Abstract
Motivation: Genome-wide association studies (GWASs) are effective for describing genetic complexities of common diseases. Phenome-wide association studies (PheWASs) offer an alternative and complementary approach to GWAS using data embedded in the electronic health record (EHR) to define the phenome. International Classification of Disease version 9 (ICD9) codes are used frequently to define the phenome, but using ICD9 codes alone misses other clinically relevant information from the EHR that can be used for PheWAS analyses and discovery.
Results: As an alternative to ICD9 coding, a text-based phenome was defined by 23 384 clinically relevant terms extracted from Marshfield Clinic’s EHR. Five single nucleotide polymorphisms (SNPs) with known phenotypic associations were genotyped in 4235 individuals and associated across the text-based phenome. All five SNPs genotyped were associated with expected terms (P < 0.02), most at or near the top of their respective PheWAS ranking. Raw association results indicate that text data performed equivalently to ICD9 coding and demonstrate the utility of information beyond ICD9 coding for application in PheWAS.
Contact: hebbring.scott@mcrf.mfldclin.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
The genetic complexities of common diseases have been well-defined by the use of the genome wide-association study (GWAS). Aside from only a few examples, GWASs have failed to identify single nucleotide polymorphisms (SNPs) that reach a threshold, where they can be used to predict, prevent or even treat complex diseases. This may be due in part to disease heterogeneity and the fact that most SNPs genotyped during a GWAS represent tag SNPs for unknown and ungenotyped causal variants (Goldstein 2009; Manolio et al., 2009; McCarthy et al., 2008; Need and Goldstein, 2010; Visscher et al., 2012). As a complementary or alternative strategy to GWAS, phenome-wide association studies (PheWASs) have demonstrated their effectiveness for rediscovering GWAS associations while also identifying novel disease-SNP correlations (Hebbring, 2014). Whereas GWAS is a phenotype-to-genotype strategy, PheWAS reverses this paradigm by exchanging the disease and genome in a GWAS with a specific genetic variant and phenome in a PheWAS. As such, PheWAS is a genotype-to-phenotype strategy.
The first PheWAS was published in 2010 by Denny et al. In this first proof-of-principle study, five disease-associated SNPs previously identified by GWAS were genotyped in a cohort of 6005 patients from Vanderbilt University Medical Center. Each SNP was associated with hundreds of International Classification of Disease version 9 (ICD9) codes that defined the ICD9-based phenome. Of the five SNPs, four demonstrated that the PheWAS technique was able to rediscover the expected associations (Denny et al., 2010). The advantage of PheWAS over GWAS is that PheWAS has the capacity to identify multiple diseases that share a common genetic etiology. For example, one of the SNPs genotyped in the original proof-of-principle study was a SNP that tags for the HLA-DRB1*1501 allele (rs3135388). This SNP is known to be associated with multiple sclerosis (MS) (Hindorff, 2012). As expected, rs3135388 was associated with the ICD9 code for MS, but was also associated with other conditions, including erythematous conditions (Denny et al., 2010). This association was subsequently validated in an independent PheWAS (Hebbring et al., 2013). Importantly, if multiple diseases share a common genetic etiology, then it may be hypothesized that drugs used to treat one disease may be repurposed to treat another. As such, PheWAS has the capacity to direct novel drug repositioning studies (Rastegar-MoJarad et al., 2015).
A prerequisite of the PheWAS strategy is the availability of in-depth phenotypic information. Not surprisingly, the majority of PheWASs conducted to date have been applied to cohorts of clinic patients linked to electronic health record (EHR) systems. An EHR system can contain longitudinal health data, including prescription records, laboratory results, physician notes and diagnostic codes, specifically ICD9 coding. The advantage of ICD9 codes is that they offer a spectrum of phenotypic information that is intuitively structured by clinical disease classifications. The first proof-of-principle PheWAS study described previously used ICD9 codes to define the phenome (Denny et al., 2010). The majority of the PheWASs conducted thereafter have also leveraged ICD9 coding (Carroll et al., 2014; Denny et al., 2011, 2013; Hebbring et al., 2013; Neuraz et al., 2013; Ritchie et al., 2013; Shameer et al., 2013); although some have applied ICD10 coding (Neuraz et al., 2013) while others have applied epidemiologic data (Pendergrass et al., 2011, 2012, 2013). In the United States, ICD9 codes are predominantly used administratively for billing. Although ICD9 coding is rooted to clinical manifestations, its reliability is known to be variable (Leone et al., 2006; Hennessy et al., 2010) and ICD9 codes may not provide adequate phenotypic information for many conditions. This is particularly relevant as many ICD9 codes define diseases as ‘other’ or ‘not elsewhere classified’. To address these limitations, other data types beyond ICD9 coding may be used as an alternative approach to define the phenome for PheWAS. Such data could include clinical documentations.
Clinical documentations are often maintained in an EHR system as text data for easy reference during clinical care and can be a powerful source of clinical information for research. Clinical documentation can provide insights into a patient’s past clinical history, current condition, prognosis and treatment. Clinical text data often contains information regarding drugs prescribed and has been mined to identify drug-drug interactions (Iyer et al., 2014), adverse drug events (Liu et al., 2012) and off-label drug use (Jung et al., 2014). Of relevance, clinical text data has also been applied to the identification of specific diseases and may be used to predict ICD9 codes (Kavuluru et al., 2013; Marafino et al., 2014).
In this study, we test the hypothesis that clinical text data can be used to extract extensive phenotypic data to create a text-based phenome. Importantly, we demonstrate that a text-based phenome can be used for a PheWAS, so that EHR systems may be more widely applied to advance genetics and precision medicine.
2 Methods
2.1 Population and genotyping
All patients genotyped come from Marshfield Clinic’s Personalized Medicine Research Project (PMRP), has been described previously (McCarty et al., 2005, 2008). In short, PMRP is a cohort of approximately 20 000 Marshfield Clinic patients over 18 years of age that reside within a 19 zip code region surrounding Marshfield, Wisconsin, USA. PMRP is 98% white/non-Hispanic with 77% claiming German ancestry. Importantly, all PMRP participants are linked to Marshfield Clinic’s extensive EHR system with most participants having over 30 years of continuous care captured in Marshfield Clinic’s EHR system. Samples genotyped include 4235 PMRP patients all over 50 years of age (mean = 74, median = 75) initially selected as a cohort for the study of high-density lipoprotein levels or cataract disease (Turner et al., 2011). The 4235 samples were previously genotyped on Illumina’s 660W BeadChip (Illumina, San Diego, CA) and previously applied to PheWAS analysis (Denny et al., 2011, 2013; Hebbring et al., 2013; Ye et al., 2014).
We analyzed five SNPs for association by text-based PheWAS. These five SNPs were selected based on reported associations by GWAS (Hindorff, 2012) and previously use as control SNPs for an ICD9-based PheWAS (Ye et al., 2014). The five SNPs include rs3135388, rs9501572, rs12678919, rs220073 and rs1061170, which are known to be associated with MS, ankylosing spondylitis, triglyceride levels, atrial fibrillation and age-related macular degeneration (AMD), respectively. Rs9501572 and rs12678919 were genotyped on the Illumina 660W BeadChip as mentioned earlier. SNPs rs3135388, rs2200733 and rs1061170 were genotyped by a Sequenom assay (Sequenom, San Diego, CA), as described previously (Hebbring et al., 2013; Ye et al., 2014).
2.2 Text-based phenome
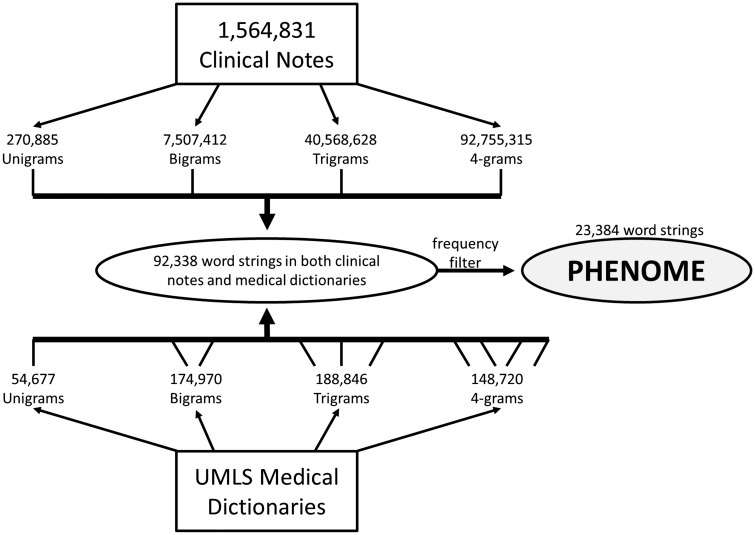
Marshfield Clinic’s EHR system dates back to 1984, with electronic maintenance of clinical documentation since 1991. A total of 1 564 831 clinical notes were extracted from the 4235 patients described earlier representing 423 537 905 unique words. On average, each patient had approximately 372 clinical notes. For each patient, all clinical notes were concatenated and scrubbed of personal identifiers using the de-identification software package ‘de-id’ (Goldberger et al., 2000; Neamatullah et al., 2008). All words were then broken down into four possible combinations. They include unigrams (one word), bigrams (two adjacent words), trigrams (three adjacent words) and 4-grams (four adjacent words). An example of these word structures can be seen in Figure 1 for a physician note indicating ‘… Patient has evidence of macular degeneration ….’ There were a total of 270 885 unigrams, 7 507 412 bigrams, 40 568 628 trigrams and 92 755 315 4-grams totaling 141 102 240 individual word strings in this sample set (Fig. 2). It is expected that these word strings can be used to identify and define clinically relevant phenotypes.
Fig. 1.
An example of unigrams, bigrams, trigrams and 4-grams extracted from the clinical phrase ‘Patient has evidence of macular degeneration’
Fig. 2.
Flow diagram of the process taken to identify the 23 384 word strings used to define the text-based phenome
To simplify the clinical text data and reduce the search space to clinically relevant terms, word strings were cross referenced with the National Library of Medicine’s Unified Medical Language System (UMLS) medical dictionary (Bodenreider, 2004; Lindberg, 1990, 1993) of disease terms (239 227 terms) and drugs (DrugBank; 34 346 terms). The UMLS medical dictionaries were developed in part to standardize biomedical vocabularies. All unigrams, bigrams, trigrams and 4-grams were extracted from these dictionaries representing over 60% of the possible terms. It was noted that only 3% of the 4-grams within the data dictionaries could be cross referenced with clinical notes. As such, word strings greater than 4-grams were not included as they did not add significant clinical information. As with the clinical text data, all extracted strings from the UMLS medical dictionaries were also broken down into their respected unigrams, bigrams, trigrams and 4-grams totaling 567 213 possible word strings. A total of 92 338 word strings were observed in both the UMLS medical dictionaries and clinical text data. A total of 23 384 strings were observed in at least 50 unique patients in the population (Fig. 2). It is these 23 384 word strings that defined the text-based phenome that consists of 97% disease terms and 3% drug terms. Individuals with a given word string were considered cases for that word string, whereas all others were considered controls. No additional word processing was conducted, such as negation analysis. Bigrams represented the largest proportion of the text-based phenome (Fig. 3). The mean and median case size was 533 and 221, respectively. All word strings extracted that define the text-based phenome are available in Supplementary Table S1.
Fig. 3.
Chart describing the number of unigrams, bigrams, trigrams and 4-grams in the text-based phenome separated by disease terms (white bars) and drug terms (black bars). Indicated are the numbers in each category
2.3 PheWAS analysis
All distinct word strings were translated into corresponding case-control groupings. For each SNP, genotypes were associated across all 23 384 word strings that defined the text-based phenome. As a comparison, ICD9 codes defining expected phenotypes, as defined by a previous PheWAS (Ye et al., 2014), were also extracted from Marshfield Clinic’s EHR system and associated with SNP genotype. Associations were measured by χ2 analysis. Phi correlation coefficients were calculated to measure correlations between cases and controls identified by clinical text data and ICD9 coding. Systematic confounding in the SNP-phenotype associations was assessed by a Q-Q plot for each SNP; none was observed (Supplementary Fig. S2). All analyses were conducted using the R statistical package.
3 Results
3.1 Text-based PheWAS results
For all five SNPs, the expected word strings were associated with the SNP genotype (P < 0.02) (Table 1). SNPs rs9501572 and rs3135388 had expected word strings nominally associated with SNP genotype including ‘spondylitis’ (P = 0.018) (Fig. 4A) and ‘MS’ (P = 0.0045) (Fig. 4B), respectively. Rs2200733, rs12678919 and rs1061170 had multiple expected word strings that were associated with SNP genotype at or near the top of their respective PheWAS (Supplementary Table S2). Rs2200733, known to be associated with atrial fibrillation, was associated with multiple word strings related to atrial fibrillation (Fig. 4C) and rs12678919, known to be associated with triglyceride levels and in strong disequilibrium with a nonsense SNP in the gene lipoprotein lipase (rs328), was associated with word strings ‘elevated triglycerides’ and ‘hypertriglyceridemia’ (Fig. 4D). Rs1061170, an AMD-associated SNP, had the strongest PheWAS results. Rs1061170 was strongly associated with the word string ‘macular degeneration’ (P = 1.8E-8) followed by the terms ‘non-exudative’ (P = 2.3E-7) and ‘exudative’ (P = 1.4E-6), which describe AMD subtypes. In addition, the term ‘visudyne’, a drug commonly prescribed to treat AMD, was also strongly associated with the rs1061170 genotype (P = 3.9E-7) suggesting that drug data may provide additional evidence when interpreting PheWAS results (Fig. 4E). Assuming independence, these associations pass a conservative SNP-dependent Bonferroni correction threshold (P < 2.1E-6 assuming α < 0.05 and 23 384 tests/phenotypes). All associations with P < 0.001 are reported in Supplementary Table S2.
Table 1.
Results and cases identified by a text-based phenome of 23 384 word strings and an ICD9-based phenome of 4841 unique codes
| Description | Cases | OR (95% CI) | Raw P-value | Unique cases (%) | Phi correlation | |
|---|---|---|---|---|---|---|
| SNP: | rs1061170—AMD | |||||
| Text | Word string: ‘macular degeneration’ | 1128 | 1.33 (1.20 to 1.46) | 1.80E-08 | 255 (23) | 0.71 |
| ICD9 | ICD9 code: 362.51 (nonexudative senile macular degeneration) | 1086 | 1.37 (1.24 to 1.51) | 5.20E-10 | 213 (20) | |
| SNP: | rs9501572—ankylosing spondylitis | |||||
| Text | Word string: ‘spondylitis’ | 94 | 1.47 (1.08 to 2.00) | 1.80E-02 | 88 (94) | 0.19 |
| ICD9 | ICD9 code: 720.8 (other inflammatory spondylopathies) | 10 | 4.56 (1.86 to 11.2) | 7.10E-04 | 4 (40) | |
| SNP: | rs3135388—MS | |||||
| Text | Word string: ‘MS’ | 255 | 1.41 (1.12 to 1.77) | 4.50E-03 | 238 (93) | 0.23 |
| ICD9 | ICD9 code: 340 (MS) | 20 | 2.55 (1.29 to 5.04) | 9.90E-03 | 3 (15) | |
| SNP: | rs2200733—atrial fibrillation | |||||
| Text | Word string: ‘of atrial’ | 1102 | 1.33 (1.15 to 1.53) | 9.50E-05 | 249 (23) | 0.75 |
| ICD9 | ICD9 code: 427.31 (atrial fibrillation) | 1001 | 1.31 (1.14 to 1.52) | 2.60E-04 | 148 (15) | |
| SNP: | rs12678919—triglyceride metabolism | |||||
| Text | Word string: ‘elevated triglycerides’ | 208 | 0.59 (0.46 to 0.76) | 4.20E-05 | 341 (67) | 0.35 |
| ICD9 | ICD9 code: 272.1 (pure hyperglyceridemia) | 322 | 0.51 (0.36 to 0.72) | 1.10E-04 | 155 (38) |
Fig. 4.
Manhattan plot graphing −log(P-values) across the text-based phenome for the SNPs known to be associated with (A) ankylosing spondylitis, (B) MS, (C) atrial fibrillation, (D) triglyceride metabolism and (E) age-related macular degeneration. Highlighted are the relevant word strings associated with SNP genotype. See Supplementary Table S2 for all phenotypes with P-values less than 0.001
3.2 Text-based versus ICD9-based associations
The top expected association results generated using text data were compared with the top expected ICD9 codes previously identified by an ICD9-based PheWAS (Ye et al., 2014). For all expected phenotypes, there were more cases identified by clinical text data than by ICD9 coding. For example, ICD9 coding identified 20 MS cases compared with 255 cases that were identified by the word string ‘MS’ observed in clinical notes. For three of the five SNPs, top association results were stronger using clinical text data. Not surprisingly, cases and controls identified by text data were variably correlated with cases and controls identified by ICD9 coding. AMD and atrial fibrillation had the strongest correlations. For all examples, text data identified more unique cases that did not overlap ICD9 coding (Table 1). Because the text-based phenome was initially filtered by word strings observed at least 50 times in the population, which may result in missed findings for rare conditions (i.e. MS and ankylosing spondylitis), rs3135388 and rs9501572 were reanalyzed without any frequency filter. No additional expected word strings were identified (data not shown).
4 Discussion
PheWASs are quickly proving effective at rediscovering and discovering new gene-disease associations. Thus far, most PheWASs have focused on genetic variants with predetermined SNP-disease associations (Hebbring, 2014), the largest of which was conducted on 3144 SNPs previously identified by GWAS (Denny et al., 2013). PheWAS exhibits the power to identify novel associations and link multiple conditions to a shared genetic etiology (Carroll et al., 2014; Denny et al., 2011, 2013; Hebbring et al., 2013; Neuraz et al., 2013; Pendergrass et al., 2013; Ritchie et al., 2013; Shameer et al., 2013). A commonality for the majority of PheWASs published to date is the use of ICD9 coding to define the phenome.
An advantage of ICD9-based phenomes is the logical architecture of the codes such that similar conditions have similar codes. For example, ICD9 codes 390–459 define diseases of the circulatory system, ICD9 codes 410–414 define different sub-types of ischemic heart disease, ICD9 codes 410.0–410.9 define acute myocardial infarctions and at the highest phenotypic resolution, ICD9 codes 410.00–410.02 define acute myocardial infarctions of anterolateral wall. It is not surprising that ICD9-based PheWASs have utilized a collapsing strategy to define cases and controls at varying levels of phenotypic resolution (Hebbring, 2014). The current text-based phenome was arranged alphabetically, and as such, lacks intuitive structure. Another challenge of a text-based phenome is the interpretation of the word strings. Cases and controls in an ICD9-based phenome are defined by the existence of a given ICD9 code. Although errors exist in the coding, it is assumed that ICD9 coding is assigned based on clinical manifestations. In comparison, phenotypes described by text data may be less clear. Cases and controls may be incompletely assigned because of misspellings or alternative word usage, such as abbreviations. Furthermore, words defining a text-based phenome may capture a term, yet context beyond the term may be lacking. For example, non-specific terms such as ‘positional’ (Supplementary Table S2) are captured in the text-based phenome. Further curation of the text-based phenome to remove such terms may improve the effectiveness of a text-based PheWAS. Using biomedical named entity recognition tools may be one approach (Leaman et al, 2008).
In addition to non-specific terms, it is conceivable that association results from clinically meaningful terms may still be difficult to interpret. Some clinical notes may indicate that a ‘patient has atrial fibrillation’, ‘patient does not have atrial fibrillation’ and/or ‘patient has a family history of atrial fibrillation’. Under these circumstances, all individuals with the word string ‘atrial fibrillation’ will be lumped together independent of meaning. This may explain why ‘atrial fibrillation’ was not the top word string for rs2200733, although it was still in the top 20 (P = 7.1E-4). The application of natural language processing techniques, such as negation analysis (Agarwal et al., 2011) and ontology-driven concept extraction (Osborne et al., 2007), may be effective when addressing these challenges. Regardless, raw association results indicate that text data performed equivalently to ICD9 coding, and in some instances, clinical text data outperformed ICD9 coding (Fig. 4 and Table 1).
Like GWAS, PheWAS is challenged by multiple comparison testing. The current standard for adjusting multiple comparison testing has been the application of a Bonferroni correction (Hebbring, 2014). Unique to PheWAS is the correlative structure in the phenotypic data. As described earlier, in an ICD9-based PheWAS, similar ICD9 codes may be correlated, especially in phenomes that use a collapsing strategy to define cases and controls at different phenotypic resolutions. Furthermore, correlations may also exist across codes (Hebbring, 2014). This text-based PheWAS is no different. A good example of the correlative structure in the text data may include the atrial fibrillation SNP rs2200733 where the bigram ‘fibrillation with’, is rooted in the trigram ‘atrial fibrillation with’, with both showing similar associations (Fig. 4C). Of all SNPs tested, the AMD SNP rs1061170 not only passed a SNP-dependent Bonferroni correction (P < 2.1E-6) but also passed an experiment-wise correction [P < 4.3E-7 assuming α < 0.05 and 116 920 total tests (total tests = 5 SNP × 23 384 phenotypes)] (Fig. 4E).
Like ICD9 codes, clinical text data can be efficiently extracted from an EHR system and can be effectively applied to a PheWAS (Fig. 4). With all of the words in a text-based phenome rooted to standard terms from a medical dictionary (Fig. 2), this approach may be easily translatable to other EHR systems. Clinical text data has the capacity to describe phenotypic information at a high resolution, while also allowing the use of drug information that may provide additional insights into the interpretation of the PheWAS result, as demonstrated by the AMD-associated SNP and its association with the AMD drug Visudyne (Fig. 4E). Because ICD9 codes are used primarily for billing, only those conditions that arise during a clinical visit are recorded. For patients who come in and out of a service area, or for conditions that predate an EHR system, ICD9 coding may not provide a comprehensive assessment of a patient’s disease history. In comparison, clinical notes may capture both current and in-depth historical health information. This may explain in part the larger case sizes for clinical text data compared with ICD9 coding. Of interest, text data and ICD9 coding demonstrated similar associations, yet both datasets had incomplete correlations between the cases and controls identified by either text data or ICD9 coding (Table 1). In general, a larger percentage of cases are unique to text data compared with cases identified by ICD9 coding. For example, cases of ankylosing spondylitis and MS were dramatically different in the text-based approached compared with the ICD9-based approach suggesting the potential for increased false positives in a text-based phenome. Conversely, SNP rs12678919, known to be associated with triglyceride metabolism, was associated with 508 patients with the word string ‘elevated triglycerides’ (P = 4.2E-5), whereas rs12678919 genotype was associated with 322 patients diagnosed with pure hyperglyceridemia (P = 1.4E-4). The correlation between cases and controls for this example was 0.35 with 67% of cases unique only to text data and 48% of cases unique only to ICD9 data (Table 1). This result may suggest that neither text data, nor ICD9 coding, can comprehensively identify all cases. Improvement in a text-based phenome may be achieved by implementing concept identification approaches, such as negation analysis, word sense disambiguation, and abbreviation expansion. Furthermore, further improvement may be achieved by combining different data types.
In conclusion, this study demonstrates for the first time that raw text data from clinical notes in an EHR system can be used effectively to define a phenome. This study also validates that clinical text data, including drug data, can be applied to PheWAS as a complementary approach to a GWAS and ICD9-based PheWAS. The future of the PheWAS strategy may not be limited to either clinical text data or ICD9 coding. A potential name for this approach might be called a text-wide association study (TextWAS). The future of the PheWAS strategy may rely on multiple structured and unstructured data types. This is particularly relevant as EHR systems become standardized, bio-repositories continue to grow and genomic medicine becomes widely applied.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the support from the Marshfield Clinic Research Foundation. The authors would also like to thank Rachel Stankowski for her assistance in editing this manuscript.
Funding
This work was supported by NCATS grant 9U54TR000021, NCRR 1UL1RR025011, NLM grant 5T15LM007359, 1K22LM011938, NHGRI 1U01HG006389, and NIGMS grant R01GM097618. Mr Majid Rastegar-Mojarad was funded through philanthropic support of Marshfield Clinic Research Foundation’s ‘Dr John Melski Endowed Physician Scientist’ Award to Dr Simon Lin.
Conflict of Interest: none declared.
References
- Agarwal S., et al. (2011) BioNOT: a searchable database of biomedical negated sentences. BMC Bioinformatics, 12, 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenreider O. (2004) The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res., 32, D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll R.J., et al. (2014) R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics, 30, 2375–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny J.C., et al. (2010) PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics, 26, 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny J.C., et al. (2011) Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am. J. Hum. Genet., 89, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny J.C., et al. (2013) Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol., 31, 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D.R., et al. (2010) Inverse association of female hormone replacement therapy with age-related macular degeneration and interactions with ARMS2 polymorphisms. Invest. Ophthalmol. Vis. Sci., 51, 1873–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feskanich D., et al. (2008) Menopausal and reproductive factors and risk of age-related macular degeneration. Arch. Ophthalmol., 126, 519–524. [DOI] [PubMed] [Google Scholar]
- Goldberger A.L., et al. (2000) PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation, 101, E215–E220. [DOI] [PubMed] [Google Scholar]
- Goldstein D.B. (2009) Common genetic variation and human traits. N. Engl. J. Med., 360, 1696–1698. [DOI] [PubMed] [Google Scholar]
- Hebbring S.J. (2014) The challenges, advantages and future of phenome-wide association studies. Immunology, 141, 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbring S.J., et al. (2013) A PheWAS approach in studying HLA-DRB1*1501. Genes Immun., 14, 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy D.A., et al. (2010) Do coder characteristics influence validity of ICD-10 hospital discharge data? BMC Health Serv. Res., 10, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L.A., et al. (2012) A Catalog of Published Genome-Wide Association Studies. www.genome.gov/gwastudies (1 June 2014, date last accessed). [Google Scholar]
- Iyer S.V., et al. (2014) Mining clinical text for signals of adverse drug-drug interactions. J. Am. Med. Inform. Assoc., 21, 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., et al. (2014) Automated detection of off-label drug use. PLoS One, 9, e89324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavuluru R., et al. (2013) Unsupervised extraction of diagnosis codes from EMRs using knowledge-based and extractive text summarization techniques. In Zaïane O., Zilles S. (eds.) Advanced in Artificial Intelligence: Lecture Notes in Computer Science, Volume 7884. Springer, Berlin, pp. 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman R., et al. (2008) Banner: an exucutable survey of advances in biomedical named entity recognition. Pac. Symp. Biocomp., 13, 652–663. [PubMed] [Google Scholar]
- Leone M.A., et al. (2006) Inter-coder agreement for ICD-9-CM coding of stroke. Neurol. Sci., 27, 445–448. [DOI] [PubMed] [Google Scholar]
- Lindberg C. (1990) The unified medical language system (UMLS) of the national library of medicine. J. Am. Med. Rec. Assoc., 61, 40–42. [PubMed] [Google Scholar]
- Lindberg D.A., et al. (1993) The unified medical language system. Methods Inf. Med., 32, 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., et al. (2012) Using temporal patterns in medical records to discern adverse drug events from indications. AMIA Summits Transl. Sci. Proc., 2012, 47–56. [PMC free article] [PubMed] [Google Scholar]
- Manolio T.A., et al. (2009) Finding the missing heritability of complex diseases. Nature, 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marafino B.J., et al. (2014) N-gram support vector machines for scalable procedure and diagnosis classification, with applications to clinical free text data from the intensive care unit. J. Am. Med. Inform. Assoc., 21, 871–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M.I., et al. (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet., 9, 356–369. [DOI] [PubMed] [Google Scholar]
- McCarty C.A., et al. (2005) Marshfield Clinic personalized medicine research project (PMRP): design, methods and recruitment for a large population-based biobank. Per. Med., 2, 49–79. [DOI] [PubMed] [Google Scholar]
- McCarty C.A., et al. (2008) Community consultation and communication for a population-based DNA biobank: the Marshfield Clinic personalized medicine research project. Am. J. Med. Genet. A, 146A, 3026–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neamatullah I. (2008) Automated de-identification of free-text medical records. BMC Med. Inform. Decis. Mak., 8, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need A.C., Goldstein D.B. (2010) Whole genome association studies in complex diseases: where do we stand? Dialogues Clin. Neurosci., 12, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuraz A., et al. (2013) Phenome-wide association studies on a quantitative trait: application to TPMT enzyme activity and thiopurine therapy in pharmacogenomics. PLoS Comput. Biol., 9, e1003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne J.D., et al. (2007) Mining biomedical data using MetaMap transfer (MMtx) and the unified medical language system (UMLS). Methods Mol. Biol., 408, 153–169. [DOI] [PubMed] [Google Scholar]
- Pendergrass S.A., et al. (2011) The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet. Epidemiol., 35, 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrass S.A., et al. (2012) Visually integrating and exploring high throughput phenome-wide association study (PheWAS) results using PheWAS-View. BioData Min., 5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrass S.A., et al. (2013) Phenome-wide association study (PheWAS) for detection of pleiotropy within the population architecture using genomics and epidemiology (PAGE) network. PLoS Genet., 9, e1003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar-Mojarad M., et al. (2015) Opportunities for drug repositioning from phenome-wide association studies. Nature Biotechnology, 33, 342–345. [DOI] [PubMed] [Google Scholar]
- Ritchie M.D., et al. (2013) Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation, 127, 1377–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shameer K., et al. (2014) A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum. Genet., 133, 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S.D., et al. (2011) Knowledge-driven multi-locus analysis reveals gene-gene interactions influencing HDL cholesterol level in two independent EMR-linked biobanks. PLoS One, 6, e19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P.M., et al. (2012) Five years of GWAS discovery. Am. J. Hum. Genet., 90, 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., et al. (2014) Phenome-wide association studies (PheWASs) for functional variants. Eur. J. Hum. Genet., 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.