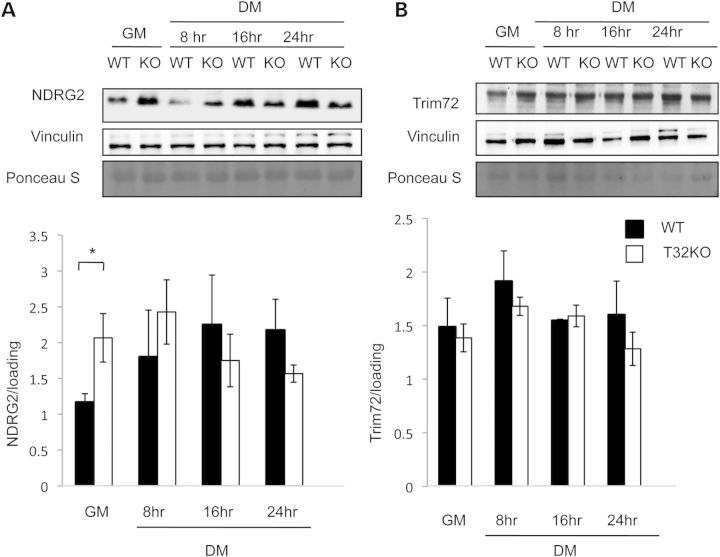
NDRG2 and TRIM72 have been previously shown to play roles in myoblast growth and differentiation (13–15). Examination of NDRG2 levels in T32KO primary myoblasts revealed its accumulation, compared with WT (Fig. 4). NDRG2 also increased in WT myoblasts during the differentiation process (Fig. 4A). TRIM72 levels in T32KO primary myoblasts or myotubes were not different from WT (Fig. 4B).
Figure 4.
NDRG2 levels increased in primary T32KO myoblasts comparing with WT cells. Western blot analysis demonstrates expression of NDRG2 (A) and Trim72 (B) proteins in primary myoblasts. Representative blots of total cell lysates are shown for each protein, in myogenic cells in GM and at different indicated times after switching to DM. Vinculin and Ponceau S stained blots are included to show the relative concentration of proteins in the cell lysates. Densitometry data were normalized to loading. Values represent the mean of at least three independent experiments ± SEM (*P < 0.05). Statistical analysis by t-test.
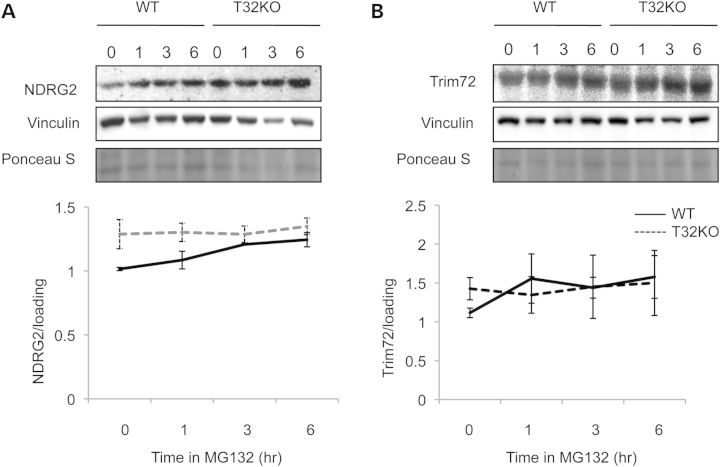
If NDRG2 and TRIM72 are in vivo substrates of TRIM32, then one would expect them to accumulate in the T32KO due to a loss of TRIM32 and, therefore, impaired ubiquitination and proteasomal degradation. To test this hypothesis, we treated primary myoblast cultures with the proteasome inhibitor MG132 and then measured NDRG2 and TRIM72 content over a time course of 3 h (Fig. 5). These studies showed that TRIM72 protein levels were not affected by MG132 treatment of WT and T32KO cells (Fig. 5B). On the other hand, NDRG2 protein was accumulated in WT myoblasts treated with MG132 (Fig. 5A) suggesting a proteasome-dependent pathway for its turnover. However, in the T32KO cells treated with MC132, NDRG2 was not additionally accumulated. Thus, the levels of NDRG2 are not affected by proteasome inhibition in the absence of TRIM32, implying that TRIM32 is necessary for controlling NDRG2 proteasome-dependent turnover.
Figure 5.
NDRG2 accumulates in WT myoblasts in the presence of proteasome inhibitor MG132. (A and B) Primary myogenic cells were treated with 50 μm of the inhibitor of proteasome activity, MG132, for 1, 3 or 6 h. Cells represented by the lane labeled ‘0’ were treated with dimethyl sulfoxide (vehicle) alone for 6 h. NDRG2 accumulated in WT myoblasts in the presence of MG132; in the T32KO cells the levels of NDRG2 were maintained, independent of the presence of MG13. Anti-vinculin blots and Ponceau S stained blots are shown as a loading control. Densitometry data normalized to loading and shown as mean ± SEM. Statistical analysis by t-test.
NDRG2 over-expression decreases cell proliferation and delays cell cycle withdrawal upon myogenic differentiation
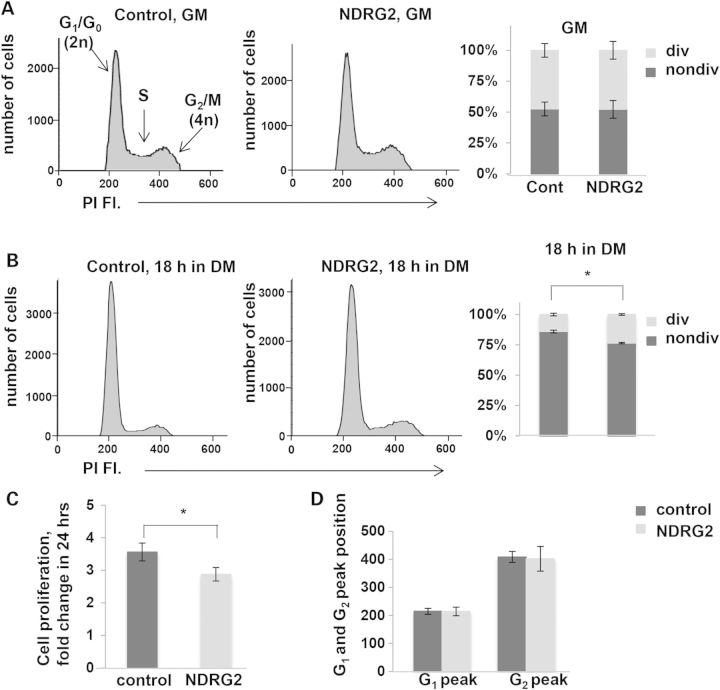
Because our prior studies demonstrated a role for TRIM32 in satellite cell cycle progression (11) and NDRG family members have been associated with control of proliferation in cancer cells (23), we tested how increased expression of NDRG2 affects muscle cell grow and proliferation. For these studies, NDRG2 was overexpressed in C2C12 myoblasts and cell cycle analysis was conducted using FACS analysis of PI-labeled cells. Unlike T32KO myoblasts, C2C12 cells overexpressing NDRG2 did not exhibit any cell cycle abnormalities under myoblast growing conditions (Fig. 6A). However, the rate of cell proliferation, determined by fold change of the cell number in 24 h, was significantly lower in NDRG2-overexpressing cells compared with control cells (Fig. 6C). Upon differentiation, the myoblast population in the S+G2/M phase was significantly larger in NDRG2-overexpressing cells, compared with control cultures (Fig. 6B) suggesting that cell cycle withdrawal was attenuated in NDRG2-overexpressing cells. These results are in line with previous observations showing that silencing of NDRG2 expression in C2C12 myoblasts promoted cell cycle exiting and the onset of differentiation (13).
Figure 6.
Over-expression of NDRG2 causes a delay in cell cycle withdrawal and slows down cell proliferation. (A and B) Cell cycle analysis of C2C12 myoblasts over-expressing NDRG2. C2C12 myoblasts were stained with PI and analyzed by flow cytometry. (A, B) Representative flow cytometric histograms showing the cell cycle distribution. (A) C2C12 control- and NDRG2-transduced cells in GM. (B) C2C12 control- and NDRG2-transduced cells were cultured for 18 h in DM. G1/G0, S and G2/M peaks are shown. Percentages of dividing (div) (S+G2/M) and non-dividing (nondiv.) (G1/G0) cells were calculated. Values represent the mean of at least three independent experiments ± SEM. (C) Effect of elevated NDRG2 levels on C2C12 myoblast proliferation. Cell proliferation index is presented as a fold change of the number of cells in 24 h. Data are summarized and presented as the mean of three independent experiments ± SEM (*P < 0.05). Statistical analysis by t-test. (D) Positions of G1 and G2 peaks are not changed on the PI fluorescence axis in NDRG2-transduced cells. Data are summarized and presented as the mean of three independent experiments ± SEM.
Previously we showed that T32KO myoblasts exhibit DNA heterochromatinization, a feature of cell senescence revealed by decreased binding of PI and decreased fluorescence intensity (leftward shifting of G1 and G2 peaks) (11). In this investigation, we did not observe any changes in PI fluorescence intensity in NDRG2-transduced C2C12 cells (Fig. 6D), indicating that NDRG2 has no effect on cellular senescence. Taken together, these data demonstrate that NDRG2 over-expression slows cell proliferation and delays cell cycle withdrawal upon differentiation of myoblasts.
Discussion
As we reported previously, senescence of muscle satellite cells and premature sarcopenia underlie the myopathic phenotype in T32KO mice (11). To gain additional insights into TRIM32's role in muscle, and to better understand the molecular pathogenesis of LGMD2H, we used a proteomic approach to reveal differentially represented proteins in muscles from T32KO and WT animals. 2D-DIGE was used to examine T32KO proteomes with the assumption that potential in vivo substrates of TRIM32 would accumulate in its absence. This analysis revealed 19 accumulated proteins on 2D-DIGE from T32KO mice.
In accordance with our earlier observations of selective type II (fast) fiber atrophy in T32KO mice (9), proteomic analysis showed reduction of the fast skeletal muscle isoform of troponin T and increased levels of RNA-editing enzyme APOBEC2, which is preferentially expressed in slow-twitch myofibers (24). Elevated levels of APOBEC2, together with increased mitochondrial aconitase and carbonic anhydrase isoform 3, iron storage protein ferritin and iron- and oxygen-binding myoglobin, could also be indicative of premature sarcopenia in T32KO muscles since similar changes were reported during rodent (25–27) and human (28) muscle aging. In ageing and disuse atrophy studies the increased levels of iron paralleled an increase in oxidative stress (29). We also found increased levels of heat shock cognate protein 71 kDa together with reduction of mitochondrial 60 kDa heat shock protein (HSP60) in T32KO muscle; both changes are concordant with age-related changes reported for human muscles (30).
Defects in both mitochondrial and SR oxidative enzymes in T32KO muscles were suggested earlier from histological analysis (9). In agreement with this hypothesis, proteomics data reveled a decrease in mitochondrial ATP synthase subunit beta and mitochondrial succinyl-CoA ligase subunit beta (Supplementary Material, Table S1). Furthermore, we found a reduction of enzymes associated with glycolysis/glycogenolysis (phosphoglucomutase-1, l-lactate dehydrogenase, pyruvate dehydrogenase, pyruvate kinase, UTP-glucose-1-phosphate uridylyltransferase) in T32KO muscles (Supplementary Material, Table S1). This decrease in glycolytic pathway enzymes (or glycolytic-to-oxidative metabolic shift) also represents one of the features of muscle aging in rodents and human (31–33). Interestingly, mouse embryonic stem cells, which are inherently immortal, have constitutively high levels of glycolysis enabling them to avoid senescence (34). Thus, this approach revealed broad changes indicative of premature sarcopenia in T32KO muscles including shifts in glycolytic-to-oxidative metabolism, impaired ion homeostasis and an altered cellular stress response. These observations are in concordance with our previously published findings of satellite cell senescence and premature sarcopenia in T32KO mice. Further studies are necessary to corroborate these data and to gain further insight into premature sarcopenia as an underlying mechanism of LGMD2H.
For the current studies, we selected two proteins with increased abundance in T32KO muscles for an in-depth analysis. NDRG2 (1.5× over WT) and TRIM72 (1.35× over WT) were focused upon, due to their putative function in myogenesis. NDRG2 belongs to a family of N-myc downstream-regulated genes whose expression is repressed by the proto-oncogenes MYCN and MYC. Although the exact functions of NDRG family members are not clearly understood, aberrant expression has been linked to carcinogenesis as well as cell proliferation, differentiation, migration, invasion and stress response (35). Foletta and coauthors investigated NDRG2's role in myoblast and skeletal muscle (13,36). NDRG2 is highly expressed in myotubes and in skeletal muscle and is responsive to metabolic signals via PGC1a/ERRa-regulated transcription (36). Importantly, silencing of NDRG2 transcription in C2C12 myoblasts promotes cell cycle exit and the onset of myogenesis (13,36).
TRIM72 similarly to TRIM32 is a tripartite motif-containing ubiquitin ligase, which plays a critical role in membrane repair after acute muscle injury (37). There have also been conflicting reports about TRIM72's involvement in myogenesis (14,15).
To our knowledge, a relationship between TRIM32 and either NDRG2 or TRIM72 has not been established. We therefore aimed to identify whether these proteins could be novel targets for TRIM32-mediated ubiquitination and degradation. Consistent with 2D-DIGE results, levels of NDRG2 and TRIM72 proteins were elevated in whole extracts of T32KO muscles (Fig. 1A and B). In vitro studies demonstrated that TRIM32 was able to ubiquitinate both NDRG2 and TRIM72 (Fig. 2A and B) with no preference toward K48 or K63-specific ubiquitinations. Interestingly, NDRG2 and TRIM72 were primarily mono-ubiquitinated by TRIM32, similar to that observed upon in vitro ubiquitination of actin (20). However, the tendency of TRIM32 to monoubiquitinate substrates may reflect the limitations of in vitro assay and/or suggest that additional co-factors such as E4 s may be required for elongation of ubiquitin chains (38). To further understand whether TRIM32 targets these proteins for proteasomal degradation in vivo, we analyzed levels of NDRG2 and TRIM72 in polyubiquitinated protein fractions of gastrocnemius muscles. A significant reduction in NDRG2 levels was observed in polyubiquitinated fractions from T32KO muscles, whereas levels of TRIM72 were not changed, further supporting a role for TRIM32 in ubiquitination and degradation of NDRG2 (but not TRIM72) in vivo.
Finally, we compared the levels of our target proteins in T32KO and WT myoblasts. Western blot analysis revealed that NDRG2 levels were significantly higher in T32KO cells than in WT counterparts (Fig. 4). To gain additional evidence of TRIM32-mediated ubiquitination and proteosomal degradation of NDRG2, primary cells were treated with the proteasome inhibitor MG132. We indeed observed accumulation of NDRG2 in WT myoblasts but not in T32KO myoblasts upon treatment with MG132 (Fig. 5A) further supporting a role for TRIM32 in degradation of NDRG2 in vivo. On the other hand, the concentrations of TRIM72 were similar in WT and T32KO cells treated with MG132 (Fig. 5B). Thus, despite the results of the proteomic analysis and in vitro ubiquitination experiments, we were not able to prove that TRIM72 is a target for TRIM32 in vivo. This is in agreement with hypothesis that TRIM32 does not participate in widespread protein degradation but rather is likely involved in the ubiquitination of a limited number of substrates (11).
Previously, we have demonstrated that primary myoblasts from T32KO muscles have impaired differentiation in vitro (11). Even though T32KO myoblasts are able to fuse following transfer to DM, the resulting myotubes are smaller and contain fewer myonuclei compared with WT cells. These changes are likely due to a decreased rate of myoblast proliferation and premature senescence in T32KO cell cultures, as was demonstrated by cell cycle analysis. The premature senescence phenotype was linked to accumulation of the known regulator of cellular senescence, E3 SUMO ligase PIAS4, which is a substrate for TRIM32 (11).
In the current studies we explored the role of TRIM32 further and identified NDRG2 as a novel substrate and demonstrated that overexpression of NDRG2 results in a decreased rate of proliferation in C2C12 myoblast cultures. The role of NDRG2 in control of cell proliferation was also reported for other cell types. For example, NDRG2 can induce cell cycle arrest in cancer cell lines (39,40). Thus, these data suggest that TRIM32 controls turnover of at least two proteins, NDRG2 and PIAS4, accumulation of either can result in decreased proliferation.
While our findings showed that overexpression of NDRG2 caused delayed cell cycle withdrawal (Fig. 6), others have demonstrated that silencing NDRG2 expression in C2C12 myoblasts promotes cell cycle exit and the onset of myogenesis (13). These findings are in concordance with recent analysis of global gene and protein expression profiles in C2C12 myotubes where enhanced expression of NDRG2 reduced protein synthesis of several muscle myosin isoforms (Myh1 and Myh4), and furthermore, partially blocked the increased protein synthesis rates elicited by a constitutively active form of estrogen-related receptor alpha, ERRα. This suggests that NDRG2 may play a role in modulating some ERRα downstream effects such as myogenesis, protein synthesis/translation and expression of contractile-type genes (36). However detailed mechanisms of biological processes influenced by NDRG2 in muscle cells await future investigation.
In conclusion, the data presented here support a role for TRIM32 in processes of myogenic cell function and differentiation. This study identified a novel target of TRIM32, NDRG2, and revealed a new role for TRIM32 in regulation of NDRG2 and myogenic precursor cell proliferation and differentiation. Finally, this study provides further support for the hypothesis of aberrant myogenic cell function as a pathogenic feature of LGMD2H.
Materials and Methods
Animal procedures
T32KO mice were described previously (9). All experimental protocols and use of animals were conducted in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and approved by the UCLA Institutional Animal Care and Use Committee.
Chemicals
All chemicals and kits used for protein solubilization and 2D-DIGE analysis were purchased from Amersham Biosciences/GE Healthcare unless indicated otherwise. Ultrapure protogel acrylamide stock solutions were from National Diagnostics (Atlanta, GA, USA). Sequencing grade-modified trypsin for peptide generation was obtained from Promega (Madison, WI, USA). Chemiluminescence substrate, protease/phosphatase inhibitors and nitrocellulose sheets were from ThermoFisher Scientific (Rockford, IL, USA) and Bio-Rad (Hercules, CA, USA), respectively. For immunoblotting, primary antibodies to the NDRG2 and Trim72 were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Cell Signalling (Danvers, MA, USA). Ultrapure lysine for quenching the DIGE labelling reaction, DNase, RNase and all general reagents were obtained from Sigma-Aldrich.
Subcellular fractionation of muscle tissue for 2D-DIGE
Gastrocnemius muscles (without solei) from 5 T32KO and 5 wild-type (WT) mice, all 1-year of age, were used for 2D-DIGE analysis. Forty milligrams of the muscle were ground to powder in a mortar and pestle under constant addition of liquid nitrogen. The powder was suspended in 15× volume of ice-cold mitobuffer [10 mm Tris–HCl, pH 7.8, 0.25 m sucrose, 0.2 mm ethylene diamine tetraacetic acid (EDTA)]. In order to eliminate excessive viscosity of the protein extract due to large amounts of muscle-derived DNA and to minimize protein degradation due to the presence of muscle-associated proteases, the solution was supplemented with DNase (2000 U/mL), RNase (750 U/mL) and protease/phosphatase inhibitor cocktail (ThermoFisher Scientific). Homogenates were centrifuged at 2000g for 8 min at 4°C and the supernatants were subjected to centrifugation at 12 000g for 15 min at 4°C followed by ultracentrifugation (130 000g for 1 h, at 4°C). Determination of protein concentration in the supernatant (cytosolic fraction) and pellet (membrane fraction) and removal of potentially interfering contaminants were performed using the 2D-Protein Concentration and 2D-Clean-Up kit from Amersham Biosciences/GE Healthcare, respectively.
Pellets collected after first low-speed centrifugation (myofibrillar fraction) were resuspended in 15× volume of homogenization buffer (20 mm Tris–HCl, pH 7.2, 100 mm KCl, 5 mm ethylene glycol tetraacetic acid, 1% Triton X-100) with protease/phosphatase inhibitor cocktail, incubated on ice for 1 h, and centrifuged at 3000g for 30 min at 4°C. After centrifugation the pellet was resuspended in 20× volume of homogenization buffer, centrifuged again under above conditions and washed twice by centrifugation in 5× volume of wash buffer [20 mm Tris–HCl, pH 7.2, 100 mm KCl, 1 mm dithiothreitol)] with protease/phosphatase inhibitor cocktail. Determination of protein concentration in the pellet and removal of potentially interfering contaminants were performed as described above. Finally, cytosolic, membrane and myofibrillar proteins were resuspended in DIGE labeling buffer (7 m urea, 2 m thiourea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propane sulfonate (CHAPS) and 25 mm Tris–HCl, pH 8.5) and adjusted to a protein concentration of 1 mg/ml.
2D gel electrophoresis, mass spectrometry analysis and protein identification
Fractionated protein samples from T32KO and WT gastrocnemius muscles were labeled with the N-hydroxysuccinimidyl ester derivatives of Cy3 and Cy5 dyes, respectively, according to standard labeling protocol (GE Healthcare). Briefly, 25 μg of protein from each sample were labeled in 15μl volume with 200 pmol of Cy3 or Cy5 dyes. The internal standard sample was prepared by pooling the equal amounts of all 10 samples (T32KO and WT) and labeled with and Cy2 dye in 15μl of DIGE labeling buffer. After 30 min on ice, the samples were quenched with 1 µl of 10 mm lysine solution and incubated for additional 10 min on ice. After labeling, protein samples were mixed together in groups of three (25 µg of each labeled with Cy2, Cy3 and Cy5). Then 150 µg of unlabeled protein samples pooled from all 10 samples were added to the labeling mixture for future picking purposes, and the rehydration solution was added up to 450 µl (7 m urea, 2 m thiourea, 4% CHAPS, 1% DTT, 0.5% non-linear pH 3–11, immobilized pH gradient (IPG) buffer, 5% glycerol, 10% isopropanol). Samples were applied to 24 cm, pH 3–11 non-linear IPG strip (GE Healthcare), and isoelectric focusing was performed for the total of 51 000 V/h for cytosolic and membrane fractions and 48 000 V/h for myofibrillar samples.
Sodium dodecyl sulfate (SDS) gel electrophoresis in large format gels, fluorescent gel imaging using Typhoon Trio Variable Mode Imager (GE Healthcare), analysis of the gel images using Decyder 2D Differential Analysis software v. 6.5 (GE Healthcare), robotic spot picking and trypsin digestion were performed as previously described (16–18).
After trypsin digestion peptides were resuspended in 8 μl of 2% acetonitrile, 0.1% formic acid solution, and using an autosampler (LC Packing Famos) and a nano-LC-2D HPLC (Eksigent) system, 4.5 μl of the sample was loaded onto a Reverse-phase C-18 (10 cm) column (19). Peptides were eluted from the column with 5–45% acetonitrile gradient in 0.1% formic acid and directly electrosprayed into the LTQ-LX linear ion trap mass spectrometer (Thermo Scientific). MS–MS acquisition of top five peptides per scan cycle was performed in the data-dependent acquisition mode and controlled by Xcalibur data system. Raw files were processed using the Proteome Discoverer software (Thermo Scientific). MASCOT v. 2.4.1 (Matrix Science, UK) database search algorithm was used for protein identification. Data were searched against SwissProt database (v. SwissProt_2014_07) with the following parameters: selected taxonomy—Mus., mass tolerance of 1.6 Da for MS and 0.8 Da for MS–MS, one missed cleavage, with carbamidomethylation of cysteine (C) as fixed modifications and oxidation of methionine (M) and as variable modifications.
Muscle protein extract preparation and western blot analysis
Forty milligram samples of the gastrocnemius muscles from T32KO and WT mice were homogenized in a Dounce homogenizer in reducing sample buffer (80 mm Tris–HCl, pH 6.8, 0.1 m DTT, 2% SDS and 10% glycerol with protease/phosphatase inhibitor cocktail; ThermoFisher Scientific). The insoluble material was removed by centrifugation (13 000g for 10 min at 4°C), and 40 μg aliquots of the proteins were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and subsequently transferred to nitrocellulose membrane for immunoblotting. Ponceau Red S staining of the membrane was carried out to confirm the efficiency of the transfer and equal loading. Membranes were destained and blocked by incubating in blocking buffer containing 3–5% nonfat, dry milk powder. Blocked and washed membranes were incubated overnight at 4°C with anti-NDRG2 and anti-TRIM72 antibodies, followed by gentle washing in blocking buffer and incubation with an appropriate dilution of peroxidase-conjugated secondary antibody for 1 h at room temperature. Blots were developed using Chemi Glow substrate (Alpha Innotech, San Leandro, CA, USA). For anti-ubiquitin staining, blots were autoclaved for 30 min before blocking. Densitometry was performed using AlphaEase FC Software Version 6.0.2 (Alpha Innotech).
Real-time polymerase chain reaction
Total RNA was isolated from mouse tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Genomic DNA contamination was removed by DNase I treatment for 30 min at 37°C. To produce cDNA, 2 μg of DNA-free RNA was used for first strand cDNA synthesis with random hexamer primers and Superscript III reverse transcriptase (Invitrogen). Analysis of relative NDRG2 and Trim72 mRNA levels in T32KO and WT gastrocnemius muscles was performed using iQ SYBR Green Supermix (Bio-Rad) in a MyIQ Single Color Real-Time Polymerase Chain Reaction (PCR) Detection System (Bio-Rad), as described previously (9). Primers for mouse NDRG2 qPCR were as follows: forward, 5′-CGCATCCTCCTGGACCAGGGAC; reverse, 5′-CATCATGGTAGGTGAATATCGC; for mouse Trim72 were as follows: forward, 5′-CACTAACCTCCAGTTGTCACGC; reverse, 5′-GCGGTCCTGCTCGCAGTAGATG. All primers were first tested in regular PCR amplifications to ensure the production of a single band in each case. Data from each sample were normalized by dividing the relative quantity of target gene cDNA by the relative quantity of house-keeping gene cDNA (18SrRNA) to correct for variability in cDNA concentration in the individual samples.
Recombinant protein expression and purification
Expression and purification of recombinant TRIM32-GST from Escherichia coli cells was described earlier (20). To obtain recombinant NDRG2 and TRIM72 proteins, mouse skeletal muscle RNA was purified using TRIzol reagent (Invitrogen). Reverse transcriptase (RT)-PCR with SuperScript II reverse transcriptase (Invitrogen) was performed to generate cDNA. Full-length mouse NDRG2 cDNA was amplified using primers: 5′-CAGGCCCATATGGCAGAACTTCAGGAGGTGCAG forward and 5′- AGGGTCTCGAGACAGGAGACTTCCATGGTGTGC reverse. Full-length mouse Trim72 cDNA was amplified using primers: 5′-TTGCTCCATATGTCGGCTGCACCCGGCCTTCTGC forward and 5′-GCAGGCTCGAGGGCCTGTTCCTGCTCCGGCCCCA reverse. Both cDNAs were cloned into pET22b+ vector.
Expression of the recombinant proteins in E. coli BL21(DE3)pLysS was induced by 0.1 mm isopropyl-β-d-thiogalactopyranoside for 4 h at 30°C. 6XHis-tagged fusion proteins were purified from the soluble fraction of bacterial cell lysates using TALON-Sepharose (Amersham).
In vitro ubiquitination assay
In vitro ubiquitination assay was performed as described earlier (20). Briefly, the ubiquitination reaction included 50 mm Tris–HCl (pH 7.5), 5 mm MgCl2, 0.5 mm DTT, 2 mm adenosine triphosphate (ATP), 100 nM E1, 250 nm E2, 34 μm His-tagged or GST-tagged ubiquitin and 250 nm recombinant TRIM32. E1, E2 s and ubiquitin were purchased from Boston Biochem. Samples were incubated at 30°C for 2 h. Reactions were stopped by addition of reducing sample buffer and boiling for 1 min. For substrate ubiquitination assay, 1 μm NDRG2 or Trim72 was added. Proteins were separated by SDS-PAGE.
Isolation of ubiquitinated fractions from gastrocnemius muscles
Forty milligram samples of the gastrocnemius muscles from T32KO and WT mice were homogenized in a Dounce homogenizer in ice-cold homogenization buffer (4 m urea in mitobuffer, see above) with protease/phosphatase inhibitor cocktail (ThermoFisher Scientific). The insoluble material was removed by centrifugation (12 000g for 10 min at 4°C), the supernatants were diluted twice with mitobuffer to bring final urea concentration to 2 m. Samples were incubated with ubiquitin-affinity resin (anti-Ubiquitin monoclonal antibody (clone FK2) covalently coupled to agarose gel, ‘MBL’) for 2 h at room temperature (RT), extensively washed by 2 m Urea in mitobuffer and bound proteins were eluted by reducing sample buffer. Eluted proteins were subjected to western blot analysis with anti-NDRG2 or anti-TRIM72 monoclonal antibody.
Cell culture and lentivirus trunsduction
C2C12 murine myoblasts were cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone Laboratories, Logan, UT) supplemented with 10% fetal bovine serum (fetal bovine serum, HyClone Laboratories) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) at 37°C with 5% CO2. For differentiation, cells grown to 70% confluence were switched to differentiation media (DMEM with 2% horse serum and then cultured for 0, 2, 4 and 6 days. The differentiation medium (DM) was changed every 2 days.
A lentiviral vector carrying the NDRG2 gene was constructed using pRRL-sin-cPPT-CMV-IRES-GFP-PGK-Puro expression plasmid (Addgene) under the control of cytomegalovirus promoter. NDRG2 sequence was cloned into XbaI/EcoRI-digested vector. Viral stocks were generated by infection of 293 T cells (21). Media containing virus were harvested from the packaging cells 48 h post-transfection and filtered using a 0.22-μm filter (Millipore). For in vitro experiments, serial dilutions of conditioned medium were used to infect 2 × 105 C2C12 cells in a six-well plate in the presence of Polybrene (8 μg/ml). Viral p24 antigen concentration was determined by immunocapture assay (Alliance; DuPont-NEN).
C2C12 myoblasts (30% confluent) were infected by lentivirus expressing NDRG2 with the MOI 40. An empty lentiviral vector was used as a control. Infection continued for 30 h at 37°C, 20% O2 and 5% CO2, before removing the virus–medium mix and replacing with fresh C2C12 growth medium (GM). The following day, 2.0 μg/ml of puromycin was added and cells were selected for 48 h prior to trypsinization, counting and re-seeding at equal numbers for each experiment. For western blotting analysis, cell pellets were resuspended in reducing sample buffer; concentration of each sample was measured by the Bio-Rad Protein assay and equal amounts of protein were separated in 9% SDS-polyacrylamide gel.
Analysis of cellular DNA content by flow cytometry
Cell cycle analysis was achieved by propidium iodide (PI) labeling of the nuclei and analyzing the fluorescence properties of the cells using fluorescence-activated cell sorting (FACS) (22). Cells were trypsinized, collected by centrifugation, washed in PBS without calcium or magnesium and resuspended at 1 × 106 cells in 1 ml of ice-cold FACS buffer (0.1% BSA in PBS). Three volumes of cold ethanol (to a final concentration of 70%) were added to the cell suspension. After overnight incubation at 4°C, fixed cells were washed in PBS and incubated with 20 μg/ml PI (Sigma-Aldrich), 0.2 mg/ml RNase A (Sigma-Aldrich) and 0.1% Triton X-100 in PBS for 30 min before analysis. Cells were analyzed on a BD FACSCalibur flow cytometer (Becton Dickinson) available at the Center for Duchenne Muscular Dystrophy Core facility, and the data were analyzed using FlowJo 9.3.1 software (Tree Star).
For the calculation of cell proliferation index, six identical plates of NDRG2- or control-transduced cells were incubated overnight and cells from three plates for each genotype were collected and counted. The remaining three plates were incubated for another 24 h before collecting and counting the cells from each plate. Cell proliferation index was calculated as a fold change in the number of cells in 24 h.
Statistical analysis
Statistical analysis of all data was carried out by Student's t-test. Differences were considered statistically significant if the P-value was <0.05. Error bars on all graphs are represented by standard errors of means (SEM).
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
This work was supported by National Institute of Arthritis, Musculoskeletal and Skin Diseases (U54AR052646, P30AR057230, RO1 AR052693, to M.S.) and the Muscular Dystrophy Association (to M.S.).
Supplementary Material
References
- 1.Nigro V., Savarese M. (2014) Genetic basis of limb-girdle muscular dystrophies: the 2014 update. Acta Myol., 33, 1–12. [PMC free article] [PubMed] [Google Scholar]
- 2.Frosk P., Weiler T., Nylen E., Sudha T., Greenberg C.R., Morgan K., Fujiwara T.M., Wrogemann K. (2002) Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putativeE3-ubiquitin-ligase gene. Am. J. Hum. Genet., 70, 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., et al. (2001) The tripartite motif family Identifies cell compartments. EMBO J., 20, 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saccone V., Palmieri M., Passamano L., Piluso G., Meroni G., Politano L., Nigro V. (2008) Mutations that impair interaction properties of TRIM32 associated with limb-girdle muscular dystrophy 2H. Hum. Mutat., 29, 240–247. [DOI] [PubMed] [Google Scholar]
- 5.Cossee M., Lagier-Tourenne C., Seguela C., Mohr M., Leturcq F., Gundesli H., Chelly J., Tranchant C., Koenig M., Mandel J.L. (2009) Use of SNP array analysis to identify a novel TRIM32 mutation in limb-girdle muscular dystrophy type 2H. Neuromuscul. Disord., 19, 255–260. [DOI] [PubMed] [Google Scholar]
- 6.Borg K., Stucka R., Locke M., Melin E., Ahlberg G., Klutzny U., Hagen M., Huebner A., Lochmuller H., Wrogemann K., et al. (2009) Intragenic deletion of TRIM32 in compound heterozygotes with sarcotubular myopathy/LGMD2H. Hum. Mutat., 30, E831–E844. [DOI] [PubMed] [Google Scholar]
- 7.Schoser B.G., Frosk P., Engel A.G., Klutzny U., Lochmuller H., Wrogemann K. (2005) Commonality of TRIM32 mutation in causing sarcotubular myopathy and LGMD2H. Ann. Neurol., 57, 591–595. [DOI] [PubMed] [Google Scholar]
- 8.Chiang A.P., Beck J.S., Yen H.J., Tayeh M.K., Scheetz T.E., Swiderski R.E., Nishimura D.Y., Braun T.A., Kim K.Y., Huang J., et al. (2006) Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc. Natl. Acad. Sci. USA, 103, 6287–6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudryashova E., Wu J., Havton L.A., Spencer M.J. (2009) Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum. Mol. Genet., 18, 1353–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen S., Zhai B., Gygi S.P., Goldberg A.L. (2012) Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J. Cell. Biol., 198, 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kudryashova E., Kramerova I., Spencer M.J. (2012) Satellite cell senescence underlies myopathy in a mouse model of limb-girdle muscular dystrophy 2H. J. Clin. Invest., 122, 1764–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicklas S., Otto A., Wu X., Miller P., Stelzer S., Wen Y., Kuang S., Wrogemann K., Patel K., Ding H., Schwamborn J.C. (2012) TRIM32 regulates skeletal muscle stem cell differentiation and is necessary for normal adult muscle regeneration. Plos One, 7, e30445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foletta V.C., Prior M.J., Stupka N., Carey K., Segal D.H., Jones S., Swinton C., Martin S., Cameron-Smith D., Walder K.R. (2009) NDRG2, a novel regulator of myoblast proliferation, is regulated by anabolic and catabolic factors. J. Physiol., 587, 1619–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C.S., Yi J.S., Jung S.Y., Kim B.W., Lee N.R., Choo H.J., Jang S.Y., Han J., Chi S.G., Park M., Lee J.H., Ko Y.G. (2010) TRIM72 negatively regulates myogenesis via targeting insulin receptor substrate-1. Cell Death Diff., 17, 1254–1265. [DOI] [PubMed] [Google Scholar]
- 15.Cai C., Masumiya H., Weisleder N., Pan Z., Nishi M., Komazaki S., Takeshima H., Ma J. (2009) MG53 regulates membrane budding and exocytosis in muscle cells. J. Biol. Chem., 284, 3314–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosessian S., Avliyakulov N.K., Mulholland D.J., Boontheung P., Loo J.A., Wu H. (2009) Analysis of PTEN complex assembly and identification of heterogeneous nuclear ribonucleoprotein C as a component of the PTEN-associated complex. J. Biol. Chem., 284, 30159–30166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh R., Avliyakulov N.K., Braga M., Haykinson M.J., Martinez L., Singh V., Parveen M., Chaudhuri G., Pervin S. (2013) Proteomic identification of mitochondrial targets of arginase in human breast cancer. PLoS One, 8, e79242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avliyakulov N.K., Rajavel K.S., Le K.M.T., Guo L., Mirsadraei L., Yong W.H., Liau L.M., Li S., Lai A., Nghiemphu P.L., et al. (2014) C-terminally truncated form of B-crystallin is associated with IDH1 R132H mutation in anaplastic astrocytoma. J. Neurooncol., 117, 53–65. [DOI] [PubMed] [Google Scholar]
- 19.Wohlchlegel J.A. (2009) Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol. Biol., 497, 33–49. [DOI] [PubMed] [Google Scholar]
- 20.Kudryashova E., Kudryashov D., Kramerova I., Spencer M.J. (2005) Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol., 354, 413–424. [DOI] [PubMed] [Google Scholar]
- 21.Naldini L., Blömer U., Gage F.H., Trono D., Verma I.M. (1996) Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA, 93, 11382–11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crissman H.A., Steinkamp J.A. (1973) Rapid, simultaneous measurement of DNA, protein, and cell volume in single cells from large mammalian cell populations. J. Cell Biol., 59, 766–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q., Li L.H., Gao G.D., Wang G., Qu L., Li J.G., Wang C.M. (2013) HIF-1α up-regulates NDRG1 expression through binding to NDRG1 promoter, leading to proliferation of lung cancer A549 cells. Mol. Biol. Rep., 40, 3723–3729. [DOI] [PubMed] [Google Scholar]
- 24.Sato Y., Probst H.C., Tatsumi R., Ikeuchi Y., Neuberger M.S., Rada C. (2010) Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J. Biol. Chem., 285, 7111–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piec I., Listrat A., Alliot J., Chambon C., Taylor R.G., Bechet D. (2005) Differential proteome analysis of aging in rat skeletal muscle. FASEB J., 19, 1143–1145. [DOI] [PubMed] [Google Scholar]
- 26.O'Connell K., Gannon J., Doran P., Ohlendieck K. (2007) Proteomic profiling reveals a severely perturbed protein expression pattern in aged skeletal muscle. Int. J. Mol. Med., 20, 145–153. [PubMed] [Google Scholar]
- 27.Jung S.H., DeRuisseau L.R., Kavazis A.N., DeRuisseau K.C. (2008) Plantaris muscle of aged rats demonstrates iron accumulation and altered expression of iron regulation proteins. Exp. Physiol., 93, 407–414. [DOI] [PubMed] [Google Scholar]
- 28.Staunton L., Zweyer M., Swandulla D., Ohlendieck K. (2012) Mass spectrometry-based proteomic analysis of middle-aged versus aged vastus lateralis reveals increased levels of carbonic anhydrase isoform 3 in senescent human skeletal muscle. Int. J. Mol. Med., 30, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofer A.M., Machen T.E. (1993) Technique for in situ measurement of calcium in intracellular inositol 1,4,5,-trisphosphatesensitive stores using the fluorescent indicator mag-fura-2. Proc. Natl. Acad. Sci. USA, 90, 2598–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joseph A.M., Adhihetty P.J., Buford T.W., Wohlgemuth S.E., Lees H.A., Nguyen L.M., Aranda J.M., Sandesara B.D., Pahor M., Manini T.M., Marzetti E., Leeuwenburgh C. (2012) The impact of aging on mitochondrial function and biogenesis pathways in skeletal muscle of sedentary high- and low-functioning elderly individuals. Aging Cell, 11, 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edstrom E., Altun M., Bergman E., Johnson H., Kullberg S., Ramirez-Leon V., Ulfhake B. (2007) Factors contributing to neuromuscular impairment and sarcopenia during aging. Physiol. Behav., 92, 129–135. [DOI] [PubMed] [Google Scholar]
- 32.Vandervoort A.A. (2002) Aging of the human neuromuscular system. Muscle Nerve, 25, 17–25. [DOI] [PubMed] [Google Scholar]
- 33.Ohlendieck K. (2011) Proteomic profiling of fast-to-slow muscle transitions during aging. Front. Physiol., 2, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kondoh H., Leonart M.E., Gil J., Beach D., Peters G. (2005) Glycolysis and cellular immortalization. Drug Discov. Today, 2, 263–267. [Google Scholar]
- 35.Melotte V., Qu X., Ongenaert M., van Criekinge W., de Bruïne A.P., Baldwin H.S., van Engeland M. (2010) The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB J., 24, 4153–4166. [DOI] [PubMed] [Google Scholar]
- 36.Foletta V.C., Brown E.L., Cho Y., Snow R.J., Kralli A., Russell A.P. (2013) Ndrg2 is a PGC-1α/ERRα target gene that controls protein synthesis and expression of contractile-type genes in C2C12 myotubes. Biochim. Biophys. Acta, 1833, 3112–3123. [DOI] [PubMed] [Google Scholar]
- 37.Cai C., Masumiya H., Weisleder N., Matsuda N., Nishi M., Hwang M., Ko J.K., Lin P., Thornton A., Zhao X., et al. (2009) MG53 nucleates assembly of cell membrane repair machinery. Nat. Cell Biol., 11, 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koegl M., Hoppe T., Schlenker S., Ulrich H.D., Mayer T.U., Jentsch S. (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell, 96, 635–644. [DOI] [PubMed] [Google Scholar]
- 39.Liu X., Niu T., Liu X., Hou W., Zhang J., Yao L. (2012) Microarray profiling of HepG2 cells ectopically expressing NDRG2. Gene, 503, 48–55. [DOI] [PubMed] [Google Scholar]
- 40.Kim Y.J., Yoon S.Y., Kim J.T., Choi S.C., Lim J.S., Kim J.H., Song E.Y., Lee H.G., Choi I., Kim J.W. (2009) NDRG2 suppresses cell proliferation through down-regulation of AP-1 activity in human colon carcinoma cells. Int. J. Cancer, 124, 7–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.