Abstract
Prader-Willi syndrome (PWS), a disorder of genomic imprinting, is characterized by neonatal hypotonia, hypogonadism, small hands and feet, hyperphagia and obesity in adulthood. PWS results from the loss of paternal copies of the cluster of SNORD116 C/D box snoRNAs and their host transcript, 116HG, on human chromosome 15q11-q13. We have investigated the mechanism of repression of the maternal SNORD116 cluster and 116HG. Here, we report that the zinc-finger protein ZNF274, in association with the histone H3 lysine 9 (H3K9) methyltransferase SETDB1, is part of a complex that binds to the silent maternal but not the active paternal alleles. Knockdown of SETDB1 in PWS-specific induced pluripotent cells (iPSCs) causes a decrease in the accumulation of H3K9 trimethylation (H3K9me3) at 116HG and corresponding accumulation of the active chromatin mark histone H3 lysine 4 dimethylation (H3K4me2). We also show that upon knockdown of SETDB1 in PWS-specific iPSCs, expression of maternally silenced 116HG RNA is partially restored. SETDB1 knockdown in PWS iPSCs also disrupts DNA methylation at the PWS-IC where a decrease in 5-methylcytosine is observed in association with a concomitant increase in 5-hydroxymethylcytosine. This observation suggests that the ZNF274/SETDB1 complex bound to the SNORD116 cluster may protect the PWS-IC from DNA demethylation during early development. Our findings reveal novel epigenetic mechanisms that function to repress the maternal 15q11-q13 region.
INTRODUCTION
Prader-Willi syndrome (PWS), a neurobehavioral disorder of genomic imprinting, is caused by the absence of a normal paternal contribution to chromosome 15q11-q13, most commonly resulting from large deletion of the ∼5000 kb imprinted region (1). PWS is characterized by neonatal hypotonia and failure to thrive, followed by hyperphagia/obesity; small stature, hands and feet; mild-to-moderate cognitive deficit; relatively low sensitivity to pain; and distinctive behavioral problems (1). Genomic imprinting regulates parent-of-origin-specific expression of genes on chromosome 15q11-q13. The bi-partite PWS-Imprinting Center (PWS-IC), a region of differential DNA methylation that encompasses the promoter and first exon of SNURF-SNRPN, controls imprinting of 15q11-q13 (2, 3). It is unmethylated on the paternal allele and methylated on the maternal allele. Several genes are exclusively expressed from the paternally inherited allele of chromosome 15q11-q13 including MAGEL2, NDN and SNURF-SNRPN and silenced on the maternal allele (4–10). A long noncoding antisense RNA (lncRNA-ATS) initiates within and upstream of the PWS-IC and extends distally to overlap UBE3A in neurons thereby silencing the paternal UBE3A allele via an antisense-mediated mechanism (8, 11). The lncRNA-ATS serves as host to several C/D box small nucleolar RNAs (snoRNAs), including the SNORD116 (HBII-85) and SNORD115 (HBII-52) clusters (8, 9). The expression and processing of the lncRNA-ATS is tissue specific. The more proximal segment of the lncRNA-ATS, encompassing the SNORD116 cluster and its processed host gene 116HG, is expressed in brain, kidney, muscle, lung and other tissues. The more distal transcripts, including the SNORD115 cluster, its host gene 115HG, and UBE3A-ATS are exclusively expressed in neurons and regulated by upstream exons of the SNURF-SNRPN gene (11–13). Moreover, it has been suggested that expression of the SNORD116 and SNORD115 clusters may be mediated by two independent transcription units that generate metabolically stable nuclear RNA species in neurons (14). In mouse, all of the downstream noncoding exons of Snurf-Snrpn are brain specific (9).
The PWS critical region (PWSCR) has been narrowed to a 108 kb region encompassing SNORD109A, the SNORD116 cluster and IPW (15–17) and to a similar region in mouse (18). While additional genes within the large chromosome 15q11-q13 deletion may contribute to the PWS phenotype (19–22), there is increasing evidence that deletion of the SNORD116 cluster alone causes the majority of the PWS clinical manifestations. In this regard, it has been reported that the processed 116HG transcript forms a subnuclear RNA cloud that modulates the expression of many neuronal transcripts (23). The 116HG RNA cloud disappears in the brains of mice with a deletion of Snord116 resulting in the dysregulation of genes including some involved in the circadian clock and in energy expenditure. These findings are the first to suggest a PWS disease mechanism and strongly implicate the loss of 116HG in PWS (23). There is sequence and gene expression heterogeneity within repeats of the SNORD116 cluster (24). Group 1 snoRNAs (SNORD116-1 to SNORD116-9) (SNOG1/host transcript 116HGG1) are expressed at least 20-fold higher in hypothalalmus than Group 2 (SNORD116-10 to SNORD116-24; SNOG2/116HGG2) and Group 3 (SNORD116-25 to SNORD116-29; SNOG3/116HGG3) snoRNAs, and therefore, SNOG1/116HGG1 appears to be most relevant to the PWS disease mechanism (24). Group 1 also appears to be the most highly expressed Snord116 in mouse brain (25).
In order to further understand the PWS disease mechanism and the mechanism of silencing of maternal alleles in PWS, we used induced pluripotent stem cells (iPSCs) from two different PWS classes: (i) the typical chromosome 15q11-q13 large deletion patient (PWS) (13) and (ii) a patient with a small deletion that encompasses the SNORD116 snoRNA cluster (PWS SD) (12, 15) to investigate the role of ZNF274, SETDB1 and trimethylation of histone H3 at lysine 9 (H3K9me3) in the repression of the maternal SNORD116 cluster . For controls, we also used Angelman syndrome (AS) iPSCs derived from a patient with a large deletion of maternal 15q11-q13, and a normal (NML) control (13). AS is caused by deletions of maternal chromosome 15q11-q13 and PWS is caused by the same deletion that occurs on the paternally inherited allele (13).
Our experiments using these cell lines reveal a novel mechanism of epigenetic regulation within the PWSCR.
RESULTS
An examination of the ENCODE database revealed binding sites for the zinc-finger protein ZNF274 mapping to the SNOG1/116HGG1 cluster in the PWS region in human embryonic stem cells (hESCs) (Supplementary Material, Fig. S1A; Fig. 1A). This was of interest since fewer than 500 ZNF274 binding sites are detected in genome-wide chromatin immunoprecipitation (ChIP) analyses (26). Binding of ZNF274 to a target chromosomal segment is hypothesized to effect chromatin silencing via recruitment of the histone methyltransferase SETDB1and the co-repressor TRIM28/KAP1. SETDB1 is a mediator of the repressive histone modification, histone H3 lysine 9 trimethylation (H3K9me3) and TRIM28 acts a molecular scaffold to recruit chromatin modification and remodeling factors that are associated with repressive chromatin (26). Interestingly, increased accumulation of H3K9me3 in the SNORD116 cluster in the hESCs is apparent upon examination of the ENCODE database (Supplementary Material, Fig. S1A).
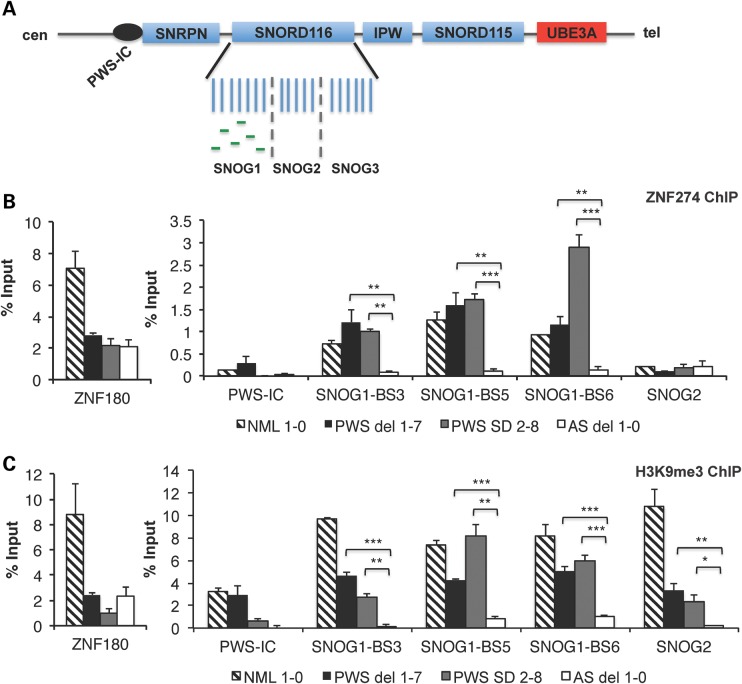
Figure 1.
ZNF274 is a novel epigenetic regulator of the silent maternal allele in PWS. (A) A map of a subregion of 15q11-q13, indicating ZNF274 binding sites in SNOG1/116HGG1 within the SNORD116 cluster. Imprinted, paternally expressed genes are denoted by blue boxes. UBE3A is biallelically expressed in iPSCs. The PWS-IC is denoted by the black circle. Green lines within SNOG1/116HGG1 represent the six ZNF274 binding sites. (B) ZNF274 ChIP assays in iPSCs. Allele specificity was determined by comparing the enrichment observed on the silent maternal allele in PWS iPSCs derived from two classes of PWS patients to the active paternal allele in AS iPSCs. Enrichment at ZNF180 was used as a positive control in these assays. Quantification of ChIPs was performed and calculated as percent input for each sample. Non-specific IgG values were subtracted from percent input values. (C) ChIP assays for the repressive histone modification H3K9me3. H3K9me3 was enriched on maternal allele in iPSCs derived from all three classes of PWS patients, when compared with AS iPSCs. *P ≤ 0.1, **P ≤ 0.05, ***P ≤ 0.01.
To investigate the possibility that H3K9me3 deposition mediated by a SETDB1-containing molecular complex could function to repress the maternal SNORD116/116HG RNAs in early development, we asked whether ZNF274 binding and H3K9me3 accumulation were allele specific using iPSCs derived from AS and PWS patients with deletions of maternal and paternal chromosome 15q11-q13, respectively. We performed ChIP assays in PWS del 1–7, PWS SD 2–8 and AS del 1-0 iPSCs in order to distinguish the maternal- and paternal-specific alleles of SNORD116/116HG. Specifically, we sought to determine whether H3K9me3 accumulation and ZNF274 binding are enriched in an allele-specific manner at the six binding sites (BS1–BS6) within SNOG1/116HGG1 observed for hESCs (Supplementary Material, Fig. S1A; Fig. 1A). ZNF274 ChIP in PWS and AS iPSCs revealed that ZNF274 binding is allele specific at all six binding sites (Fig. 1B; Supplementary Material, Fig. S1B) with significant enrichment on the silent SNOG1/116HGG1 alleles in PWS but not on the active paternal alleles in AS iPSCs (Fig. 1B; Supplementary Material, Fig. S1B). ZNF274 binding is also detected at the six PWSCR binding sites in NML iPSCs (Fig. 1B; Supplementary Material, Fig. S1B) and, as a control, at a previously characterized ZNF274 binding site (26) in the 3′UTR of ZNF180, located on chromosome 19 (Fig. 1B). We also investigated the allele specificity of the repressive H3K9me3 modification at the ZNF274 binding sites SNOG1-BS3, SNOG1-BS5 and SNOG1-BS6 within SNORD116 and observed significant enrichment for H3K9me3 at the repressed maternal SNOG1/116HGG1 allele in PWS iPSCs, but not on the active paternal alleles in AS iPSCs (Fig. 1C). We also observed some spreading of H3K9me3 accumulation to the more distal SNOG2/116HGG2 (Fig. 1C), a finding consistent the diffuse pattern of the H3K9me3 signal in hESCs (Supplementary Material, Fig. S1A). We conclude that ZNF274 binding and the associated H3K9me3 deposition at SNOG1/116HGG1 are allele specific and function as a novel epigenetic signature at the maternal PWSCR.
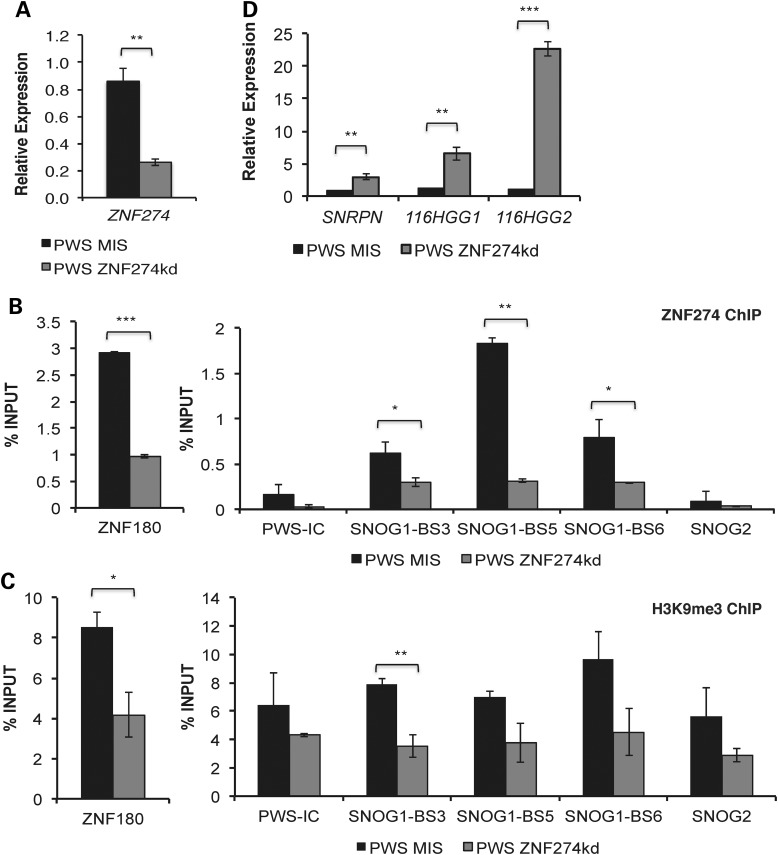
In order to further investigate the functional role of this putative PWSCR repressive complex, we knocked down the expression of ZNF274 by RNA interference in PWS del 1–7 iPSCs using a pool of three siRNA constructs directed against ZNF274 mRNA or a mismatch (MIS) negative control. The expression levels of ZNF274 were reduced by 70% in the PWS del 1–7 ZNF274 knockdown (ZNF274kd) iPSCs relative to the MIS control (Fig. 2A). To determine if ZNF274 knockdown was correlated with a decrease in ZNF274 binding within SNORD116, we performed ChIP analysis in PWS ZNF274kd iPSCs. ZNF274 enrichment was decreased within SNOG1 binding sites in the PWS ZNF274kd iPSCs relative to MIS control (Fig. 2B). Accumulation of the repressive histone modification H3K9me3 was decreased in PWS ZNF274kd iPSCs at the control locus ZNF180 and at SNOG1-BS3 (Fig. 2C). It should be noted that although a trend was observed for a reduction in H3K9me3 levels at other sites within 15q11-q13, these changes did not achieve statistical significance. These results suggest that ZNF274 may be involved in the allele-specific deposition of the repressive H3K9me3 mark at the maternal SNORD116 cluster. The reduction of ZNF274 binding at SNOG1 in PWS ZNF274kd iPSCs also resulted in an increase in SNRPN, 116HGG1 and 116HGG2 expression levels, when compared with MIS controls (Fig. 2D). However, we estimate that the SNORD116 expression levels are ∼1000× less than those of active paternal alleles. Nevertheless, these results raise the possibility that expression of the silent maternal PWSCR can be re-activated by disrupting ZNF274 and potentially other interacting partners.
Figure 2.
Functional role of ZNF274 complex in the silencing of the maternal allele in PWS. (A) Gene expression of ZNF274 in PWS iPSCs following transfection with control siRNAs or specific siRNAs targeting ZNF274. GADPH was used as an endogenous control. Data were normalized to PWS MIS. ChIP assays for ZNF274 (B) and H3K9me3 (C), respectively, in PWS ZNF274kd iPSCs. Enrichment at ZNF180 was used as a control in these assays. (D) Gene expression analysis of SNRPN and 116HG in PWS ZNF274kd iPSCs. GADPH was used as an endogenous control and data were normalized to PWS MIS. * P ≤ 0.1, ** P ≤ 0.05, *** P ≤ 0.01.
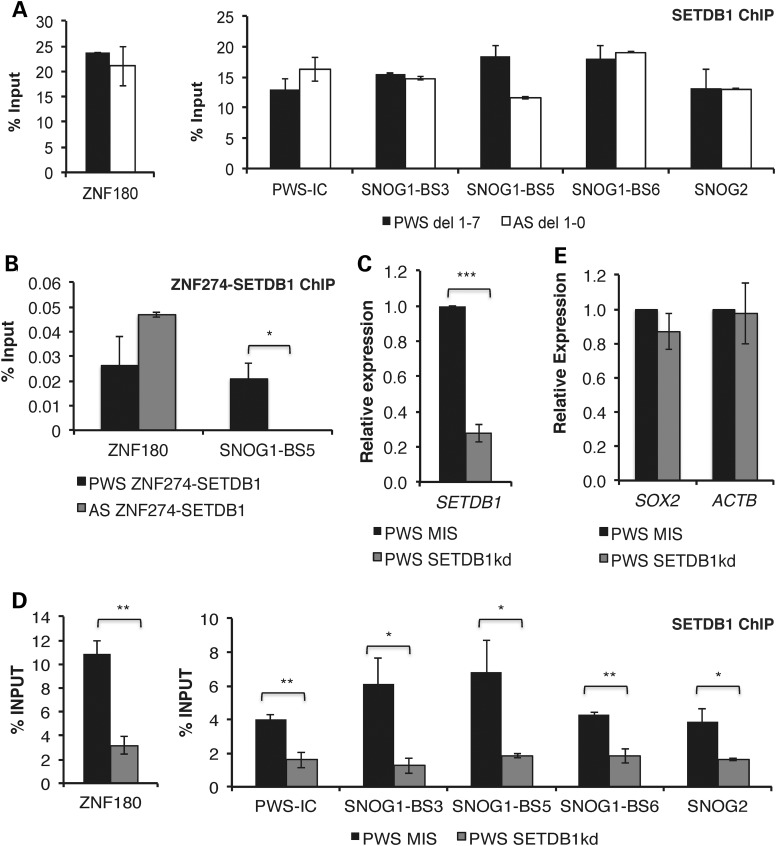
Given the report that ZNF274 recruits the histone methyltransferase SETDB1 to specific regions within the genome (26), we asked whether the binding of SETDB1 within 15q11-q13 was also allele specific. In contrast to ZNF274 and H3K9me3, we found that SETDB1 is enriched on both maternal and paternal alleles of SNOG1/116HG1 (Fig. 3A). To determine whether SETDB1 was in a complex with ZNF274 on the silent maternal allele, we used a sequential ChIP assay, similar to that used previously to investigate co-occupancy of ZNF274 and SETDB1 at ZNF274 binding sites within 3′UTR of ZNF genes on chromosome 19 (26). We first performed ChIP with the ZNF274 antibody, eluted the protein-DNA complexes, and then immunoprecipitated the resulting sample with the SETDB1-specific antibody. We then performed qPCR using primers for SNOG1-BS5 and the control locus ZNF180 with the DNA isolated from the ZNF274/SETDB1 co-immunoprecipitate (Fig. 3B). We found that SETDB1 was enriched in the ZNF274 immunoprecipitated fraction bound to SNOG1/116HGG1 in PWS, but not AS iPSCs. SETDB1-ZNF274 binding was enriched at ZNF180 in both cell types. These findings suggest that SETDB1 co-occupies the maternal allele-specific ZNF274 binding sites at SNOG1/116HGG1.
Figure 3.
SETDB1 associates with ZNF274 on the silent maternal allele within SNORD116. (A) Enrichment of SETDB1 within 15q11-q13 by ChIP analyses, in PWS del 1–7 and AS del 1-0 iPSCs. (B) Sequential ChIP assays in PWS del 1–7 and AS del 1-0 iPSCs within SNORD116. Enrichment at ZNF180 was used as a control in these assays. ZNF274 ChIP samples were sequentially immunoprecipitated using ZNF274 antibody followed by SETDB1 antibody. (C) Gene expression of SETDB1 in PWS 1–7 iPSCs following lentiviral transduction with control or specific shRNA constructs targeting SETDB1. GAPDH was used as an endogenous control and data were normalized to PWS MIS iPSCs. (D) ChIP analyses for enrichment of SETDB1 in PWS SETDB1kd iPSCs, relative to MIS control. Enrichment at ZNF180 was used as a control in these assays. (E) Expression of genes outside of 15q11-q13 in PWS SETDB1kd iPSCs used as negative controls. *P ≤ 0.1, **P ≤ 0.05, ***P ≤ 0.01.
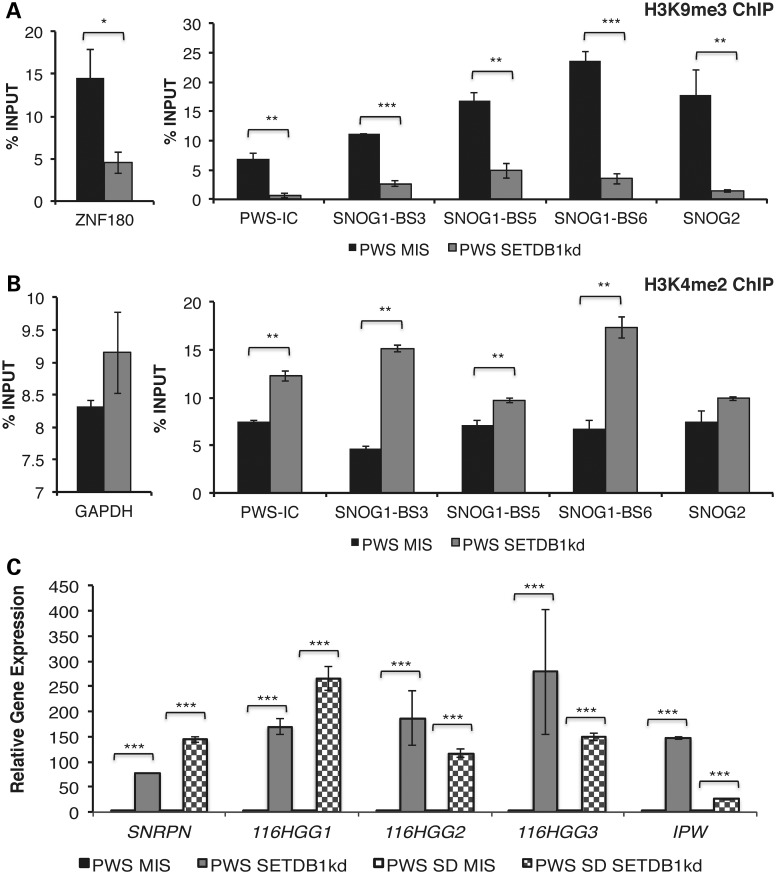
To further examine the role of SETDB1 in repression of the maternal SNORD116 cluster, we performed constitutive knockdown of SETDB1 in PWS 1–7 iPSCs using a lentiviral vector expressing a small hairpin RNA (shRNA) directed against SETDB1 mRNA or a MIS negative control. Knockdown of SETDB1 in PWS 1–7 iPSCs was efficient resulting in a reduction of ∼70% in mRNA levels relative to the MIS control (Fig. 3C). We also performed ChIP to confirm the efficiency of the knockdown and observed decreased enrichment of SETDB1 within the SNORD116 cluster (Fig. 3D). We next investigated the effect of SETDB1 on the maternal allele-specific enrichment of H3K9me3 in the SNORD116 cluster. There was a significant decrease in H3K9me3 accumulation at SNOG1/116HGG1 and SNOG2/116HGG2 in SETDB1kd relative to MIS iPSCs (Fig. 4A). Conversely, the levels of the active chromatin mark, dimethylated histone H3 lysine 4 (H3K4me2), increased at the PWS locus in SETDB1kd relative to MIS iPSCs (Fig. 4B). The findings summarized in Figure 4A and B suggest that knockdown of SETDB1 derepresses the maternal SNORD116 alleles. To further test this hypothesis, we performed qRT-PCR for SNRPN, 116HGG1, 116HGG2, 116HGG3 and IPW. We observed a 75- to 300-fold increase in expression of these maternally repressed genes in PWS SETDB1kd iPSCs relative to the control MIS iPSCs (Fig. 4C). The SNORD115 host transcript 115HG is expressed exclusively in neuronal cells and is not detectable in iPSCs (12, 13). We did not observe expression of 115HG in these experiments and, thus, elements that regulate the brain-specific portions of the transcripts driven by the PWS-IC (i.e. 115HG and UBE3A-ATS) do not appear to be perturbed upon SETDB1 knockdown. We also examined the expression of genes outside of 15q11-q13 to examine the specificity of the knockdown of SETDB1. Knockdown of SETDB1 did not alter the expression of the β-actin (ACTB) and SOX2 in PWS 1–7 iPSCs (Fig. 3E).
Figure 4.
Reactivation of the silent maternal allele in PWS. ChIP analyses for H3K9me3 (A) and H3K4 dimethylation (H3K4me2) (B) in PWS SETDB1kd iPSCs, relative to PWS MIS iPSCs. Enrichment at ZNF180 and GAPDH was used as a control in these assays, respectively. (C) Analysis of SNRPN and 116HG expression levels in PWS SETDB1kd and PWS SD SETDB1kd iPSCs. GADPH was used as an endogenous control and data were normalized to their respective MIS controls. *P ≤ 0.1, **P ≤ 0.05, ***P ≤ 0.01.
To further verify that knockdown of SETDB1 mediates derepression of maternally repressed transcripts in the PWSCR, SETDB1 was knocked down in the PWS SD 2–8 iPSC line (12, 15), using the same shRNA lentiviral vector approach as described above. The expression of SETDB1 was reduced ∼80% in the PWS SD SETDB1kd iPSCs, relative to the PWS SD MIS control (Supplementary Material, Fig. S2A). A marked increase was again observed in the expression of SNRPN, 116HGG1, 116HGG2, 116HGG3 and IPW in the PWS SD SETDB1kd iPSCs relative to PWS SD MIS iPSCs (Fig. 4C). Although the RNA levels of maternal PWSCR transcripts are markedly increased here (Fig. 4C) in comparison to those observed upon ZNF274kd (Fig. 2D), we estimate that they remain significantly lower (1–35%) than those of active paternal alleles. Similarly to the case of PWS 1–7 SETDB1kd, no significant differences were observed for expression of the ACTB and SOX2 (Supplementary Material, Fig. S2B) in PWS SD SETDB1kd. Thus, changes in gene expression were observed in two PWS iPSC lines derived from different PWS patients with distinct genotypes, indicating that the knockdown of SETDB1 can reactivate expression the silent maternal alleles in the PWSCR.
Since ZNF274 is thought to mediate chromatin silencing via recruitment of both SETDB1and the co-repressor TRIM28/KAP1 (26), we determined whether the binding of TRIM28 was allele specific within 15q11-q13. Similar to SETDB1, we found that TRIM28 is enriched on both maternal and paternal alleles of SNOG1/116HGG1 (Supplementary Material, Fig. S3A). We also used a sequential ChIP assay to determine if TRIM28 is found in the ZNF274 repressive complex on the silent maternal allele in PWS. We first performed ChIP with the ZNF274 antibody, eluted the protein-DNA complexes, and immunoprecipitated the resulting sample with the TRIM28-specific antibody. We examined their association at SNOG1-BS5 and the control locus ZNF180 with the DNA isolated from the ZNF274/KAP1 co-immunoprecipitate (Supplementary Material, Fig. S3B). We found that TRIM28 was enriched in the ZNF274 immunoprecipitated fraction bound to SNOG1/116HGG1 at both SNOG1-BS5, as well as the control locus ZNF180, in PWS iPSCs. These findings suggest that TRIM28 co-occupies the maternal allele-specific ZNF274 binding sites at SNOG1/116HGG1. Next, we knocked down the expression of TRIM28 in PWS 1–7 iPSCs, via RNA interference using siRNA constructs directed against TRIM28 or a negative mismatch control. Although the expression levels of TRIM28 were reduced by 80%, relative to the negative control (Supplementary Material, Fig. S3C), the knock down of TRIM28 mRNA did not have a detectable effect on 116HGG1 expression (Supplementary Material, Fig. S3D). These results suggest that our knockdown approach may not be sufficiently disrupting a putative ZNF274/TRIM28/SETDB1-chromatin complex to cause transcriptional re-activation of the silent maternal PWSCR transcripts. Alternatively, the repressive complex that includes SETDB1 to deposit H3K9me3 in the PWSCR does not contain TRIM28.
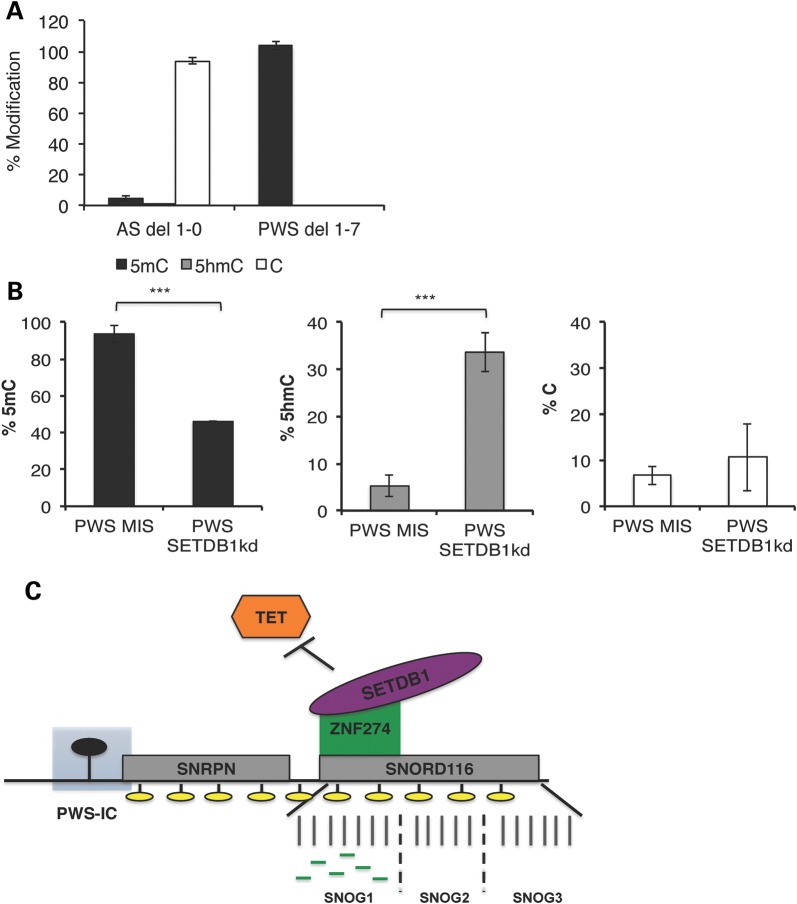
Upon SETDB1 knockdown in PWS 1–7 iPSCs, we observe a decrease in the repressive modification H3K9me3 and corresponding increase in the accumulation of H3K4me2 at the PWS-IC (Fig. 4A and B). To investigate the possibility that the re-activation of the maternal PWSCR is associated with a change in DNA methylation at the PWS-IC, we used a restriction endonuclease assay that measures the relative levels 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC), a DNA demethylation intermediate that can be converted to unmodified cytosine (C). In this assay, genomic DNA is treated with T4-β-glucosyltransferase, an enzyme that specifically tags 5hmC with a glucose moiety. This glucose tag prevents the modified base from being digested by the restriction endonuclease, MspI, and thus distinguishes between 5hmC and 5mC. As expected, the PWS-IC displays almost complete methylation (5mC) of the maternal allele with almost complete absence of methylation of the paternal allele (Fig. 5A), in untreated PWS and AS iPSCs, respectively. SETDB1kd in PWS 1–7 iPSCs has a marked impact on the levels of 5mC at the PWS-IC where a decrease of roughly 50% is observed. The loss of 5mC at the PWS-IC upon SETDB1kd is associated with a corresponding 50% increase in 5hmC levels at the PWS-IC (Fig. 5B), suggesting that re-activation of the repressed maternal alleles may be associated with a conversion of 5mC to 5hmC at the PWS-IC. To confirm this surprising result, we used a second assay, methylated DNA immunoprecipitation (MeDIPs), to analyze the levels of 5mC and 5hmC at the PWS-IC in PWS and AS iPSCs (Supplementary Material, Fig. S4A) and in PWS 1–7 SETDB1kd iPSCs and MIS controls (Supplementary Material, Fig. S4B). MeDIPs with a 5-methylcytosine (5mC) antibody showed the expected preferential 5mC methylation at the PWS-IC on the silent maternal allele in PWS relative to the active paternal allele in AS iPSCs (Supplementary Material, Fig. S4A). Low levels of 5hmC were observed at the PWS-IC in both the PWS and AS iPSCs (Supplementary Material, Fig. S4A). Although the 5mC levels by MeDIP were unchanged, an increase in enrichment of 5hmC at the PWS-IC in both the PWS SETDB1kd iPSCs relative to the MIS control was observed (Supplementary Material, Fig. S4B) consistent with the results summarized in Figure 5B.
Figure 5.
DNA methylation levels change when the silent maternal allele in PWS is reactivated. Content of 5hmC at the PWS-IC, quantified by subtraction of 5hmC from the total methylated cytosine levels, determined by HpaII sensitivity as a percentage of total cytosine in PWS del 1–7 and AS del 1-0 iPSCs (A) or PWS SETDB1kd iPSCs, relative to PWS MIS iPSCs (B). ***P ≤ 0.01. (C) Model describing the role of the ZNF274/SETDB1 complex in the establishment/maintenance of silencing of the maternal allele of 15q11-q13. Boxes represent: SNRPN (small nuclear ribonucleoprotein polypeptide N) and SNORD116 (small nucleolar RNA, C/D box 116). Filled oval represents presence of 5mC methylation at the PWS-IC. ZNF274 and SETDB1 potentially in association with other proteins recognize the tandemly repeated SNOG1 maternal alleles at early times after fertilization. Green lines within SNOG1/116HGG1 represent the six ZNF274 binding sites. Binding of the repressive complex to paternal SNOG1 alleles may be prevented by active transcription through the PWSCR at early stages of development. Binding of the complex to the maternal SNOG1 alleles results in deposition of the repressive H3K9me3 (indicated by yellow ovals) and stabilizes epigenetic silencing of the PWSCR in early development. The ZNF274/SETDB1 complex may also protect the 5mC mark at the maternal PWS-IC from oxidative modification by TET proteins that are highly expressed at the blastocyst and epiblast stages (27).
DISCUSSION
There are several novel findings reported here that are relevant to the mechanism of silencing at the PWSCR: (i) ZNF274 binds specifically to the repressed maternal SNORD116 allele cluster, a locus that defines the PWSCR (Fig. 1B; Supplementary Material, Fig. S1B); (ii) consistent with other reports of ZNF274 being associated with the repressive H3K9me3 mark, we observe H3K9me3 accumulation at the maternal SNORD116 cluster (Fig. 1C); (iii) knockdown of the histone methyltransferase SETDB1, which associates with ZNF274 at the PWSCR (Fig. 3B), not only results in the loss of the H3K9 repressed chromatin mark but also in the accumulation of the H3K4me2 active histone mark (Fig. 4A and B); (iv) SETDB1 knockdown also results in the transcriptional reactivation of the PWSCR transcripts SNRPN, 116HG and IPW observed in both PWS iPSC cell lines (Fig. 4C); and (v) Knockdown of SETDB1 alters relative levels of 5mC and 5hmC at the maternal PWS-IC (Fig. 5B; Supplementary Material, Fig. S4B). These results suggest that SETDB1, in a complex that likely includes ZNF274, plays an essential role in the imprinting/silencing of the maternal PWSCR during early development, as modeled in iPSCs.
Genome-wide ZNF274 ChIP studies annotated in the UCSC browser (26) reveal that ZNF274 binds to the six PWSCR binding sites in established cell lines (lymphoblasts, K562, HeLa and HEPG2), although the level of enrichment is variable and generally decreased relative to that observed in hESCs and in another stem cell line, NTD2. The observation of increased ZNF274 binding at BS1–BS6 in stem cells is what led us to focus on iPSCs as models early development and we did not examine the binding of ZNF274 in neurons differentiated from PWS or AS iPSCs, using our standard protocols (12, 13). We expect that the extent of ZNF274 binding at BS1–BS6 in fibroblasts and in differentiated neurons will be reduced as is observed in other somatic cells. Moreover, we would predict that the maternal allele specificity of ZNF274 binding should be maintained.
ChIP analyses of ZNF274 binding in established cell lines also reveal a relatively limited number of binding sites (∼200–500) in the human genome with the majority on chromosome 19 (26). The ZNF274 binding sites are enriched in the 3′-UTR of C2H2 ZNF genes that are clustered on chromosome 19 (26). The 3′UTR of this subset of ZNFs is marked by accumulation of H3K9me3 and co-localization of SETDB1 and the co-repressor TRIM28/KAP1 leading to the elegant model for SETDB1 recruitment via ZNF274- and TRIM28-mediated transcriptional repression (26). Further studies found that although the ZNF274/SETDB1/TRIM28 complex is associated with H3K9me3 accumulation at the 3′-end of ZNF genes located on chromosome 19, these regions also enriched for histone H3 lysine 36 trimethylation (H3K36me3), a mark associated with transcriptional elongation (28). How these contradictory chromatin signatures function to regulate transcription of ZNF genes on chromosome 19 is not completely understood. There are parallels between the studies of ZNF274 binding to chromosome 19 and to the PWSCR as reported here. The binding of ZNF274 and the accumulation of H3K9me3 is associated with tandem repeat in the 3′UTR of ZNF genes (28) and with the SNORD116 repeat gene cluster on chromosome 15 (Fig. 1B and C; Supplementary Material, Fig. S1B). Frietze et al. (26) identified a number of motifs that were highly enriched in the ZNF274 binding sites. However, it is not known if all fingers in the multi-finger DNA binding region contact the DNA, or whether ZNF274 works as a monomer, homodimer or as a heterodimer with another factor (26). Due to the number of motifs identified in ZNF274 binding sites, there is also the possibility that ZNF274 may use different fingers to bind to different motifs (26). The most common motifs termed ZNF274M1 and ZNF274M11 were derived by examination of the ZNF274 binding at the 3′UTR of the chromosome 19 ZNF gene cluster (26), and are not present in the SNOG1 binding sites. Other conserved motifs termed ZNF274W12, ZNF274W16, ZNF274W5 and ZNF274W9 as reported by Frietze et al. (26) are, however, found within all six of the ZNF274 binding sites within SNOG1 and are six nucleotides each. ZNF274W16 and ZNF274W9 overlap an identical region in each of the binding sites, as do ZNF274W12 and ZNF274W5. A report by Khurana et al. (29) found that the ZNF274 binding sites within the PWSCR possess a high fraction of rare variants comparable with that of coding sequences (29). In the Khurana et al. study (29), the ZNF274 distal binding sites at SNOG1/116HGG1 are classified as ultrasensitive, i.e. display enrichment for functional variants but depletion of common variants. Disruptive mutations of SNOG1/116HGG1 binding sites for ZNF274 are, thus, under strong selection at a level similar to that of the exome. These findings support our hypothesis that ZNF274 binding within the PWSCR is of high functional significance.
Co-occupancy of ZNF274 and SETDB1 is demonstrated by sequential ChIP assay for chromosome 19 (26) and for PWSCR ZNF274 binding sites (Fig. 3B), suggesting that ZNF274 also recruits SETDB1 to the SNOG1 locus. Moreover, the role of ZNF274 and SETDB1 in maintaining H3K9me3 at the PWSCR is confirmed in our knockdown experiments (Figs 2C and 4A). However, this does not rule out the possibility that SETDB1 may be also recruited to the region by additional factors/complexes. We also performed siRNA-mediated knockdown of TRIM28 in PWS 1–7 iPSCs but, in contrast to what was observed for SETDB1, we could not detect re-activation of expression of PWSCR transcripts (Supplementary Material, Fig. S3). DNA methylation at imprinted gene loci is robustly maintained during early embryonic mouse development by several maternal factors including Trim28. In maternal Trim28-null embryos, stochastic demethylation of differentially methylated regions within imprinting centers is observed, including Snrpn (30, 31). Despite the defined role of Trim28 in the maintenance of imprinting in postfertilization stages in mouse, the knockdown of TRIM28 in human iPSCs does not result in the stochastic demethylation of the PWS-IC, raising the possibility that the mechanism by which imprinting is maintained at the PWS-IC is different in human compared with mouse. Another possibility is that the developmental window for which TRIM28 is necessary for the maintenance of imprinting at the PWS-IC has passed, suggesting that human iPSCs may represent a time point during development upon which TRIM28 is no longer necessary. There are temporal and developmental requirements for the PWS-IC that become dispensable during later stages of development (32).
In PWS 1–7 iPSCs, knockdown of SETDB1 not only decreases the accumulation of H3K9me3 at sites within the PWSCR (Fig. 4A) but also results in an increase in the levels of the active H3K4me2 mark at these sites (Fig. 4B) and a corresponding transcriptional activation of the SNRPN, 116HGG1, 116HGG2, 116HGG3 and IPW transcripts (Fig. 4C). The levels of PWSCR transcript expression in SETDB1kd PWS iPSCs do not, however, attain those of the active paternal allele in NML control iPSC. For example, we estimate that the highest level of derepression is detected for 116HGG1 RNA in PWS SD SETDB1kd iPSCs, with the expression level corresponding to only ∼35% that of the paternal allele expression. The low levels of expression of PWSCR transcripts observed upon SETDB1 knockdown could result from the residual complex bound to PWSCR chromatin in PWS iPSCs or from re-expression from only a subset of cells. It is difficult to predict what levels of expression SNRPN, 116HG and IPW are required to rescue the PWS disease etiology and further study is needed to determine whether wild-type levels need to be fully restored. SETDB1 knockdown does not appear to increase the expression levels of 115HG and UBE3A-ATS in PWS iPSCs. An open question is whether transcription of UBE3A-ATS would increase in neurons upon perturbation of the ZNF274/SETDB1 complex, thereby repressing the maternal UBE3A allele, and causing an AS-like disorder.
We also investigated whether knockdown of SETDB1 alters CG methylation at the PWS-IC, by examining the levels of 5mC and 5hmC at the PWS-IC in PWS SETDB1kd iPSCs relative to MIS iPSCs (Fig. 5; Supplementary Material, Fig. S4). Interestingly, the knockdown of SETDB1 resulted in a roughly 50% conversion of 5mC to 5hmC at the PWS-IC (Fig. 5B), suggesting that altered CG methylation at the PWS-IC may be responsible for derepression of PWSCR transcripts. The fact that 5mC is not completely converted to 5hmC and to unmodified cytosine could explain partial derepression of the PWSCR upon SETDB1 knockdown (Fig. 4B and C). The very limited conversion of 5mC to unmodified cytosine could, in turn, be associated with a decrease in cell proliferation that we observe in SETDB1kd iPSCs (our unpublished observation) since it is known that one pathway for conversion of 5hmC to unmodified cytosine involves replication-dependent dilution (33).
The correlation between altered CG methylation at the PWS-IC and loss of H3K9me3 at the PWSCR also raises the possibility that the SETDB1/ZNF274 complex plays a role in the maintenance of imprinting at the PWS-IC. In this model (Fig. 5C), the SETDB1/ZNF274 complex binds to the maternal SNOG1/116HGG1 locus early in embryonic development resulting in the accumulation and spreading of H3K9me3. This epigenetic event could function to protect the PWS-IC from the global waves of demethylation in pre- and postimplantation development perhaps interfering with the binding to the PWSCR of 10–11 translocation (TET) dioxygenases, proteins that are expressed at high levels in human pluripotent stem cells (34). SETDB1 knockdown and the resulting loss of H3K9me3 might thereby allow TET binding to the PWSCR and a TET-mediated conversion of 5mC to 5hmC at the PWS-IC (Fig. 5B). Thus, the ZNF274/SETDB1 complex may be required to establish/maintain epigenetic states in the early embryo. This is consistent with previous studies that indicate that there are temporal and developmental requirements for the PWS-IC, which become dispensable during later stages of development (32). The model proposed in Figure 5C is also consistent with the recent finding that ZNF274 and SETDB1 are expressed at the 4-cell stage and at later stages of zygotic development, although these molecules are not expressed in oocytes so are unlikely to function in the establishment of imprinting at the PWS-IC (35). Our suggestion that a complex acts in early development to protect 5mC imprinting mark is consistent with earlier studies indicating that the establishment of imprinting at the PWS-IC extends to the period after fertilization in humans (36, 37).
SETDB1 and TRIM28 also associate with the active paternal allele in AS iPSCs. The association of SETDB1 at transcriptionally active genes is a feature of bivalent chromatin in ES cells. In mouse ESCs, Setdb1 is also found to be associated with transcriptionally active, as well as transcriptionally repressed genes (38). Setdb1-occupied genes encompass a subset of ‘bivalent’ genes, which contain both H3K4me3 and H3K27me3 modifications. Setdb1 association at transcriptionally active genes is thought to contribute to the maintenance of ES cell state (38). Moreover, repressed genes were co-occupied by Setdb1 and other factors involved in transcriptional repression including Polycomb group proteins (38). We speculate that SETDB1, and perhaps TRIM28, may act in a similar context on 15q11-q13 and may have different functions, depending on their binding partners. SETDB1 and TRIM28 may have unknown role in the regulation of transcription of paternal 15q11-q13, in addition to their role in the silencing of the maternal allele. Thus, SETDB1 is required and not sufficient to promote silencing on the maternal allele. Additional factors are also required to silence the maternal allele.
In summary, we have identified ZNF274 and SETDB1 as novel epigenetic signatures of the PWSCR. The complex formed by these molecules and possibly other constituents potentially play a role in protecting the 5mC mark at the PWS-IC during early human development and disruption of the complex could be a target of therapeutic intervention in Prader-Willi syndrome (PWS). It should be noted that strategies would need to be developed to target ZNF274 and SETDB1 specifically at the SNORD116 locus since the knockdown approach used here undoubtedly has off-target effects. Nevertheless, our study highlights a mechanistic link between the association of ZNF274 and SETDB1 within SNORD116 on the maternal allele of human 15q11-q13 and repression of SNRPN, 116HG, and IPW on the silent maternal allele.
MATERIALS AND METHODS
Cell lines and culture conditions
PWS del 1–7, PWS SD 2–8, AS del 1-0 and NML 1-0 iPSC lines used in this study were cultured, as described (12, 13). The iPSC lines were cultured in hESC medium, i.e. Dulbecco's modified Eagle's medium/F-12 containing 20% knockout serum replacer, 0.1 mm nonessential amino acids, 1 mm l-glutamine (all from Life Technologies), 0.1 mm β-mercaptoethanol (Sigma-Aldrich), supplemented with 4 ng/ml bFGF (Millipore, Billerica, MA, USA).
To knockdown ZNF274, ZNF274 siRNAs (Stealth Select RNAi, Life Technologies: HSS116638, HSS116639, HSS116640) or non-specific control si-GLO RISC-Free (Dharmacon; catalog # D-001600-01) were used as described (26). PWS del 1–7 iPSCs were transfected with 36 pmol ZNF274 siRNAs, using Lipofectamine RNAiMAX as described (39). PWS del 1–7 iPSCs were transfected a second time 48 h following the first transfection. Cells were harvested 72 h after the initial transfection. The lentiviral shRNA construct used to knockdown SETDB1 was purchased from Sigma-Aldrich (Clone number TRCN0000148112), as well as a MIS control (catalog # SHC002). PWS del 1–7 iPSCs or PWS SD 2-8 iPSCs were transduced with either the SETDB1 KD or MIS virus, and selected with puromycin as described (40). To knockdown TRIM28, TRIM28 siRNA (Silencer® Select RNAi, Life Technologies: s19778) or Silencer® Select RNAi Negative control (Life Technologies: 4390846) were used. PWS del 1–7 iPSCs were transfected with 12.5 pmol of the siRNA constructs, using Lipofectamine RNAiMAX. PWS del 1–7 iPSCs were transfected a second time 48 h following the first transfection. Cells were harvested 72 h after the initial transfection.
qRT-PCR
RNA was isolated from cells using RNA-Bee (Tel Test, Inc.), and cDNA was synthesized using High Capacity cDNA Reverse Transcription Kit (Life Technologies) per the manufacturer's instructions. Gene expression was examined using TaqMan® Gene Expression Assays, and GAPDH Endogenous Control TaqMan® Assay was used as an endogenous control. Each reported value represents the average of at least three independent experiments, each analyzed in triplicate by quantitative PCR. Data were presented as means and SEM of independent experiments. Student's t-test was performed to determine significance.
Chromatin immunoprecipitations
ChIP assays were performed as described (40). The antibodies used in these assays were anti-ZNF274 (Novus Biologicals; catalog # H00010782-M01), anti-SETDB1 (Proteintech; catalog # 11231-1-AP), anti-KAP1 (Abcam; catalog # ab10483), anti-trimethyl histone H3 (Lys9) (H3K9me3; Millipore, catalog # 07-442) and anti-dimethyl histone H3 (Lys4) (H3K4me2; Millipore, catalog # 07-030). The normal rabbit IgG or normal mouse IgG was used as the isotype controls. Quantification of ChIPs was performed using SYBR Green quantitative PCR. PCR primers used to amplify the purified DNA can be found in Supplementary Material, Table S1. The enrichment of the DNA was calculated as described (40). Data were calculated as percent input, and non-specific IgG values were subtracted from percent input values. Each reported value represents the average of at least two independent ChIP experiments, each analyzed in triplicate by quantitative PCR. Data were presented as means and SEM of independent experiments. Student's t-test was performed to determine significance.
Sequential ChIP assays
Sequential ChIP assays were performed using the Re-CHIP-IT® kit (Active Motif; catalog # 53016) per the manufacturer's instructions. Quantification of the sequential ChIPs was performed using SYBR Green quantitative PCR, using PCR primers listed in Supplementary Material, Table S1. The enrichment of the DNA was calculated as described above. Each reported value represents the average of at least three independent ChIP experiments, each analyzed in triplicate by quantitative PCR.
Detection of 5hmC levels
Percentages of 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC) and unmodified cytosine (C) in DNA were assessed using the EpiMark Kit (New England Biolabs, catalog # E3317S), per the manufacturer's instructions. This technique has been previously described in detail by Ficz et al. (41) and Doege et al. (42). Briefly, genomic DNA was treated with T4-β-glucosyltransferase, which specifically tags 5hmC with a glucose moiety and thus distinguishes between 5hmC and 5mC. Genomic DNA was then digested with MspI, HpaII or no enzyme (mock digestion). MspI can digest 5hmC, 5mC and C but cannot digest glucosyl-5hmC. When 5hmC occurs in the context of CCGG, this modification converts a cleavable MspI site to a noncleavable one. Therefore, resistance to MspI, or percentage of Glucosyl-5hmC, translates to percentage of 5hMC. HpaII digestion is blocked by 5hmC and 5mC and translates to total CpG methylation (5mC + 5hmC). The percentage of 5mC was obtained by subtracting the 5hmC contribution (MspI digestion) from the total methylation (5mC + 5hmC) or HpaII resistance. Data were normalized to mock digested genomic DNA. QPCR primers used in these assays are denoted in Supplementary Material, Table S1. Each reported value represents the average of at least two independent experiments, each analyzed in triplicate by quantitative PCR. Data were presented as means and SEM of independent experiments. Student's t-test was performed to determine significance.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the Brazilian Council for Scientific and Technological Development, CNPq (to E.C.), the Foundation for Prader-Willi Syndrome (to M.L. and K.M.T.) and Connecticut Stem Cell Research Grant (11SCA01 to K.M.T.). The contents in this work are solely the responsibility of the authors and do not necessarily represent the official views of the state of Connecticut.
Supplementary Material
ACKNOWLEDGEMENTS
We also thank members of the lab for their helpful discussions and Claire Rougeulle and Janine LaSalle for critical reading of the manuscript. We are grateful to Susan Holder, Alexandra Blakemore, Adam de Smith, Philippe Froguel, David S. Rosenblatt and Gail Dunbar for patient clinical evaluation and information, and for providing skin biopsies/fibroblasts.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Cassidy S.B., Schwartz S., Miller J.L., Driscoll D.J. Prader-Willi syndrome. Genet. Med. 2012;14:10–26. doi: 10.1038/gim.0b013e31822bead0. doi:10.1038/gim.1030b1013e31822bead31820. [DOI] [PubMed] [Google Scholar]
- 2.Reis A., Dittrich B., Greger V., Buiting K., Lalande M., Gillessen-Kaesbach G., Anvret M., Horsthemke B. Imprinting mutations suggested by abnormal DNA methylation patterns in familial Angelman and Prader-Willi syndromes. Am. J. Hum. Genet. 1994;54:741–747. [PMC free article] [PubMed] [Google Scholar]
- 3.Saitoh S., Buiting K., Rogan P.K., Buxton J.L., Driscoll D.J., Arnemann J., Konig R., Malcolm S., Horsthemke B., Nicholls R.D. Minimal definition of the imprinting center and fixation of chromosome 15q11-q13 epigenotype by imprinting mutations. Proc. Natl. Acad. Sci. USA. 1996;93:7811–7815. doi: 10.1073/pnas.93.15.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boccaccio I., Glatt-Deeley H., Watrin F., Roeckel N., Lalande M., Muscatelli F. The human MAGEL2 gene and its mouse homologue are paternally expressed and mapped to the Prader-Willi region. Hum. Mol. Genet. 1999;8:2497–2505. doi: 10.1093/hmg/8.13.2497. [DOI] [PubMed] [Google Scholar]
- 5.Jay P., Rougeulle C., Massacrier A., Moncla A., Mattei M.G., Malzac P., Roeckel N., Taviaux S., Lefranc J.L., Cau P., et al. The human necdin gene, NDN, is maternally imprinted and located in the Prader-Willi syndrome chromosomal region. Nat. Genet. 1997;17:357–361. doi: 10.1038/ng1197-357. [DOI] [PubMed] [Google Scholar]
- 6.Cattanach B.M., Barr J.A., Evans E.P., Burtenshaw M., Beechey C.V., Leff S.E., Brannan C.I., Copeland N.G., Jenkins N.A., Jones J. A candidate mouse model for Prader-Willi syndrome which shows an absence of Snrpn expression. Nat. Genet. 1992;2:270–274. doi: 10.1038/ng1292-270. [DOI] [PubMed] [Google Scholar]
- 7.Glenn C.C., Porter K.A., Jong M.T., Nicholls R.D., Driscoll D.J. Functional imprinting and epigenetic modification of the human SNRPN gene. Hum. Mol. Genet. 1993;2:2001–2005. doi: 10.1093/hmg/2.12.2001. [DOI] [PubMed] [Google Scholar]
- 8.Runte M., Huttenhofer A., Gross S., Kiefmann M., Horsthemke B., Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 9.Cavaille J., Buiting K., Kiefmann M., Lalande M., Brannan C.I., Horsthemke B., Bachellerie J.P., Brosius J., Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wevrick R., Kerns J.A., Francke U. Identification of a novel paternally expressed gene in the Prader-Willi-syndrome region. Hum. Mol. Genet. 1994;3:1877–1882. doi: 10.1093/hmg/3.10.1877. [DOI] [PubMed] [Google Scholar]
- 11.Rougeulle C., Cardoso C., Fontes M., Colleaux L., Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat. Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 12.Martins-Taylor K., Hsiao J.S., Chen P.F., Glatt-Deeley H., De Smith A.J., Blakemore A.I., Lalande M., Chamberlain S.J. Imprinted expression of UBE3A in non-neuronal cells from a Prader-Willi syndrome patient with an atypical deletion. Hum. Mol. Genet. 2014;23:2364–2373. doi: 10.1093/hmg/ddt628. doi:10.1093/hmg/ddt628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chamberlain S.J., Chen P.F., Ng K.Y., Bourgois-Rocha F., Lemtiri-Chlieh F., Levine E.S., Lalande M. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc. Natl. Acad. Sci. USA. 2010;107:17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vitali P., Royo H., Marty V., Bortolin-Cavaille M.-L., Cavaille J. Long nuclear-retained non-coding RNAs and allele-specific higher-order chromatin. J. Cell Sci. 2010;123:70–83. doi: 10.1242/jcs.054957. doi:10.1242/jcs.054957. [DOI] [PubMed] [Google Scholar]
- 15.de Smith A.J., Purmann C., Walters R.G., Ellis R.J., Holder S.E., Van Haelst M.M., Brady A.F., Fairbrother U.L., Dattani M., Keogh J.M., et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum. Mol. Genet. 2009;18:3257–3265. doi: 10.1093/hmg/ddp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahoo T., Bacino C.A., German J.R., Shaw C.A., Bird L.M., Kimonis V., Anselm I., Waisbren S., Beaudet A.L., Peters S.U. Identification of novel deletions of 15q11q13 in Angelman syndrome by array-CGH: molecular characterization and genotype-phenotype correlations. Eur. J. Hum. Genet. 2007;15:943–949. doi: 10.1038/sj.ejhg.5201859. [DOI] [PubMed] [Google Scholar]
- 17.Sahoo T., del Gaudio D., German J.R., Shinawi M., Peters S.U., Person R.E., Garnica A., Cheung S.W., Beaudet A.L. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding F., Li H.H., Zhang S., Solomon N.M., Camper S.A., Cohen P., Francke U. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS ONE. 2008;3:e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muscatelli F., Abrous D.N., Massacrier A., Boccaccio I., Le Moal M., Cau P., Cremer H. Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Hum. Mol. Genet. 2000;9:3101–3110. doi: 10.1093/hmg/9.20.3101. [DOI] [PubMed] [Google Scholar]
- 20.Schaaf C.P., Gonzalez-Garay M.L., Xia F., Potocki L., Gripp K.W., Zhang B., Peters B.A., McElwain M.A., Drmanac R., Beaudet A.L., et al. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat. Genet. 2013;45:1405–1408. doi: 10.1038/ng.2776. doi:1410.1038/ng.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devos J., Weselake S.V., Wevrick R. Magel2, a Prader-Willi syndrome candidate gene, modulates the activities of circadian rhythm proteins in cultured cells. J. Circadian Rhythms. 2011;9:12. doi: 10.1186/1740-3391-9-12. doi:10.1186/1740-3391-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rieusset A., Schaller F., Unmehopa U., Matarazzo V., Watrin F., Linke M., Georges B., Bischof J., Dijkstra F., Bloemsma M., et al. Stochastic loss of silencing of the imprinted Ndn/NDN allele, in a mouse model and humans with Prader-Willi syndrome, has functional consequences. PLoS Genet. 2013;9:e1003752. doi: 10.1371/journal.pgen.1003752. doi:1003710.1001371/journal.pgen.1003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell W.T., Coulson R.L., Crary F.K., Wong S.S., Ach R.A., Tsang P., Alice Yamada N., Yasui D.H., Lasalle J.M. A Prader-Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum. Mol. Genet. 2013;22:4318–4328. doi: 10.1093/hmg/ddt281. doi:4310.1093/hmg/ddt4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castle J.C., Armour C.D., Lower M., Haynor D., Biery M., Bouzek H., Chen R., Jackson S., Johnson J.M., Rohl C.A., et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS ONE. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen M., Eyras E., Wu J., Khanna A., Josiah S., Rederstorff M., Zhang M.Q., Stamm S. Direct cloning of double-stranded RNAs from RNase protection analysis reveals processing patterns of C/D box snoRNAs and provides evidence for widespread antisense transcript expression. Nucleic Acids Res. 2011;39:9720–9730. doi: 10.1093/nar/gkr684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frietze S., O'Geen H., Blahnik K.R., Jin V.X., Farnham P.J. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS ONE. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohn F., Weber M., Schubeler D., Roloff T.C. Methylated DNA immunoprecipitation (MeDIP) Methods Mol. Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. doi:10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 28.Blahnik K.R., Dou L., Echipare L., Iyengar S., O'Geen H., Sanchez E., Zhao Y., Marra M.A., Hirst M., Costello J.F., et al. Characterization of the contradictory chromatin signatures at the 3′ exons of zinc finger genes. PLoS ONE. 2011;6:e17121. doi: 10.1371/journal.pone.0017121. doi:17110.11371/journal.pone.0017121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khurana E., Fu Y., Colonna V., Mu X.J., Kang H.M., Lappalainen T., Sboner A., Lochovsky L., Chen J., Harmanci A., Das J., et al. Integrative annotation of variants from 1092 humans: application to cancer. Science. 2013;342:1235587. doi: 10.1126/science.1235587. doi:10.1231126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messerschmidt D.M., de Vries W., Ito M., Solter D., Ferguson-Smith A., Knowles B.B. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- 31.Messerschmidt D.M. Should I stay or should I go: protection and maintenance of DNA methylation at imprinted genes. Epigenetics. 2012;7:969–975. doi: 10.4161/epi.21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DuBose A.J., Smith E.Y., Johnstone K.A., Resnick J.L. Temporal and developmental requirements for the Prader-Willi imprinting center. Proc. Natl. Acad. Sci. USA. 2012;109:3446–3450. doi: 10.1073/pnas.1115057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen L., Zhang Y. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Curr. Opin. Cell Biol. 2013;25:289–296. doi: 10.1016/j.ceb.2013.02.017. doi:210.1016/j.ceb.2013.1002.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H., Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. doi:10.1016/j.cell.2013.1012.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorthongpanich C., Cheow L.F., Balu S., Quake S.R., Knowles B.B., Burkholder W.F., Solter D., Messerschmidt D.M. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science. 2013;341:1110–1112. doi: 10.1126/science.1240617. doi:1110.1126/science.1240617. [DOI] [PubMed] [Google Scholar]
- 36.El-Maarri O., Buiting K., Peery E.G., Kroisel P.M., Balaban B., Wagner K., Urman B., Heyd J., Lich C., Brannan C.I., et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat. Genet. 2001;27:341–344. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- 37.Xin Z., Allis C.D., Wagstaff J. Parent-specific complementary patterns of histone H3 lysine 9 and H3 lysine 4 methylation at the Prader-Willi syndrome imprinting center. Am. J. Hum. Genet. 2001;69:1389–1394. doi: 10.1086/324469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bilodeau S., Kagey M.H., Frampton G.M., Rahl P.B., Young R.A. SetDB1 contributes to repression of genes encoding developmental regulators and. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. doi:10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Y., Jin J., Dong C., Cheng E.C., Lin H., Huang Y., Qiu C. High-efficiency siRNA-based gene knockdown in human embryonic stem cells. RNA. 2010;16:2564–2569. doi: 10.1261/rna.2350710. doi:2510.1261/rna.2350710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martins-Taylor K., Schroeder D.I., Lasalle J.M., Lalande M., Xu R.H. Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics. 2012;7:71–82. doi: 10.4161/epi.7.1.18750. doi:10.4161/epi.4167.4161.18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ficz G., Branco M.R., Seisenberger S., Santos F., Krueger F., Hore T.A., Marques C.J., Andrews S., Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. doi:310.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 42.Doege C.A., Inoue K., Yamashita T., Rhee D.B., Travis S., Fujita R., Guarnieri P., Bhagat G., Vanti W.B., Shih A., et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. doi:610.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.