Abstract
The emergence of avian H7N9 viruses has raised concerns about its pandemic potential and prompted vaccine trials. At present, it is unknown whether there will be sufficient cross-reactive hemagglutinin (HA)–specific CD4 T-cell memory with seasonal influenza to facilitate antibody production to H7 HA. There has also been speculation that H7N9 will have few CD4 T-cell epitopes. In this study, we quantified the potential of seasonal influenza to provide memory CD4 T cells that can cross-reactively recognize H7 HA–derived peptides. These studies have revealed that many humans have substantial H7-reactive CD4 T cells, whereas up to 40% are lacking such reactivity. Correlation studies indicate that CD4 T cells reactive with H7 HA are drawn from reactivity generated from seasonal strains. Overall, our findings suggest that previous exposure of humans to seasonal influenza can poise them to respond to avian H7N9, but this is likely to be uneven across populations.
Keywords: influenza, CD4 T cells, H7N9, vaccines
In 2013, an avian H7N9 virus was reported to infect individuals in China, with the number of deaths thus far approaching 200. This avian virus, like the H5N1 viruses causing ongoing sporadic infection in humans, has caused great concern because of its apparently high mortality rate. Unlike avian H5N1 viruses, H7N9 causes low pathogenicity in poultry and is difficult to track. Based on sequence analyses and phylogenetic relationships [1–4] the H7N9 virus detected in humans may have evolved from as many as 4 origin species, including domestic ducks (H7N3), migratory wild birds (H7N9), and domestic poultry (H9N2). The number of individuals infected, the novel composition, and recent findings indicating that this virus may spread in droplets between ferrets [5, 6] have all raised concerns that the virus has human pandemic potential.
The possibility of the spread or adaptation of H7N9 to humans [7–9] has raised the issue of protective H7N9 vaccination [10, 11]. There is considerable uncertainty regarding whether the typical human has sufficient influenza-specific cross-reactive immunity to respond to H7N9. During infection, CD4 and CD8 T cells cross-reactive to heterosubtypic viruses can be important in protection and lead to more rapid clearance and diminished morbidity. Memory CD4 T cells can contribute to this protection in multiple ways (reviewed in references 12–14), including through up-regulation of the innate response in the lung [15, 16] and direct cytotoxicity [13]. Most importantly, for protection prior to infection, high-affinity neutralizing antibody responses are needed, for which CD4 T cells are essential [17].
Our laboratory has shown the correlation between the neutralizing antibody response to vaccination with a novel influenza strain and expansion of CD4 T cells [18]. In addition, correlations were noted between increases in CD4 T cells with markers associated with follicular helper cells and neutralizing antibody responses to seasonal vaccination [19]. When we addressed whether the specificity of CD4 T cells influences their ability to help B cells during infection or vaccination, we found evidence for linked specificities between CD4 T-cell help and antibody responses: CD4 T cells specific for hemagglutinin (HA) were better correlated with the neutralizing antibody response to HA than were CD4 T cells specific for nucleocapsid protein (NP) and/or M1 [18, 20]. Therefore, although the conserved internal viral proteins such as NP might provide epitopes for some CD4 T-cell functions, help for neutralizing antibody responses to viral infection or vaccination is likely to be delivered most effectively by HA-specific CD4 T cells.
In this study, because of the importance of HA-specific CD4 T cells in neutralizing antibody production, we have directly quantified the potential for cross-reactive recognition of CD4 T cells to H7 HA–derived epitopes. Using human subjects, we found strong evidence for recognition of H7-derived epitopes in individuals exposed only to seasonal influenza. We anticipate that the degree and abundance of cross-reactive CD4 T cells in humans will be a strong correlate of the ability to respond to H7N9 vaccines and infection by production of neutralizing antibodies.
MATERIALS AND METHODS
Synthetic Peptides and Libraries
We used 17mer peptides overlapping by 11 amino acids to encompass the entire sequences of the viral proteins, obtained from the Biodefense and Emerging Infections Research Repository, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) or made in our own facility and reconstituted as described elsewhere [21]. The peptide arrays used included influenza virus A/New Caledonia/20/99 (H1N1) HA protein (NR-2602), influenza virus A/California/04/09 (H1N1) HA protein (NR-15433), influenza virus A/New York/384/2005 (H3N2) HA protein (NR-2603), and influenza virus A/New York/348/2003 (H1N1) NP (NR-2611).
Isolation of Peripheral Blood Mononuclear Cells From Human Blood
Blood samples were obtained from a group of healthy adult subjects (aged 18–50 years) who had provided informed consent. This study was approved by the University of Rochester Biosafety Committee. Plasma was removed, and the peripheral blood mononuclear cells (PBMCs) were collected, washed and resuspended at 15 million PBMCs per milliliter in fetal calf serum (Gibco) containing 10% dimethyl sulfoxide (Sigma-Aldrich), and frozen. Aliquots of frozen PBMCs were thawed and diluted in Roswell Park Memorial Institute 1640 medium (Gibco) containing 10% fetal calf serum, washed, and rested overnight at 37°C and 5% carbon dioxide. They were then harvested from culture, washed, and depleted of CD8 cells by using MACS CD8 microbeads, according to the manufacturer's instructions (Miltenyi Biotec).
Vaccine Recipients
A group of healthy adults received a single 45-µg dose of subvirion H7N7 vaccine. The PBMCs were collected on day 0 and on day 14 after vaccination. Samples were generously provided by Kanta Subbarao, MD (NIH/NIAID) and John Treanor, MD (University of Rochester) through an ongoing trial (ClinicalTrials.gov identifier NCT 01534468). Samples were frozen and used as described above.
Human Enzyme-Linked Immunospot Assays
Enzyme-linked immunospot (ELISPOT) assays were performed as described elsewhere [18, 22]. Briefly, CD8-depleted PBMCs were cultured with peptides on plates coated with anti-human interferon (IFN) γ for 36 hours at 37°C in 5% carbon dioxide. After incubation, cells were removed from the plates by washing. Biotinylated anti-human IFN-γ was added in wash buffer with 10% fetal bovine serum and incubated at room temperature for 2 hours. The plates were washed, and then streptavidin-conjugated alkaline phosphatase was added, incubated for 30 minutes, washed, and developed using Vector Blue substrate kit III (Vector Laboratories) prepared in 100 mmol/L Tris (pH 8.2). Cytokine-secreting cells were quantified with an Immunospot reader (series 2A), using Immunospot software (version 5.0.9.19).
RESULTS
Potential for Cross-reactivity in CD4 T Cells Specific for Seasonal and Avian-Derived H7 HA Proteins
To consider the possibility that there may be cross-reactive CD4 T-cell recognition of HA, the amino acid sequences of HA from recently circulation H1N1 and H3N2 viruses were compared to H7-HA (Supplementary Figure 1). If one calculates the number of identical amino acids in homologous positions in the HA proteins, H1 HA proteins have approximately 40% sequence identity to H7 HA, compared with approximately 46% for the H3 HA proteins. However, for T-cell cross-reactivity, contiguous segments are most important. The minimal peptide that can activate CD4 T cells via major histocompatibility complex (MHC)–restricted recognition is a 9mer, but MHC class II molecules are typically occupied by at least a 12mer peptides because the binding pocket extends from P2 to P10 (reviewed in [23]). Of the “core” amino acids, only 3–4 amino acids (P2, P3, P5, P7, or P8) are solvent exposed and most commonly contact T-cell receptors, whereas amino acids at P1, P4, P6, and P9 anchor the peptide to the MHC class II protein (reviewed in [23, 24]). Therefore, for 2 homologous peptides (eg, H7 vs H1) to be recognized by the same CD4 T cells, they must bind to the same MHC class II molecule in the same “frame”, share the key anchor residues, and display at least 2 or 3 of the same T-cell receptor contact residues. Accordingly, a stretch of 8–9 identical amino acids might confer full cross-reactivity, whereas 4–5 identical amino acids might confer partial cross-reactivity.
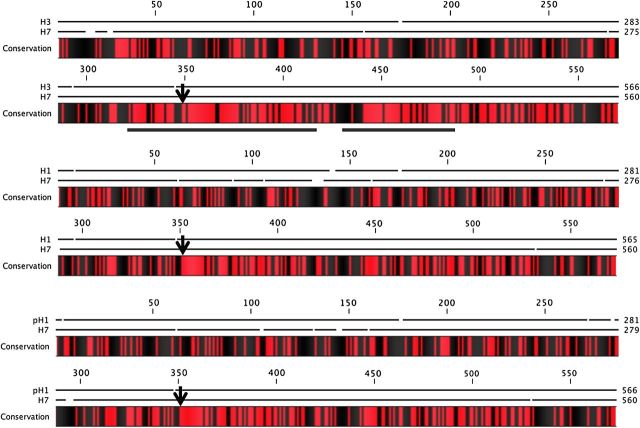
Therefore, we scanned for segments of H7 that might be sufficient for CD4 T-cell cross-reactivity. In Figure 1, the coding sequence of H7 HA was compared with the HA of seasonal strains. We noted that the amino terminal half of the HAs, while collectively amounting to substantial “sequence identity,” primarily consist of very small stretches of identical amino acids (<4), thus probably not sufficient for T-cell cross-reactivity. However, beginning at approximately position 310 (indicated by an underline), longer stretches of identity between seasonal and avian HA were identified. The H3 HA showed the most extensive homology to H7 HA, consistent with their phylogenetic assignment into group 2 [25]. Overall, this analysis indicated that there might be CD4 T-cell cross-reactivity between H7 and seasonal HA proteins, if these sequence-shared regions of HA constituted CD4 T-cell epitopes.
Figure 1.
Amino acid sequence alignment reveals the highest degree of homology between H3N2 hemagglutinin (HA) and H7N9 HA. Pairwise alignment of H3 A/New York/384/05 (top), H1 A/New Caledonia/20/99 (middle), and H1 A/California/04/09 (bottom) with the amino acid sequence of HA from H7 A/Anhui/02/13 is shown; alignment was performed using CLC Sequence Viewer software (version 6.8.2) (CLC bio, a QIAGEN Company). Conservation between 2 compared sequences is indicated by red shading, and black indicates nonidentity. Gaps in the black lines above the sequences indicate any amino acid gaps in the alignment. Arrow indicates HA0 cleavage site. Underlined sections of the alignment represent coverage of synthesized H7 peptides used in the T-cell enzyme-linked immunospot assays (Supplementary Table 1; Figures 2 and 3). Reference numbers (GenBank/PubMed and EpiFlu/ Global Initiative on Sharing Avian Influenza Data) for aligned sequences are as follows: H1 New Caledonia (ACF41878); H1 California (ACQ55359); H3 New York (AAZ79974); H7 Anhui (EPI439507).
Human CD4 T-Cell Reactivity to H7 HA–Derived Epitopes
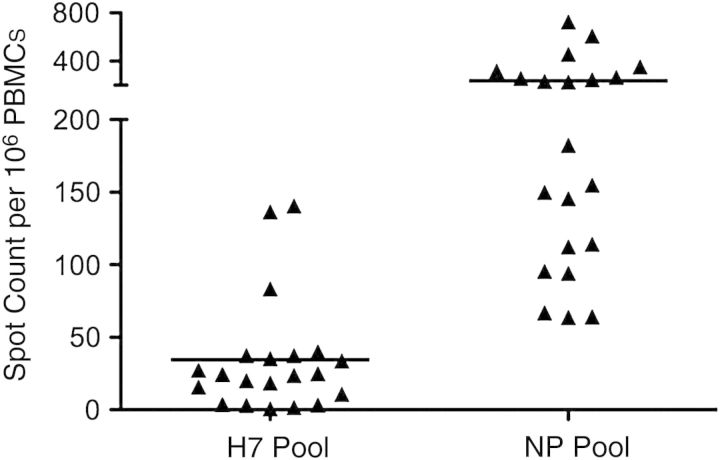
To begin to assess the potential for human CD4 T cells to recognize the avian H7 HA, we synthesized 17mer peptides representing the most highly conserved segments of H7 HA from the Anhui strain relative to the seasonal strains (Supplementary Table 1). These 26 overlapping peptides were pooled and tested for their ability to activate human CD4 T cells from healthy donors. We tested PBMCs depleted of CD8 T cells for the ability to produce IFN-γ in response to the H7 HA–derived peptides. Reactivity to NP-derived peptides was used as a control. Figure 2 shows that many individuals (15 of 21) were able to recognize H7-derived peptides, albeit at a low level. Reactivity toward H7 HA epitopes ranged from 0 to 140 reactive CD4 T cells per million tested. In contrast, NP reactivity in these subjects ranged from 62 to 750 reactive CD4 T cells per million. These numbers are probably an underestimate, because when large pools of peptides are added to CD4 T-cell activation assays, there can be suppression due to solvents in the peptides [26]. Collectively, our data indicate that many individuals are likely to have some CD4 T-cell reactivity to H7 HA and that a few seem to have quite high reactivity to H7-derived HA peptides. We also find that CD4 T cells can respond to purified recombinant H7 HA protein, indicating that these circulating memory CD4 T cells can recognized naturally processed peptides (data not shown).
Figure 2.
CD4 T cells from healthy human donors display reactivity to H7 A/Anhui/02/13 influenza hemagglutinin (HA). Twenty-one donors not exposed to H7N9 were tested for reactivity to a pool of semiconserved H7 HA peptide epitopes (shown in Supplementary Table 1) in interferon (IFN) γ enzyme-linked immunospot assays. CD8-depleted peripheral blood mononuclear cells (PBMCs) were cocultured with peptide pools with each peptide present at 1 µmol/L at 37°C for 36 hours. The reactivity to HA peptides is compared with the response to nucleocapsid protein (NP)–derived peptide pools. The response is shown as the number of IFN-γ–producing cells per 106 CD8-depleted PBMCs, with the background subtracted, and the mean response of the population is shown as a black line.
We next examined individual H7-derived peptides from the carboxy-terminal region of HA for their ability to activate human CD4 T cells. Each individual is represented by a different symbol in Figure 3A, and the average response per peptide is shown in Figure 3B. The sequence of each peptide indicated below the Figure is listed in Supplementary Table 1. These studies revealed that many H7 HA–derived peptides were able to activate human CD4 T cells. Frequencies ranged from 5 epitope-specific cells per million to >150 reactive cells per million.
Figure 3.
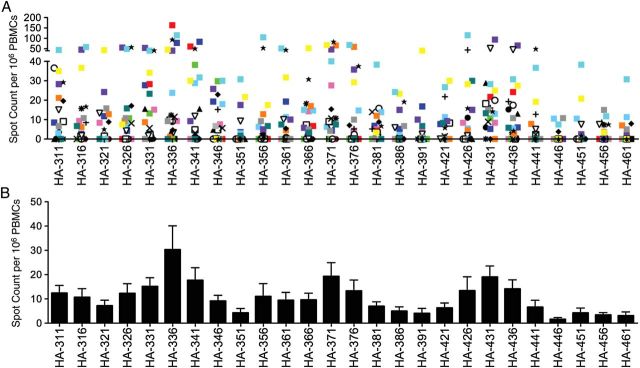
CD4 T cells in the peripheral blood of H7N9-naive individuals are able to respond to individual H7 hemagglutinin (HA) epitopes in the carboxy terminal end of the HA molecule. CD8-depleted peripheral blood mononuclear cells (PBMCs) (400 000/well) were restimulated with individual H7 HA peptides (10 µmol/L) directly ex vivo. The peptide sequences are shown in Supplementary Table 1. Data are shown as the average number of interferon γ–producing cells per million CD8 depleted PBMCs (60%–80% CD4 T cells), with background subtracted. A, Range of reactivity, with each individual (n = 21) represented as a different symbol or color. B, Mean response of the population to each peptide is shown as a bar graph.
Of particular note were several segments of H7 HA (336–357, 371–392, and 426–452) that could activate a relatively high frequency of CD4 T cells in many of the donors. These segments included the most highly conserved regions of HA, suggesting that, indeed, these regions of the HA2 domain of H7 HA can generate peptides recognizable by human CD4 T cells. Interestingly, there were some individuals whose CD4 T cells recognized many different H7 HA–derived peptides at a high frequency, whereas others recognized only a limited subset, though also at a high frequency. There was also a subset of individuals (approximately 25%) who displayed a very low frequency (<100 total cytokine-producing cells) in response to H7 HA. These disparities in recognition may be due to either differences in influenza confrontation in the subjects or expression of particular HLA class II molecules that vary in the number of HA epitopes they can present to CD4 T cells.
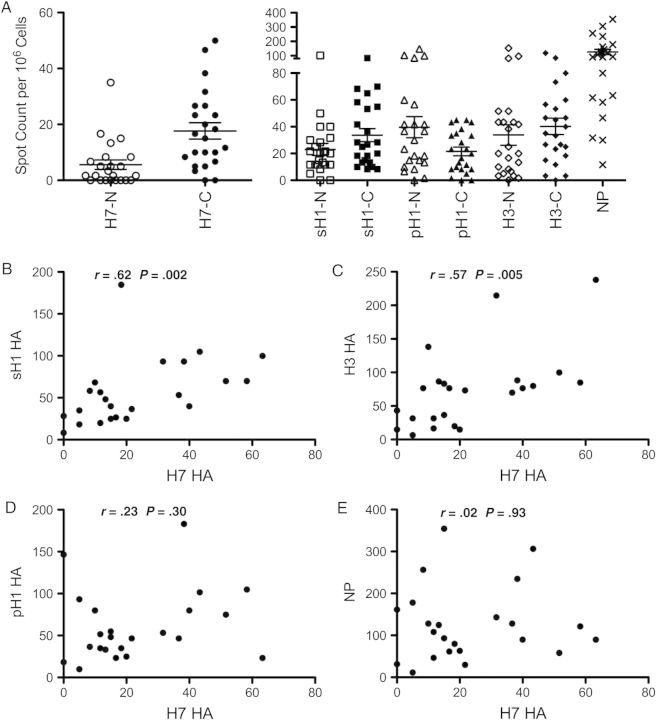
To examine the relationship between reactivity to H7-derived HA epitopes and epitopes encountered from seasonal vaccines and viruses, we examined a new set of healthy donors for CD4 T-cell reactivity to HA epitopes from pH1, sH1, sH3, and H7 HA proteins. Reactivity to NP was used as a control. Figure 4A shows the reactivity to these different viral proteins, using cytokine ELISPOT assays and peptide pools representing each of these viral proteins. For HA, we divided the pools of peptides into the NH2 versus carboxy-terminal halves of the protein. Reactivity toward H7 HA was found to be variable, as before, and heavily skewed toward the conserved HA2 domain. Reactivity to seasonal HA and pandemic H1N1 HA proteins was more substantial and was much more equally distributed across the entire HA protein. Strikingly, when the correlation between reactivity among the different viral proteins among the subjects was evaluated, there was a strong correlation between H7 reactivity to the older seasonal H1N1 (Figure 4B) and seasonal H3 HA (Figure 4C). There was a very modest correlation between pH1 and H7 (Figure 4D) and no detectable correlation among different subjects between the abundance of CD4 T cells specific for H7 and the number specific for NP (Figure 4E).
Figure 4.
CD4 T cells from healthy donors focus on epitopes in the genetically conserved HA2 domain that were probably elicited by seasonal influenza. A, New cohort of healthy donors (n = 23) were tested for CD4 T-cell reactivity to influenza hemagglutinin (HA) and nucleocapsid protein (NP) using interferon γ enzyme-linked immunospot assays and pools of overlapping peptides (each 1 µmol/L) encompassing the NH2 (N) and the carboxy-terminal (C) portions of the HA protein from seasonal H1N1 A/New Caledonia/20/99 (sH1-N [amino acids {aa} 1–320], sH1-C [aa 310–565]), pandemic H1N1 A/California/04/09 (pH1-N [aa 1–323], pH1-C [aa 309–566]), seasonal H3N2 A/New York/384/05 (H3-N [aa 1–319], H3-C [aa 309–566]), H7N9 A/Anhui/1/13 (H7-N [aa 1–322], H7-C [aa 311–560]) and NP from H1N1 A/New York/348/03. B–D, Correlation between H7 reactivity and seasonal HA proteins. E, No correlation between NP and H7 reactivity. All r and P values were calculated using the Spearman rank correlation test.
We were also able to study a small group of human subjects vaccinated with an H7N7 subvirion vaccine (see “Materials and Methods” section). The reactive CD4 T cells were quantified by cytokine ELISPOT assays, wherein the number of protein specific spots detected before vaccination was subtracted from the reactivity detected at day 14 after vaccination (Figure 5). The elicited response to H7 HA was modest, as expected, with approximately half of the subjects' responses after vaccination detectable in the range of 25–150 cytokine-producing cells per million (Figure 5A, left). In all subjects, baseline NP reactivity was higher at day 0, as expected, and gained substantially in response to vaccination (Figure 5A, right). The elicited response to NP is consistent with our earlier studies showing that clinical vaccines contain sufficient NP to promote CD4 T-cell recruitment [18, 27].
Figure 5.
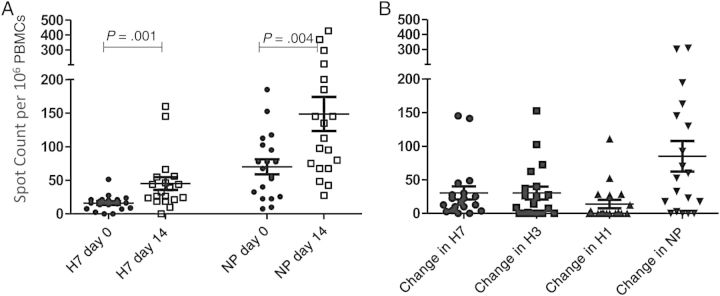
Responses to vaccination with a H7N7 monovalent influenza subvirion vaccine. A, CD4 T-cell responses against H7 and nucleocapsid protein (NP) peptide pools at baseline (day 0) and approximately 14 days after vaccination, determined using interferon γ enzyme-linked immunospot assays. Horizontal lines represent mean of response; error bars, standard error of the mean. P values were determined with the Wilcoxon signed rank test. B, Change (from day 0 to day 14) in CD4 T-cell reactivity to H7, H3, H1, or NP peptide pools (1 µmol/L) after vaccination. Horizontal lines represent mean change in CD4 T-cell response; error bars, standard error of the mean.
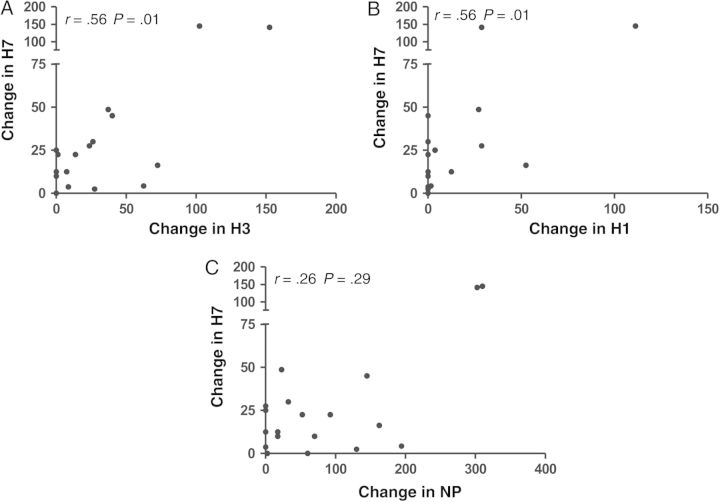
In coordination with the gains in reactivity to H7-derived peptides, there were also modest gains in reactivity to H1 and H3 derived peptides after vaccination (Figure 5B). These gains are presumably due to cross-reactivity in some of the peptide epitopes, because the vaccine did not contain any seasonal H3 or H1 HA proteins. When correlations between responses to different viral proteins were tested (Figure 6), positive correlations with H7 were noted for both H1 and H3. This was rarely observed with reactivity of the CD4 T cells with NP and H7. This argues that the correlations in reactivity among different HA epitopes does not reflect the general magnitude of the immune response to vaccination. Rather, these data support the view that some human subjects not previously exposed to avian viruses or vaccines can draw on cross-reactive CD4 T-cell memory to respond to novel pandemic strains of avian influenza. The degree of cross-reactivity will probably reflect the peptides within the related HA proteins that are selected by host HLA class II molecules during responses to seasonal infection and vaccination.
Figure 6.
Relationship between development of a CD4 T-cell response to H7 peptides and CD4 T-cell response to H1, H3, and nucleocapsid protein (NP) peptides. Interferon (IFN) γ enzyme-linked immunospot assays were performed on peripheral blood mononuclear cells (PBMCs) from subjects at day 0 and day 14 after vaccination with a monovalent inactivated subvirion H7N7 vaccine, and the change in CD4 T-cell reactivity to H7, H3, H1, or NP peptide pools (1 µmol/L) between baseline and day 14. There is a statistically significant correlation between the development of a response to H7 and H3 peptides (A) and H7 and H1 peptides (B), but no correlation was observed when H7 reactivity and reactivity to the NP pool were compared (C). Reactivity is defined as the number of IFN-γ producing cells per million CD8-PBMCs. All r and P values were determined with the Spearman rank correlation test.
DISCUSSION
In this study, we have evaluated the potential of preexisting CD4 T-cell immunity generated by seasonal influenza viruses to recognize HA-derived epitopes from the avian H7N9 virus that has recently emerged in humans. Because CD4 T-cell help can be limiting in antibody responses, cross-reactive recognition by HA-specific CD4 T cells might provide sufficient “help” to facilitate neutralizing antibody responses to H7N9 influenza. We have recently shown that HA-specific CD4 T cells are likely to be the most potent in providing help in the cognate CD4 T-cell–B-cell interactions needed for high-affinity, neutralizing antibody production by HA-specific B cells [18, 20].
Sequence comparison of H7 HA with seasonal HA showed significant sequence divergence, but we did note several stretches of amino acid identity in the carboxy-terminal HA2 domain that theoretically might provide peptides for CD4 T-cell cross-reactivity. Empirical testing of reactivity of human CD4 T cells revealed that many but not all of these segments were recognized. Importantly, reactivity to H7 HA–derived peptides both at baseline and during responses to vaccination correlated strongly with reactivity to seasonal H3 and H1 proteins. We do not know the previous exposure history of the subjects analyzed in this study, but, based on serological analyses using enzyme-linked immosorbent assays, the high versus low responders to H7 HA proteins do not seem distinct with regard to existing antibodies to seasonal influenza strains.
Our results raise several intriguing questions. The first is whether neutralizing antibody responses to H7N9 vaccines will be predictable based on preexisting CD4 T cells to H7 HA–derived peptides. Previous vaccine trials with H7 viruses [28–30] have proved disappointing thus far and revealed an overall poor “immunogenicity” of H7 vaccines, particularly when the vaccine does not contain adjuvants, as is typical in the United States. It is possible that individuals who display substantial cross-reactivity may be most able to respond to limiting doses or unadjuvanted versions of avian vaccines. It is not yet clear whether there is a threshold of memory CD4 T cells that must be elicited by the vaccine to potentiate antibody responses, and, if so, whether they must possess a defined phenotype associated with recruitment into the draining lymph node and participation in help for the germinal center response. Our current studies are evaluating this issue. We have also evaluated the effects of prepandemic strategies to further enrich for reactivity to avian HA proteins, in a recent H5N1 vaccine trial. We find that long-lived circulating CD4 T-cell memory reactive with avian HA proteins can be potentiated by vaccination years earlier [31]. This strategy might be useful for those individuals with low circulating cross-reactive memory to avian viruses caused by exposure to seasonal vaccines.
The second issue raised by this study is the mechanism that underlies the variable H7 HA CD4 T-cell cross-reactivity in humans. The mouse models studied in our laboratory thus far provide at least a partial explanation. It is well accepted that host MHC class II molecules select the epitopes recognized by CD4 T cells [23, 24, 32, 33]. We have found highly variable MHC class II–dependent focus on the HA protein [21, 26, 34]. In humans, the options for class II diversity are greater than in mice, but the presenting HLA class II molecules available to elicit CD4 T cells in any given human are still limited to a few of the large constellation of possible HLA-class II alleles. Therefore, only some humans are likely to be able to bind to the sequence-conserved HA peptides and select the cross-reactive CD4 T cells during seasonal exposure to influenza.
The other parameter probably responsible for the variability in responses to H7 HA derived epitopes or preferences for one strain of seasonal virus epitopes is the complex vaccination/infection history of individual humans. With currently available technology, this variable is difficult to document with certainty, owing to life-long exposure and uncertainty regarding the strains to which a given individual has been exposed. However, we have noted that serum antibodies reactive with seasonal HA proteins do not differ between subjects with high CD4 reactivity to H7 derived epitopes and those with low reactivity, so by this criterion, they do not show distinct patterns of exposure to seasonal H1N1 and H3N2 viruses and vaccines.
Based on the use of predictive algorithms for MHC class II peptide binding, H7 HA proteins have been proposed to contain a paucity of T-cell epitopes [24, 35–37]. Although not yet confirmed experimentally, this possibility suggests that recalling memory CD4 T cells generated from H3N2 and H1N1 viruses may be particularly important in generating CD4 T-cell help for responses to this avian virus. Whether or not there is a generalized defect in H7 HA to generate peptides that elicit CD4 T cells, it is clear that recall of memory CD4 T cells offers considerable advantages in immune responses. These cells exist at a higher frequency, are more readily activated to expand, and are more independent of costimulation than naive T cells [38–42]. Therefore, we would anticipate that memory CD4 cells will have a considerable advantage in potentiating the vaccine response. Our studies have revealed that cross-reactive recognition will be variable, can be recruited into the response to H7-based vaccines, and will probably depend on both influenza exposure history and the specific HLA class II molecules expressed in the host.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the New York Influenza Center of Excellence clinical core for recruitment of donors and sample handling, and we thank Dr Kanata Subbarao, MD, for generously sharing the H7N7 vaccine samples. Inactivated subvirion H7N7 vaccine was manufactured by Sanofi Pasteur and was provided by the Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH).
Financial support. This work was supported by the NIH (contracts HHSN27220201200005C, HHSN266200700008C, and R01AI51542 to A. J. S., by the Divisions of Intramural Research, NIAID, and by the Biomedical Advanced Research and Development Authority, US Department of Health and Human Services (under contract HHSN272200900026C).
Potential conflicts of interest. J. J. T. has professional relationships or consultancies with the following: Novartis Vaccines and Immune Targeting Systems, Abt Associates, Farmak PJSC, Novartis, Sanofi, GlaxoSmithKline, Pfizer, Takeda Pharmaceutical Co Ltd, Romark Laboratories, and Protein Sciences Corp. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Wang Y, Dai Z, Cheng H, et al. Towards a better understanding of the novel avian-origin H7N9 influenza A virus in China. Sci Rep 2013; 3:2318–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 2013; 368:1888–97. [DOI] [PubMed] [Google Scholar]
- 3.Kageyama T, Fujisaki S, Takashita E, et al. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill 2013; 18:20453. [PMC free article] [PubMed] [Google Scholar]
- 4.Liu D, Shi W, Shi Y, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 2013; 381:1926–32. [DOI] [PubMed] [Google Scholar]
- 5.Zhu H, Wang D, Kelvin DJ, et al. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 2013; 341:183–6. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Q, Shi J, Deng G, et al. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 2013; 341:410–4. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe T, Kiso M, Fukuyama S, et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature 2013; 501:551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morens DM, Taubenberger JK, Fauci AS. H7N9 avian influenza A virus and the perpetual challenge of potential human pandemicity. MBio 2013; 4–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinhauer DA. Influenza: pathways to human adaptation. Nature 2013; 499:412–3. [DOI] [PubMed] [Google Scholar]
- 10.Jennings LC. Influenza vaccines: an Asia-Pacific perspective. Influenza Other Respir Viruses 2013; 7(suppl 3):44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osterholm MT, Ballering KS, Kelley NS. Major challenges in providing an effective and timely pandemic vaccine for influenza A(H7N9). JAMA 2013; 309:2557–8. [DOI] [PubMed] [Google Scholar]
- 12.Sant AJ, McMichael A. Revealing the role of CD4+ T cells in viral immunity. J Exp Med 2012; 209:1391–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4+ T cells in immunity to viruses. Nat Rev Immunol 2012; 12:136–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J, Braciale TJ. Role of T cell immunity in recovery from influenza virus infection. Curr Opin Virol 2013; 3:425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strutt TM, McKinstry KK, Swain SL. Control of innate immunity by memory CD4 T cells. Adv Exp Med Biol 2011; 780:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teijaro JR, Verhoeven D, Page CA, Turner D, Farber DL. Memory CD4 T cells direct protective responses to influenza virus in the lungs through helper-independent mechanisms. J Virol 2010; 84:9217–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma CS, Deenick EK, Batten M, Tangye SG. The origins, function, and regulation of T follicular helper cells. J Exp Med 2012; 209:1241–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nayak JL, Fitzgerald TF, Richards KA, Yang H, Treanor JJ, Sant AJ. CD4+ T-cell expansion predicts neutralizing antibody responses to monovalent, inactivated 2009 pandemic influenza A(H1N1) virus subtype H1N1 vaccine. J Infect Dis 2013; 207:297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bentebibel SE, Lopez S, Obermoser G, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci Transl Med 2013; 5:176ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alam S, Knowlden ZA, Sangster MY, Sant AJ. CD4 T cell help is limiting and selective during the primary B cell response to influenza virus infection. J Virol 2014; 88:314–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards KA, Chaves FA, Sant AJ. Infection of HLA-DR1 transgenic mice with a human isolate of influenza a virus (H1N1) primes a diverse CD4 T-cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza virus. J Virol 2009; 83:6566–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards KA, Topham D, Chaves FA, Sant AJ. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J Immunol 2010; 185:4998–5002. [DOI] [PubMed] [Google Scholar]
- 23.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol 2006; 24:419–66. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen M, Lund O, Buus S, Lundegaard C. MHC class II epitope predictive algorithms. Immunology 2010; 130:319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nabel GJ, Fauci AS. Induction of unnatural immunity: prospects for a broadly protective universal influenza vaccine. Nat Med 2010; 16:1389–91. [DOI] [PubMed] [Google Scholar]
- 26.Nayak JL, Richards KA, Chaves FA, Sant AJ. Analyses of the specificity of CD4 T cells during the primary immune response to influenza virus reveals dramatic MHC-linked asymmetries in reactivity to individual viral proteins. Viral Immunol 2010; 23:169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards KA, Chaves FA, Alam S, Sant AJ. Trivalent inactivated influenza vaccines induce broad immunological reactivity to both internal virion components and influenza surface proteins. Vaccine 2012; 31:219–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulligan MJ, Bernstein DI, Winokur P, Rupp R, et al. Serological responses to an avian influenza A/H7N9 vaccine mixed at the point-of-use with MF59 adjuvant: a randomized clinical trial. JAMA 2014; 312:1409–19. [DOI] [PubMed] [Google Scholar]
- 29.Rudenko L, Kiseleva I, Naykhin AN, Erofeeva M, et al. Assessment of human immune responses to H7 avian influenza virus of pandemic potential: results from a placebo-controlled, randomized double-blind phase I study of live attenuated H7N3 influenza vaccine. PLoS One 2014; 9:e87962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couch RB, Patel SM, Wade-Bowers CL, Nino D. A randomized clinical trial of an inactivated avian influenza A (H7N7) vaccine. PLoS One 2012; 7:e49704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nayak JL, Richards KA, Yang H, Treanor JJ, Sant AJ. Effect of influenza A(H5N1) vaccine prepandemic priming on CD4+ T-cell responses. J Infect Dis 2015; 211:1408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y, Ponomarenko J, Zhu Z, Tamang D, et al. Immune epitope database analysis resource. Nucleic Acids Res 2012; 40:W525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huseby ES, White J, Crawford F, et al. How the T cell repertoire becomes peptide and MHC specific. Cell 2005; 122:247–60. [DOI] [PubMed] [Google Scholar]
- 34.Richards KA, Chaves FA, Krafcik FR, Topham DJ, Lazarski CA, Sant AJ. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J Virol 2007; 81:7608–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Groot AS, Ardito M, Terry F, et al. Low immunogenicity predicted for emerging avian-origin H7N9: implication for influenza vaccine design. Hum Vaccin Immunother 2013; 9:950–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaves FA, Lee AH, Nayak JL, Richards KA, Sant AJ. The utility and limitations of current Web-available algorithms to predict peptides recognized by CD4 T cells in response to pathogen infection. J Immunol 2012; 188:4235–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P, Sidney J, Kim Y, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics 2010; 11:568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers PR, Dubey C, Swain SL. Qualitative changes accompany memory T cell generation: faster, more effective responses at lower doses of antigen. J Immunol 2000; 164:2338–46. [DOI] [PubMed] [Google Scholar]
- 39.MacLeod MK, Kappler JW, Marrack P. Memory CD4 T cells: generation, reactivation and re-assignment. Immunology 2010; 130:10–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.London CA, Lodge MY, Abbas AK. Functional responses and costimulator dependence of memory CD4+ T cells. J Immunol 2000; 164:265–72. [DOI] [PubMed] [Google Scholar]
- 41.Lai W, Yu M, Huang MN, Okoye F, Keegan AD, Farber DL. Transcriptional control of rapid recall by memory CD4 T cells. J Immunol 2011; 187:133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar R, Ferez M, Swamy M, et al. Increased sensitivity of antigen-experienced T cells through the enrichment of oligomeric T cell receptor complexes. Immunity 2011; 35:375–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.