Abstract
Background. We investigated whether children with a higher respiratory syncytial virus (RSV) genomic load are at a higher risk of more-severe bronchiolitis.
Methods. Two multicenter prospective cohort studies in the United States and Finland used the same protocol to enroll children aged <2 years hospitalized for bronchiolitis and collect nasopharyngeal aspirates. By using real-time polymerase chain reaction analysis, patients were classified into 3 genomic load status groups: low, intermediate, and high. Outcome measures were a length of hospital stay (LOS) of ≥3 days and intensive care use, defined as admission to the intensive care unit or use of mechanical ventilation.
Results. Of 2615 enrolled children, 1764 (67%) had RSV bronchiolitis. Children with a low genomic load had a higher unadjusted risk of having a length of stay of ≥3 days (52%), compared with children with intermediate and those with high genomic loads (42% and 51%, respectively). In a multivariable model, the risk of having a length of stay of ≥3 days remained significantly higher in the groups with intermediate (odds ratio [OR], 1.43; 95% confidence interval [CI], 1.20–1.69) and high (OR, 1.58; 95% CI, 1.29–1.94) genomic loads. Similarly, children with a high genomic load had a higher risk of intensive care use (20%, compared with 15% and 16% in the groups with low and intermediate genomic loads, respectively). In a multivariable model, the risk remained significantly higher in the group with a high genomic load (OR, 1.43; 95% CI, 1.03–1.99).
Conclusion. Children with a higher RSV genomic load had a higher risk for more-severe bronchiolitis.
Keywords: bronchiolitis, infants, children, genomic load, respiratory syncytial virus, hospitalization, length of stay, intensive care unit, mechanical ventilation, cohort study
Bronchiolitis is a significant public health problem in the United States and worldwide [1–3]. In the United States, bronchiolitis is the leading cause of hospitalization for infants, accounting for 18% of all infant hospitalizations [4], and it creates an even greater outpatient burden [5]. Among many infectious pathogens, respiratory synctial virus (RSV) is the most common pathogen associated with severe bronchiolitis (ie, bronchiolitis requiring hospitalization), accounting for 50%–80% of all cases [5, 6].
Despite the substantial burden of RSV bronchiolitis, treatment remains largely supportive, and no antiviral agents are available for immunocompetent children with RSV bronchiolitis. One of the reasons for the lack of RSV treatment options is insufficient existing evidence of RSV pathogenesis that might support the putative efficacy of anti-RSV agents [7]; that is, if disease caused by RSV is the direct result of viral replication rather than the result of RSV triggering delayed and aberrant pathogenic immune response [7], then reducing RSV loads with antiviral agents might decrease RSV morbidity [8]. The limited literature about RSV-related diseases from adult and pediatric populations is conflicting, with the severity among those with higher viral load found to be no different from [9–12] or greater than [7, 13–18] that among those with a lower viral load. Moreover, data are scarce on the role of viral load in acute severity in the most relevant population: children with bronchiolitis [18–21].
To address this knowledge gap, we investigated the impact of RSV genomic load on bronchiolitis severity, using the data from 2 multicenter prospective cohort studies of children hospitalized for bronchiolitis. We used genomic load as an indirect measure of viral load. We hypothesized that children with a higher RSV genomic load have worse severity, as demonstrated by a longer length of hospital stay (LOS), admission to the intensive care unit (ICU), and increased use of mechanical ventilation.
METHODS
Study Design, Setting, and Participants
The present analysis combines the data from 2 multicenter prospective cohort studies of children hospitalized for bronchiolitis. The studies used a similar protocol, with one performed in the United States and the other performed in Finland. Both studies were performed as part of the Multicenter Airway Research Collaboration (MARC). The MARC is a program of the Emergency Medicine Network (EMNet; http://www.emnet-usa.org), a collaboration with >225 participating hospitals. The design, setting, participants, and methods of data collection used in the studies have been reported previously [6, 22–27].
In summary, using a standardized protocol [6, 22], we conducted the US study (MARC-30 USA; Supplementary Table 1) at 16 sites across 12 US states during the 2007–2010 winter seasons. The Finland study (MARC-30 Finland) was conducted at 3 Finnish sites during the 2008–2010 winter seasons. We enrolled children aged <2 years who were hospitalized for an attending physician's diagnosis of bronchiolitis and had a parent/guardian with the ability to give informed consent. We excluded children with previous enrollment and those who were transferred to a participating hospital >48 hours after the original hospitalization. Site investigators enrolled a target number of consecutive patients from the inpatient wards and the ICU, with a goal of enrolling 20% of the total sample from the ICU. To oversample children in the ICU, the ward and ICU enrollments were separate. Once the site reached their target enrollment for that month, the investigators would stop enrollment until the beginning of the following month. All patients were treated at the discretion of the treating physician. The institutional review board at each of the participating hospitals approved the study. Informed consent was obtained from the parents or guardians.
Data Collection
Investigators conducted a structured interview that assessed patients' demographic characteristics, medical and environmental history, symptom duration, and details of the acute illness. Emergency department and daily hospital chart review provided further clinical data, including vital signs, physical examinations, medical management, and disposition. We also collected follow-up data by telephone interview 2 weeks after the hospital discharge. These data were reviewed at the EMNet coordinating center, and site investigators were queried about missing data and discrepancies identified by manual data checks.
Nasopharyngeal Aspirate (NPA) Sample Collection and Virologic Testing
NPAs were collected using a standardized protocol [6, 22]. Designated site personnel were trained using a lecture, written instructions, and video. All sites used the same collection equipment (Medline Industries, Mundelein, Illinois) and collected the samples ≤24 hours after a child's arrival on the medical ward or ICU. For the collection, the child was placed supine, 1 mL of normal saline was instilled into one naris, and mucus was removed by means of an 8 French suction catheter. This procedure was performed once on each nostril. After sample collection from both nares, 2 mL of normal saline was suctioned through the catheter to clear the tubing and ensure that a standard volume of aspirate was obtained. Once collected, the NPA sample was added to transport medium at a 1:1 ratio. The samples were immediately placed on ice within 1 hour of collection and then stored at −80°C within 24 hours of collection. Frozen samples were shipped in batches on dry ice to Baylor College of Medicine, where they were stored at −80°C.
Polymerase Chain Reaction (PCR) Assay
Viral identification was performed by using singleplex or duplex 2-step real-time PCR analysis. Real-time reverse transcription PCR analysis was performed to detect RNA respiratory viruses, such as RSV types A and B; rhinovirus; coronaviruses NL-63, HKU1, OC43, and 229E; enterovirus; human metapneumovirus; influenza virus type A, type B, and 2009 novel A(H1N1), and parainfluenza virus types 1–3. Real-time PCR was used for the detection of DNA pathogens which included adenovirus, Mycoplasma pneumoniae, and Bordetella pertussis.
These tests are routinely conducted in the laboratory of one of the investigators (P. A. P.), and details of the primers and probes have been described elsewhere [28–30]. All real-time PCR assays were tested in duplicate, and samples with incongruent values (ie, those for which 1 well was positive) were retested. To reduce carryover contamination, sample preparation, RNA/DNA extraction, complementary DNA and amplification were performed in separate areas. All PCR runs had extraction and reagent positive and negative controls. These methods allowed us to detect viral pathogens in 2418 of 2615 enrolled children (93%) with a 28% prevalence of viral coinfection in the 2 original cohorts [6, 22].
RSV genomic load was quantified by real-time reverse transcription PCR as the number of amplification cycles needed for a positive PCR test (ie, the threshold cycle [CT] value). CT values provide a semiquantitative measure of viral load, with a highly significant inverse linear relationship between viral load and CT value [16, 31]. Therefore, the lower the CT value, the higher the amount of genomic material, and vice versa. For reference, a CT value of 17 equals approximately 6.4 log10 plaque-forming units (PFU)/mL of infectious RSV, a CT value of 35 equals approximately 1 log10 PFU/mL of infectious RSV, and a CT value of ≥40 is considered negative.
Outcome Measures
The primary outcome measure was a LOS of ≥3 days, defined using the median LOS of 2 days and similar to the approach used in prior studies [6, 22], as LOS was not normally distributed in the study population. The secondary outcome measure was intensive care use, defined as admission to ICU and/or use of mechanical ventilation (continuous positive airway pressure and/or intubation during inpatient stay, regardless of location) at any time during the index hospitalization [26].
Statistical Analyses
Our analyses focused on RSV, the most commonly detected virus in children with severe bronchiolitis. For the primary analysis, we combined CT values from RSV-A with those from RSV-B since the clinical distinction between the 2 subtypes of RSV was unremarkable. We categorized CT values into tertiles to classify patients into 3 RSV genomic load status groups: low, ≥24.3; intermediate, 20.8–24.2; and high, <20.8. In cases in which a subject was coinfected with RSV-A and RSV-B (n = 15), the lowest CT value (indicative of larger viral loads) was used.
We compared patient demographic characteristics, medical history, and hospital course, by RSV load status, using χ2 or Kruskal-Wallis tests as appropriate. We used a nonparametric technique for continuous variables, as these were not normally distributed in the study population. To examine the association of RSV genomic load status with the outcomes, we constructed 2 logistic regression models. First, we fitted an unadjusted model that included only RSV genomic load status as the independent variable. Second, we constructed a multivariable model controlling for 10 patient-level variables (ie, age, sex, race/ethnicity, gestational age, maternal smoking during pregnancy, history of wheezing, history of eczema, comorbid medical disorder, duration of difficulty breathing before hospitalization, and viral coinfection status [RSV plus rhinovirus and RSV plus nonrhinovirus pathogens]). We chose these potential confounders on the basis of clinical plausibility and a priori knowledge [6, 22, 23]. We did not adjust for markers of acute severity (eg, vital signs and retractions) because these were considered intermediate factors in the association of interest. In both models, we used generalized estimating equations to account for patient clustering at the hospital level.
We performed a series of sensitivity analyses to assess the robustness of our findings. First, we examined the association of RSV genomic load and the primary outcome, modeling the CT value as a continuous variable, in the US and Finnish cohort separately. Second, after confirming a similar association in the both cohorts, we combined the US and Finnish data set and then repeated the analysis by using a more restrictive definition of children with bronchiolitis (ie, those younger than 12 months with a gestational age of ≥37 weeks) [6]. Third, we repeated the analysis by classifying patients into 4 RSV genomic load groups, using CT values split into quartiles, as follows: 1 (lowest quartile), ≥25.9; 2, 22.5–25.8; 3, 20.2–22.4; and 4 (highest quartile), <20.2. Fourth, we stratified the analysis by RSV subtype (RSV-A and RSV-B). Last, we repeated the analysis in the subgroup of children with RSV monoinfection. All analyses were performed using SAS 9.3 (SAS Institute, Cary, NC). Results are presented as proportions with 95% confidence intervals (CIs), medians with interquartile ranges (IQRs), and odds ratios (ORs) with 95% CIs. All P values were 2-tailed, with a P value of < .05 considered statistically significant.
RESULTS
Patient Characteristics
Of 2615 enrolled children with severe bronchiolitis from 19 sites, 1764 (67%; 1589 in the US study and 175 in the Finnish study) had RSV bronchiolitis and were eligible for the current analysis. Among this analytic cohort, the median age was 3 months (IQR, 2–7 months), 58% were male, and 44% were non-Hispanic white. Additionally, 1219 children (69%) had bronchiolitis with RSV only, and 545 (31%) had bronchiolitis with RSV and another virus. More specifically, 297 (17%) had RSV plus rhinovirus and 248 (14%) had RSV plus nonrhinovirus pathogens. The median LOS was 2 days (IQR, 1–4 days).
Of the 1764 children in the analytic cohort, 587 (33%) were categorized into the low RSV genomic load group, 598 (34%) into the intermediate genomic load group, and 579 (33%) into the high genomic load group. Demographic characteristics differed across the genomic load groups (Table 1). Compared with children with low genomic load, children with high genomic load were younger and more likely to be male and non-Hispanic white (P < .01 for both comparisons). In contrast, other factors did not differ across the groups.
Table 1.
Demographic Characteristics and Medical History of Children Hospitalized with Respiratory Syncytial Virus (RSV) Bronchiolitis, by RSV Genomic Load Tertile
| Characteristic | RSV Genomic Load,a Children, No. (%) |
P Valueb | ||
|---|---|---|---|---|
| Low (n = 587) | Intermediate (n = 598) | High (n = 579) | ||
| Age, moc | <.001 | |||
| <2 | 165 (28) | 195 (33) | 208 (36) | |
| 2–5.9 | 225 (38) | 198 (33) | 231 (40) | |
| 6–11.9 | 115 (20) | 133 (22) | 85 (15) | |
| 12–23.9 | 82 (14) | 72 (12) | 55 (10) | |
| Male sex | 321 (55) | 341 (57) | 369 (64) | .005 |
| Race/ethnicity | <.001 | |||
| Non-Hispanic white | 201 (34) | 270 (45) | 301 (52) | |
| Non-Hispanic black | 123 (21) | 109 (18) | 110 (19) | |
| Hispanic | 238 (41) | 193 (32) | 142 (25) | |
| Other | 25 (4) | 26 (4) | 26 (5) | |
| Insurance | <.001 | |||
| Nonprivate | 443 (75) | 389 (65) | 381 (66) | |
| Private | 144 (25) | 209 (35) | 198 (34) | |
| Family history of asthma | .23 | |||
| Neither parent | 411 (70) | 396 (66) | 390 (67) | |
| Either mother or father | 143 (24) | 168 (28) | 163 (28) | |
| Both parents | 22 (4) | 25 (4) | 14 (2) | |
| Unknown/missing | 11 (2) | 9 (2) | 12 (2) | |
| Maternal smoking during pregnancy | 76 (13) | 93 (16) | 96 (17) | .19 |
| Gestational age, wk | ||||
| <32 | 29 (5) | 18 (3) | 20 (4) | |
| 32–36 | 83 (14) | 103 (17) | 95 (16) | |
| ≥37 or full term | 453 (77) | 465 (78) | 453 (78) | |
| Is or was breast-fed | 378 (64) | 386 (65) | 377 (65) | .98 |
| History of wheezing | 113 (19) | 105 (18) | 115 (20) | .58 |
| History of eczema | 94 (16) | 96 (16) | 71 (12) | .11 |
| History of intubation | 42 (7) | 44 (7) | 39 (7) | .91 |
| Major, relevant, comorbid medical disorderd | 87 (15) | 103 (17) | 88 (15) | .45 |
| Cohort | .004 | |||
| United States | 546 (93) | 538 (90) | 505 (87) | |
| Finland | 41 (7) | 60 (10) | 74 (13) | |
Percentages may not equal 100, because of rounding.
a Threshold cycle tertiles for RSV genomic loads were defined as follows: low, ≥24.3; intermediate, 20.8–24.2; and high, <20.8.
b By the χ2 test.
c Spearman correlation was also performed (P < .001).
d Defined by respiratory, cardiac, neurologic, gastrointestinal, and immunologic diseases.
Table 2 summarizes the clinical course, by RSV genomic load status. There was no statistically significant difference in the duration of symptoms and no clinically significant difference in vital signs across the groups. By contrast, children with a high genomic load were more likely to have apnea and lower weight at presentation (P ≤ .01 for both comparisons). Similarly, children with high genomic load were more likely to have sole RSV infection, particularly RSV-A infection; by contrast, they were less likely to have coinfections with rhinovirus (P < .001 for both comparisons).
Table 2.
Clinical Course of Children Hospitalized with Respiratory Syncytial Virus (RSV) Bronchiolitis, by RSV Genomic Load Tertile
| Characteristic | RSV Genomic Loada |
P Valueb | ||
|---|---|---|---|---|
| Low (n = 587) | Intermediate (n = 598) | High (n = 579) | ||
| Onset of difficulty breathing, d before hospitalizationc | .93 | |||
| ≥1 | 141 (24) | 135 (23) | 136 (24) | |
| <1 | 432 (74) | 446 (75) | 425 (73) | |
| No difficulty | 14 (2) | 17 (3) | 18 (3) | |
| Apnea documented in medical record | 42 (7) | 27 (5) | 51 (9) | .01 |
| Weight, kg | 5.9 (4.3–8.2) | 5.6 (4.1–8.1) | 5.5 (3.8–7.4) | .004d |
| Pulse, beats/min | 163 (148–177) | 160 (147–175) | 160 (145–174) | .03d |
| Respiratory rate, breaths/min | 48 (40–60) | 48 (40–60) | 48 (40–60) | .32d |
| Oxygen saturation, %, by pulse oximetry or ABG test | .15 | |||
| <90 | 64 (11) | 56 (9) | 54 (9) | |
| 90–93.9 | 114 (19) | 106 (18) | 83 (14) | |
| ≥94 | 401 (68) | 423 (71) | 428 (74) | |
| Retractions | .48 | |||
| None | 130 (22) | 107 (18) | 122 (21) | |
| Mild | 256 (44) | 263 (44) | 241 (42) | |
| Moderate or severe | 169 (29) | 184 (31) | 173 (30) | |
| Missing | 32 (6) | 44 (7) | 43 (7) | |
| Oral intake | .50 | |||
| Adequate | 270 (46) | 254 (43) | 257 (44) | |
| Inadequate | 21 (43) | 271 (45) | 245 (42) | |
| Missing | 66 (11) | 73 (12) | 77 (13) | |
| Coinfectione,f | <.001 | |||
| No coinfection | 388 (66) | 391 (65) | 440 (76) | |
| Rhinovirus | 121 (21) | 113 (19) | 63 (11) | |
| Nonrhinovirus | ||||
| Overall | 78 (13) | 94 (16) | 76 (13) | |
| Coronavirus (NL-63, HKU1, OC43, 229E) | 29 (5) | 56 (9) | 30 (5) | .002 |
| Enterovirus | 24 (4) | 21 (4) | 15 (3) | .36 |
| Human metapneumovirus | 13 (2) | 5 (1) | 6 (1) | .09 |
| Influenza virus (type A, type B, 2009 novel H1N1) | 2 (<1) | 2 (<1) | 0 (0) | .38 |
| Parainfluenza virus (types 1–3) | 5 (1) | 6 (1) | 6 (1) | .94 |
| Adenovirus | 38 (6) | 47 (8) | 27 (5) | .08 |
| M. pneumoniae | 3 (1) | 2 (<1) | 4 (1) | .69 |
| B. pertussis | 0 (0) | 0 (0) | 0 (0) | ND |
| RSV subtypeg | <.001 | |||
| RSV-A | 315 (54) | 371 (62) | 377 (65) | |
| RSV-B | 278 (47) | 232 (39) | 206 (36) | |
| Outcome | ||||
| LOS, dc | 2 (1–4) | 3 (1–4) | 3 (1–4) | .007d |
| LOS ≥3 dh | 244 (42) | 302 (51) | 300 (52) | <.001 |
| Intensive care use | 87 (15) | 93 (16) | 115 (20) | .03 |
| Intubation and/or CPAP during hospitalization | 35 (6) | 29 (5) | 55 (10) | .003 |
| ICU admission | 83 (14) | 90 (15) | 110 (19) | .06 |
Data are no. (%) of children or median value (interquartile range). Percentages may not equal 100, because of rounding.
Abbreviations: ABG, arterial blood gas; B. pertussis, Bordetella pertussis; CPAP, continuous positive airway pressure; CT, threshold cycle; ICU, intensive care unit; LOS, length of hospital stay; M. pneumoniae, Mycoplasma pneumoniae; ND, not done.
a CT tertiles for RSV genomic loads were defined as follows: low, ≥24.3; intermediate, 20.8–24.2; and high, <20.8.
b By the χ2 test, unless otherwise indicated.
c Spearman correlation was also performed with duration of difficulty breathing (P = .98) and LOS (P = .001).
d By the Kruskal-Wallis test.
e Median CT values for rhinovirus, human metapneumovirus, and adenovirus did not differ across the RSV genomic load categories (all P > .40).
f Children may have had ≥3 detected.
g Fifteen children had RSV-A and RSV-B infection.
h The proportion of children with a LOS of ≥3 days did not differ significantly between the US and Finnish cohorts (P = .11).
Risk of Longer LOS
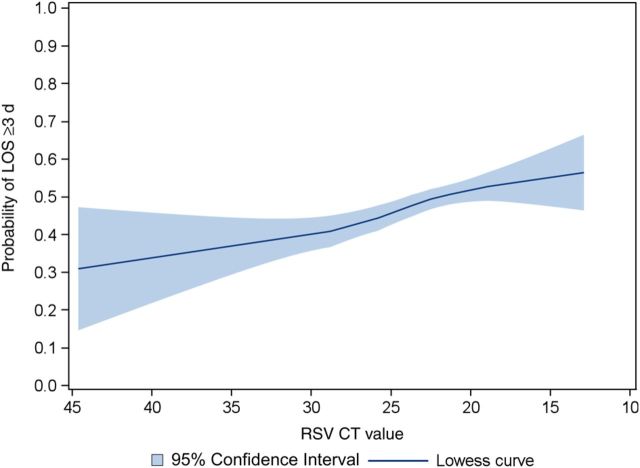
Overall, there was a positive relationship between RSV genomic load (an inverse of the CT value) and risk of a LOS of ≥3 days (Figure 1); this positive, nonlinear relationship was observed in both the US and Finnish cohort (Supplementary Figures 1 and 2). Compared with children with a low genomic load, the unadjusted risk of a LOS of ≥3 days was higher in children with an intermediate genomic load (42% vs 51%) and those with a high genomic load (42% vs 52%; P < .001 for both comparisons; Table 2). In the multivariable model controlling for 10 patient characteristics and clustering at the hospital level, the risk remained significantly higher in both children with an intermediate genomic load (OR, 1.43; 95% CI, 1.20–1.69) and those with a high genomic load (OR, 1.58; 95% CI, 1.29–1.94; P < .001 for both comparisons; Table 3). Likewise, in sensitivity analyses, the significant association between genomic load and a risk of a LOS of ≥3 days persisted with the use of the stricter definition of bronchiolitis (n = 1223; Table 3), different categories of RSV genomic load (Table 4), stratification by RSV subtype (Table 5), and in the subgroup of children with sole RSV infection (n = 1219; Table 6).
Figure 1.
Unadjusted association of respiratory syncytial virus genomic load with the risk of a length of hospital stay (LOS) of ≥3 days in children hospitalized for bronchiolitis. The fitted line represents locally weighted scatterplot smoothed (lowess) curve. Lower threshold cycle (CT) values indicate higher RSV genomic loads. There was a positive relationship between RSV genomic load (an inverse of the CT value) and the risk of a LOS of ≥3 days (P < .001).
Table 3.
Unadjusted and Multivariable Associations of Respiratory Syncytial Virus (RSV) Genomic Loads With Bronchiolitis Outcomes
| Outcome, RSV Genomic Load Categorya | Unadjusted Modelb |
Adjusted Modelc |
Sensitivity Analysisd |
|||
|---|---|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | |
| LOS ≥3 d | ||||||
| Low | Reference | Reference | Reference | |||
| Intermediate | 1.42 (1.23–1.65) | <.001 | 1.43 (1.20–1.69) | <.001 | 1.40 (1.16–1.69) | <.001 |
| High | 1.55 (1.31–1.83) | <.001 | 1.58 (1.29–1.94) | <.001 | 1.46 (1.09–1.94) | .01 |
| Intensive care use | ||||||
| Low | Reference | Reference | Reference | |||
| Intermediate | 1.09 (.87–1.36) | .46 | 1.06 (.84–1.34) | .62 | 1.24 (.85–1.82) | .26 |
| High | 1.49 (1.14–1.95) | .003 | 1.43 (1.03–1.99) | .03 | 1.36 (.91–2.03) | .14 |
Abbreviations: CI, confidence interval; LOS, length of hospital stay; OR, odds ratio.
a Threshold cycle tertiles for RSV genomic loads were defined as follows: low, ≥24.3; intermediate, 20.8–24.2; and high, <20.8.
b Unadjusted model controlling for clustering of patients within the sites, using the generalized estimating equations.
c Multivariable model controlling for 10 patient-level variables (age, sex, race/ethnicity, gestational age, maternal smoking during pregnancy, history of wheezing, history of eczema, comorbid medical disorder, duration of difficulty breathing before hospitalization, viral coinfection status [RSV plus rhinovirus and RSV plus nonrhinovirus pathogens]) and clustering of patients within the sites.
d Multivariable model using a restrictive definition of children with bronchiolitis (ie, those younger than 12 months with gestational age of ≥37 weeks; n = 1223).
Table 4.
Unadjusted and Multivariable Associations of Respiratory Syncytial Virus (RSV) Genomic Loads With Bronchiolitis Outcomes
| Outcome, RSV Genomic Load Quartilea | Unadjusted Modelb |
Adjusted Modelc |
||
|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | |
| LOS ≥3 d | ||||
| 1 | Reference | … | Reference | … |
| 2 | 1.29 (.93–1.78) | .13 | 1.26 (.92–1.74) | .15 |
| 3 | 1.65 (1.35–2.00) | <.001 | 1.65 (1.33–2.04) | <.001 |
| 4 | 1.69 (1.31–2.19) | <.001 | 1.73 (1.29–2.32) | <.001 |
| Intensive care use | ||||
| 1 | Reference | … | Reference | … |
| 2 | 1.09 (.77–1.53) | .64 | 1.07 (.77–1.48) | .69 |
| 3 | 1.15 (.78–1.70) | .49 | 1.07 (.70–1.64) | .76 |
| 4 | 1.60 (1.11–2.31) | .01 | 1.51 (1.00–2.28) | .048 |
Abbreviations: CI, confidence interval; LOS, length of hospital stay; OR, odds ratio.
a Threshold cycle quartiles for RSV genomic loads were defined as follows: 1 (lowest quartile), ≥25.9; 2, 22.5–25.8; 3, 20.2–22.4; and 4 (highest quartile), <20.2.
b Unadjusted model controlling for clustering of patients within the sites, using the generalized estimating equations.
c Multivariable model controlling for 10 patient-level variables (age, sex, race/ethnicity, gestational age, maternal smoking during pregnancy, history of wheezing, history of eczema, comorbid medical disorder, duration of difficulty breathing before hospitalization, and viral coinfection status [RSV plus rhinovirus and RSV plus nonrhinovirus pathogens]) and clustering of patients within the sites.
Table 5.
Unadjusted and Multivariable Associations of Respiratory Syncytial Virus (RSV) Genomic Loads with Bronchiolitis Outcomes, According to RSV Subtype
| Outcome, RSV Genomic Load Tertilea | RSV-A |
RSV-B |
||||||
|---|---|---|---|---|---|---|---|---|
| Unadjusted Modelb |
Adjusted Modelc |
Unadjusted Modelb |
Adjusted Modelc |
|||||
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | |
| LOS ≥3 d | ||||||||
| Low | Reference | Reference | Reference | Reference | ||||
| Intermediate | 1.33 (1.02–1.73) | .04 | 1.32 (.97–1.80) | .08 | 1.54 (1.14–2.07) | .004 | 1.57 (1.09–2.26) | .02 |
| High | 1.41 (1.05–1.91) | .03 | 1.47 (1.02–2.10) | .04 | 1.77 (1.24–2.52) | .002 | 1.85 (1.32–2.58) | <.001 |
| Intensive care use | ||||||||
| Low | Reference | Reference | Reference | Reference | ||||
| Intermediate | 0.93 (.73–1.19) | .57 | 0.89 (.66–1.20) | .44 | 1.31 (.82–2.12) | .26 | 1.39 (.82–2.36) | .22 |
| High | 1.15 (.90–1.45) | .26 | 1.09 (.81–1.47) | .57 | 2.13 (1.16–3.93) | .02 | 2.12 (1.05–4.27) | .04 |
Abbreviations: CI, confidence interval; LOS, length of hospital stay; OR, odds ratio.
a Threshold cycle tertiles for RSV genomic loads were defined as follows: low, ≥24.3; intermediate, 20.8–24.2; and high, <20.8.
b Unadjusted model controlling for clustering of patients within the sites, using the generalized estimating equations.
c Multivariable model controlling for 10 patient-level variables (age, sex, race/ethnicity, gestational age, maternal smoking during pregnancy, history of wheezing, history of eczema, comorbid medical disorder, duration of difficulty breathing before hospitalization, and viral coinfection status [RSV plus rhinovirus and RSV plus nonrhinovirus pathogens]) and clustering of patients within the sites.
Table 6.
Unadjusted and Multivariable Associations of Respiratory Syncytial Virus (RSV) Genomic Loads With Bronchiolitis Outcomes in Children With RSV Monoinfection
| Outcome, RSV Genomic Load Tertile | Unadjusted Modela |
Adjusted Modelb |
||
|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | |
| LOS ≥3 d | ||||
| Low | Reference | Reference | ||
| Intermediate | 1.53 (1.17–1.99) | .002 | 1.43 (1.06–1.95) | .02 |
| High | 1.76 (1.36–2.28) | <.001 | 1.70 (1.30–2.23) | <.001 |
| Intensive care use | ||||
| Low | Reference | Reference | ||
| Intermediate | 1.37 (1.03–1.82) | .03 | 1.24 (.93–1.65) | .15 |
| High | 1.87 (1.37–2.56) | <.001 | 1.69 (1.21–2.36) | .002 |
Abbreviations: CI, confidence interval; LOS, length of hospital stay; OR, odds ratio.
a Threshold cycle tertiles for RSV genomic loads were defined as follows: low, ≥24.3; intermediate, 20.8–24.2; and high, <20.8.
b Unadjusted model controlling for clustering of patients within the sites, using the generalized estimating equations.
c Multivariable model controlling for 9 patient-level variables (age, sex, race/ethnicity, gestational age, maternal smoking during pregnancy, history of wheezing, history of eczema, comorbid medical disorder, and duration of difficulty breathing before hospitalization) and clustering of patients within the sites.
Risk of Intensive Care Use
Similar to the analysis of primary outcome, compared with children with a low RSV genomic load, those with a high genomic load had a higher risk of intensive care use (15% vs 20%; P = .003). In the multivariable model, the risk remained significantly higher in children with a high genomic load (OR, 1.43; 95% CI, 1.03–1.99; P = .03; Table 3). In sensitivity analyses with a limited statistical power, children with a high genomic load had a nonsignificantly increased odds of intensive care use among those with the more restrictive definition of bronchiolitis (Table 3) and those with RSV-A infection (Table 5). With the use of different categories of genomic load, among children with RSV-B infection and those with sole RSV infection, the adjusted association remained significant (Tables 4–6).
DISCUSSION
In this multicenter analysis based on 2 multiyear prospective cohort studies of children hospitalized for bronchiolitis in the United States and Finland, we identified 1764 children with RSV infection. In this analytic cohort, using a standardized approach to NPA sample collection and relative quantification of viral load, we found that children with high RSV genomic loads had a significantly higher risk of a longer LOS and of intensive care use than their counterparts with low genomic loads. To our knowledge, this is the largest study to have examined the association between RSV genomic load and acute severity in children with bronchiolitis. Our data corroborate and build on previous reports about the association between viral load and bronchiolitis outcomes [18, 20, 21], a finding of clinical and research importance.
Previous studies on acute respiratory infection with RSV in children [10–13, 15–18], adults [19, 20], and healthy volunteers [7] have reported inconsistent associations between RSV loads and severity of illness. Moreover, data are scarce in children with bronchiolitis. Single-center studies with small sample sizes (77–219 children) have shown that the severity of RSV bronchiolitis with higher viral loads is no different from [19] or greater than [18, 20, 21] that for those with lower viral loads. Potential explanations of the inconsistency across studies include differences in study design, patient populations, and/or outcomes. Furthermore, the studies that did not find a significant association may have been confounded by the nonstandardized technique used in specimen collections within the studies [11, 12, 19] and the use of different analytic techniques (eg, quantitative culture) for viral load measurement [19]. Real-time PCR has added another dimension to the quantitation of virus replication. Genomic load might be a better marker than quantitative culture of virus replication in the respiratory tract because it is not impacted by neutralizing activity found in respiratory secretions [7]. In contrast, the validity of our findings is buttressed by the use of a standardized protocol for NPA collection and processing [6, 22] and by the use of real-time PCR for measurement of RSV genomic loads.
While it may seem intuitive that children with a higher RSV load would have a more severe clinical course, diseases caused by RSV have long been thought to be the result of RSV triggering of an exaggerated T-helper type 2 (Th2) response, based on the results of rodent model studies [32, 33]. However, a human experimental model of RSV infection demonstrated that the RSV loads are positively correlated not only with the amount of non-Th2 proinflammatory markers (eg, interleukin 6 and interleukin 8), but also with disease severity [7]. Similarly, a recent clinical trial in healthy adults experimentally infected with RSV also demonstrated that treatment with an antiviral agent (GS-5806) reduced both viral load and symptom score [8]. In studies of infants with RSV bronchiolitis, researchers reported a strong activation of the interferon system (types I and II) [34] and positive associations between high RSV genomic load and expressions of interferon-stimulated gene 56 (ISG56) in the airways and a longer LOS [21]. Our multicenter prospective studies, with a sample size many times larger than any other prior study on this topic, corroborate these findings and extend them by demonstrating the robustness of the associations between RSV genomic load and acute severity of illness in the real-world setting of children hospitalized with bronchiolitis. Now, several rigorously designed studies have arrived at similar conclusions, despite the apparent inconsistencies in the literature. Collectively, these observations argue against a pathogenesis hypothesis involving RSV triggering delayed pathogenic immune response and, conversely, lend significant support to the concept that reductions of viral load with a robust antiviral agent may decrease disease severity in children with bronchiolitis.
We acknowledge several potential limitations. First, bronchiolitis is a clinical diagnosis without a common international definition [3, 35]. Thus, it is possible that we enrolled children with other respiratory disorders. However, the significant association of high genomic load with a longer LOS persisted even when the data were restricted to children with classic bronchiolitis (ie, those younger than 12 months with a gestational age of ≥37 weeks). Second, we evaluated RSV genomic loads only in the upper respiratory tract. However, previous studies have shown that RSV load in nasal wash among intubated infants closely resembles that in tracheal aspirate [36, 37]. Therefore, the nasopharyngeal RSV load in children may well be indicative of the viral load in the lower respiratory tract. Third, we measured genomic loads in the first 24 hours of hospitalization. Longitudinal studies are warranted to address questions regarding the relationship between persistence of RSV nucleic acids and clinical course. However, prior studies indicate that the peak RSV loads in hospitalized infants are seen shortly after hospitalization [18, 38]. Therefore, the genomic loads measured in our 2 cohorts likely reflect the maximum viral loads. Fourth, we did not adjust the RSV CT values on the basis of the quality of the sample. It is likely that poor-quality samples will result in genomic CT values that underestimate their true genomic CT value. Poor-quality samples will add to the variability in CT values, making it more difficult to identify a significant association between genomic load and severity of RSV bronchiolitis, thereby biasing our conclusions toward the null. Furthermore, we attempted to minimize the collection of poor-quality samples by training site personnel with lecture, written instructions, and video to collect NPA by means of a standardized protocol. Fifth, in these multicenter observational studies, we did not use standard criteria for hospital discharge or intensive care use. Therefore, institutional variability in care is possible. However, the association of RSV genomic load with LOS was observed in the both cohorts, and the significant association persisted after adjustment for clustering at the hospital level. Last, our study populations consisted of children hospitalized for bronchiolitis in academic centers. Therefore, our inferences may not be generalizable to children with bronchiolitis in community medical centers or those in the ambulatory care setting. Nevertheless, our inferences are of direct relevance to hundreds of thousands of children with severe bronchiolitis every year [4].
In summary, on the basis of 2 multicenter prospective studies of 1764 children hospitalized for RSV bronchiolitis in the United States and Finland, we found that children with high genomic loads had a significantly higher risk of a longer LOS and of intensive care use. For clinicians, evidence to predict the disease course in children with severe bronchiolitis remains limited [4], which is well documented by marked variability in acute management and disposition in this population [39]. Our findings suggest a new line of research using genomic loads as a means to improve the ability of clinicians to predict the disease course and outcomes in children with bronchiolitis. Furthermore, our data support the concept that reductions of RSV (genomic) load may decrease disease severity in children with bronchiolitis. Our findings, therefore, should facilitate further research to develop robust antiviral agents that reduce viral load and could thereby decrease bronchiolitis morbidity.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Disclaimer. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases (NIAID) or the National Institutes of Health (NIH).
Financial support. This work was supported by the National Institutes of Health (grant U01 AI-67693). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or NIH.
Potential conflicts of interest. J. M. M. has provided bronchiolitis-related consultation for Regeneron. P. A. P. has received grants from Gilead and provided bronchiolitis-related consultation for Regeneron, Alios, Gilead, and Teva. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Nair H, Nokes DJ, Gessner BD, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 2010; 375:1545–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zorc JJ, Hall CB. Bronchiolitis: recent evidence on diagnosis and management. Pediatrics 2010; 125:342–9. [DOI] [PubMed] [Google Scholar]
- 3.Hasegawa K, Mansbach JM, Camargo CA., Jr Infectious pathogens and bronchiolitis outcomes. Expert Rev Anti Infect Ther 2014; 12:817–28. [DOI] [PubMed] [Google Scholar]
- 4.Hasegawa K, Tsugawa Y, Brown DF, Mansbach JM, Camargo CA., Jr Trends in bronchiolitis hospitalizations in the United States, 2000–2009. Pediatrics 2013; 132:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasegawa K, Tsugawa Y, Brown DF, Mansbach JM, Camargo CA., Jr Temporal trends in emergency department visits for bronchiolitis in the United States, 2006–2010. Pediatr Infect Dis J 2014; 33:11–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansbach JM, Piedra PA, Teach SJ, et al. Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis. Arch Pediatr Adolesc Med 2012; 166:700–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeVincenzo JP, Wilkinson T, Vaishnaw A, et al. Viral load drives disease in humans experimentally infected with respiratory syncytial virus. Am J Respir Crit Care Med 2010; 182:1305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeVincenzo JP, Whitley RJ, Mackman RL, et al. Oral GS-5806 activity in a respiratory syncytial virus challenge study. N Engl J Med 2014; 371:711–22. [DOI] [PubMed] [Google Scholar]
- 9.Walsh EE, Peterson DR, Kalkanoglu AE, Lee FE, Falsey AR. Viral shedding and immune responses to respiratory syncytial virus infection in older adults. J Infect Dis 2013; 207:1424–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin ET, Kuypers J, Wald A, Englund JA. Multiple versus single virus respiratory infections: viral load and clinical disease severity in hospitalized children. Influenza Other Respir Virus 2012; 6:71–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franz A, Adams O, Willems R, et al. Correlation of viral load of respiratory pathogens and co-infections with disease severity in children hospitalized for lower respiratory tract infection. J Clin Virol 2010; 48:239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansen RR, Schinkel J, Dek I, et al. Quantitation of respiratory viruses in relation to clinical course in children with acute respiratory tract infections. Pediatr Infect Dis J 2010; 29:82–4. [DOI] [PubMed] [Google Scholar]
- 13.Buckingham SC, Bush AJ, Devincenzo JP. Nasal quantity of respiratory syncytical virus correlates with disease severity in hospitalized infants. Pediatr Infect Dis J 2000; 19:113–7. [DOI] [PubMed] [Google Scholar]
- 14.Duncan CB, Walsh EE, Peterson DR, Lee FE, Falsey AR. Risk factors for respiratory failure associated with respiratory syncytial virus infection in adults. J Infect Dis 2009; 200:1242–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houben ML, Coenjaerts FE, Rossen JW, et al. Disease severity and viral load are correlated in infants with primary respiratory syncytial virus infection in the community. J Med Virol 2010; 82:1266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Do LA, van Doorn HR, Bryant JE, et al. A sensitive real-time PCR for detection and subgrouping of human respiratory syncytial virus. J Virol Methods 2012; 179:250–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuller JA, Njenga MK, Bigogo G, et al. Association of the CT values of real-time PCR of viral upper respiratory tract infection with clinical severity, Kenya. J Med Virol 2013; 85:924–32. [DOI] [PubMed] [Google Scholar]
- 18.El Saleeby CM, Bush AJ, Harrison LM, Aitken JA, Devincenzo JP. Respiratory syncytial virus load, viral dynamics, and disease severity in previously healthy naturally infected children. J Infect Dis 2011; 204:996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright PF, Gruber WC, Peters M, et al. Illness severity, viral shedding, and antibody responses in infants hospitalized with bronchiolitis caused by respiratory syncytial virus. J Infect Dis 2002; 185:1011–8. [DOI] [PubMed] [Google Scholar]
- 20.Fodha I, Vabret A, Ghedira L, et al. Respiratory syncytial virus infections in hospitalized infants: association between viral load, virus subgroup, and disease severity. J Med Virol 2007; 79:1951–8. [DOI] [PubMed] [Google Scholar]
- 21.Scagnolari C, Midulla F, Selvaggi C, et al. Evaluation of viral load in infants hospitalized with bronchiolitis caused by respiratory syncytial virus. Med Microbio Immunol 2012; 201:311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jartti T, Aakula M, Mansbach JM, et al. Hospital length-of-stay is associated with rhinovirus etiology of bronchiolitis. Pedatr Infect Dis J 2014; 33:829–34. [DOI] [PubMed] [Google Scholar]
- 23.Mansbach JM, Piedra PA, Stevenson MD, et al. Prospective multicenter study of children with bronchiolitis requiring mechanical ventilation. Pediatrics 2012; 130:e492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroeder AR, Mansbach JM, Stevenson M, et al. Apnea in children hospitalized with bronchiolitis. Pediatrics 2013; 132:e1194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasegawa K, Mansbach JM, Teach SJ, et al. Multicenter study of viral etiology and relapse in hospitalized children with bronchiolitis. Pediatr Infect Dis J 2014; 33:809–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasegawa K, Pate BM, Mansbach JM, et al. Risk factors for requiring intensive care among children admitted to ward with bronchiolitis. Acad Pediatr 2014. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santiago J, Mansbach JM, Chou SC, et al. Racial/Ethnic differences in the presentation and management of severe bronchiolitis. J Hosp Med 2014; 9:565–72. [DOI] [PubMed] [Google Scholar]
- 28.Beckham JD, Cadena A, Lin J, et al. Respiratory viral infections in patients with chronic, obstructive pulmonary disease. J Infect 2005; 50:322–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knorr L, Fox JD, Tilley PA, Ahmed-Bentley J. Evaluation of real-time PCR for diagnosis of Bordetella pertussis infection. BMC Infect Dis 2006; 6:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winchell JM, Thurman KA, Mitchell SL, Thacker WL, Fields BS. Evaluation of three real-time PCR assays for detection of Mycoplasma pneumoniae in an outbreak investigation. J Clin Microbiol 2008; 46:3116–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borg I, Rohde G, Loseke S, et al. Evaluation of a quantitative real-time PCR for the detection of respiratory syncytial virus in pulmonary diseases. Eur Respir J 2003; 21:944–51. [DOI] [PubMed] [Google Scholar]
- 32.Openshaw PJ, Tregoning JS. Immune responses and disease enhancement during respiratory syncytial virus infection. Clin Microbio Rev 2005; 18:541–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tripp RA. Pathogenesis of respiratory syncytial virus infection. Viral Immunol 2004; 17:165–81. [DOI] [PubMed] [Google Scholar]
- 34.Bucasas KL, Mian AI, Demmler-Harrison GJ, et al. Global gene expression profiling in infants with acute respiratory syncytial virus broncholitis demonstrates systemic activation of interferon signaling networks. Pediatr Infect Dis J 2013; 32:e68–76. [DOI] [PubMed] [Google Scholar]
- 35.American Academy of Pediatrics Subcommittee on Diagnosis and Management of Bronchiolitis. Diagnosis and management of bronchiolitis. Pediatrics 2006; 118:1774–93. [DOI] [PubMed] [Google Scholar]
- 36.Malley R, DeVincenzo J, Ramilo O, et al. Reduction of respiratory syncytial virus (RSV) in tracheal aspirates in intubated infants by use of humanized monoclonal antibody to RSV F protein. J Infect Dis 1998; 178:1555–61. [DOI] [PubMed] [Google Scholar]
- 37.Malley R, Vernacchio L, Devincenzo J, et al. Enzyme-linked immunosorbent assay to assess respiratory syncytial virus concentration and correlate results with inflammatory mediators in tracheal secretions. Pediatr Infect Dis J 2000; 19:1–7. [DOI] [PubMed] [Google Scholar]
- 38.Gerna G, Campanini G, Rognoni V, et al. Correlation of viral load as determined by real-time RT-PCR and clinical characteristics of respiratory syncytial virus lower respiratory tract infections in early infancy. J Clin Virol 2008; 41:45–8. [DOI] [PubMed] [Google Scholar]
- 39.Christakis DA, Cowan CA, Garrison MM, Molteni R, Marcuse E, Zerr DM. Variation in inpatient diagnostic testing and management of bronchiolitis. Pediatrics 2005; 115:878–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.