Abstract
Motivation: The goal of large-scale metabolite profiling is to compare the relative concentrations of as many metabolites extracted from biological samples as possible. This is typically accomplished by measuring the abundances of thousands of ions with high-resolution and high mass accuracy mass spectrometers. Although the data from these instruments provide a comprehensive fingerprint of each sample, identifying the structures of the thousands of detected ions is still challenging and time intensive. An alternative, less-comprehensive approach is to use triple quadrupole (QqQ) mass spectrometry to analyze predetermined sets of metabolites (typically fewer than several hundred). This is done using authentic standards to develop QqQ experiments that specifically detect only the targeted metabolites, with the advantage that the need for ion identification after profiling is eliminated.
Results: Here, we propose a framework to extend the application of QqQ mass spectrometers to large-scale metabolite profiling. We aim to provide a foundation for designing QqQ multiple reaction monitoring (MRM) experiments for each of the 82 696 metabolites in the METLIN metabolite database. First, we identify common fragmentation products from the experimental fragmentation data in METLIN. Then, we model the likelihoods of each precursor structure in METLIN producing each common fragmentation product. With these likelihood estimates, we select ensembles of common fragmentation products that minimize our uncertainty about metabolite identities. We demonstrate encouraging performance and, based on our results, we suggest how our method can be integrated with future work to develop large-scale MRM experiments.
Availability and implementation: Our predictions, Supplementary results, and the code for estimating likelihoods and selecting ensembles of fragmentation reactions are made available on the lab website at http://pattilab.wustl.edu/FragPred.
Contact: gjpattij@wustl.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Large-scale metabolite profiling aims to compare the relative concentrations of as many metabolites as possible between two or more groups of biological samples (Patti et al., 2012). This is generally done using high-resolution, high-mass accuracy quadrupole time-of-flight (QTOF) or Orbitrap mass spectrometers to measure abundances of thousands of ions. The rate-limiting step when using these instruments is establishing metabolite identifications. Obtaining fragmentation data are often necessary for metabolite identification, but there are challenges with both acquiring a breadth of high-quality fragmentation data on targeted precursor ions, and with identifying structures given the resulting fragmentation data. As such, very few of the thousands of ions detected in large-scale metabolite profiling experiments are structurally identified.
Several frameworks exist for using fragmentation data collected with QTOF or Orbitrap instruments to structurally identify detected ions. The most robust approach is to match the fragmentation spectrum of a research sample to the fragmentation spectrum of an authentic standard. This can be done in cases where an experimental spectrum matches a reference spectrum in a database (Tautenhahn et al., 2012), or when the correct reference spectrum can be generated in the lab. When there is no matching reference spectrum, computational methods can be applied to prioritize the structures most likely to have generated the experimental fragmentation spectrum. These methods fall into two categories: those that first precompute a spectrum for every structure in a database and then match the experimental spectrum against the in silico generated spectra (Allen et al., 2014; Kangas et al., 2012; Kind et al., 2013), and those that predict the structural features that likely generated the observed spectrum, and then prioritize putatively matched database structures on the basis of structural similarity (Heinonen et al., 2012; Shen et al., 2014; Wolf et al., 2010).
An alternate, widely used approach for metabolite profiling relies on triple quadrupole (QqQ) mass spectrometers. Metabolite profiling with QqQ mass spectrometers is accomplished by performing targeted analysis on a relatively small number of compounds. This can be employed in a discovery context to assay whether the concentration of a targeted list of metabolites changes between biological samples. Although QqQ-based metabolite profiling provides information about only a limited number of compounds, a major advantage of this approach is that metabolite identifications can be made efficiently once QqQ methods are established. To design a QqQ method, researchers generally purchase commercial standards and use them to identify precursor-to-product ion transitions that can be readily measured by using the QqQ in either multiple reaction monitoring (MRM) (Bajad et al., 2006; Jain et al., 2012) or fragmentation product scanning (Han et al., 2012) mode. The objective is to identify combinations of precursor-fragment pairs that are specific to the metabolite being profiled, although the specificity is often not assessed explicitly.
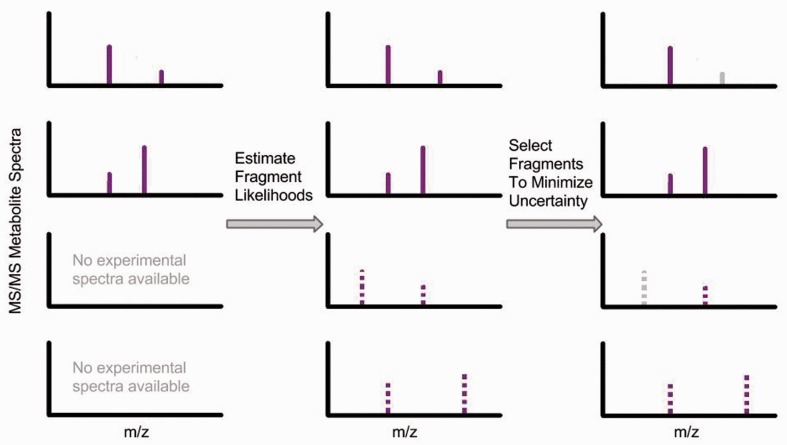
To date, QqQ-based methods have not been applied to profile more than several hundred metabolites. Here, we explore the possibility of extending the use of QqQ-based experiments to analyze the portion of the thousands of detected ions that match a precursor mass in the METLIN metabolite database. First, we identify common fragmentation products from the experimental data in METLIN. Then, we model the likelihoods of each precursor structure in METLIN producing each common fragmentation product. With these likelihood estimates, we select ensembles of common fragmentation products that minimize our uncertainty about metabolite identities. On the basis of these results, we suggest how our method can be integrated with future work to develop large-scale MRM experiments (Figure 1).
Fig. 1.
Schematic for our approach to using QqQ mass spectrometry for large-scale metabolite profiling. Here, we demonstrate our workflow applied to four different representative metabolites, each shown in a different row. First, we use the metabolites with experimental spectra in METLIN to estimate the likelihoods of common fragment production. Then, using these likelihoods we propose a method for selecting a subset of the most informative fragments to design MRM experiments. Dashed spectra represent estimated likelihoods, while grayed-out fragments represent fragments not selected for MRM experiments
2 Methods
QqQ instruments allow sensitive and efficient profiling of ions present in a sample. The major advantage afforded by using the QqQ is that instead of profiling all fragmentation products produced by a specific ion, it is possible to profile select subsets of precursor-product pairs that are sufficient for ion identification. Historically, these precursor-product pairs have been selected by profiling metabolite standards in the lab, without the use of databases, and are assumed to be specific signatures of metabolite structures. We would like to select informative precursor-product transitions using a database, explicitly quantifying our uncertainties about the identities of a detected ion's fragments.
2.1 Uncertainty about structure identity given fragmentation data
We quantify our uncertainty about the metabolite identity given fragmentation data using conditional entropy:
where are the unique structures in METLIN, are the N fragments being considered, each with two states denoting the presence or absence of a fragment, and are the precursor masses. Although future work could focus on specific relative intensities of the fragments, as well as the expected chromatographic retention time of the structures, here we restrict ourselves to just the masses of the precursors and fragments. The data for each metabolite are therefore a precursor mass coupled with a binary string of length N indicating the presences and absences of product ions.
To compute the conditional entropy, we require the joint probability of the structure and the mass spectrometry data, and the conditional probability of the structure given the data. If there was a database containing fragmentation data for every expected metabolite, we could empirically estimate these probabilities using counts. Currently, METLIN contains fragmentation data for only a portion of the expected metabolite structures. To overcome this limitation, we first model the likelihoods that each structure in METLIN produces each relevant fragment, and then approximate the joint and posterior probabilities of those likelihoods.
Because we restrict our analysis of metabolites to those present in the METLIN database, we express conditional probability of the structure given the fragmentation data as:
To compute this probability, we factor the joint probabilities:
where indicates the likelihood of a metabolite ionizing and producing fragments given the structure representation s, is the likelihood of randomly selecting the structure representation s from all representations that have the same precursor mass, and is the prior probability of observing a particular precursor mass.
To obtain the likelihood of observing a pattern of common fragments, we assume that the likelihoods of the selected fragments are conditionally independent, and obtain:
This is not true in general, but a common and convenient simplifying assumption, which has in the past been applied to analysis of text (Ng et al., 2002) and transcription factor binding motifs (Bailey et al., 1995). Additionally, if we greedily select N fragments based on information gain, the fragments added to the ensemble on each iteration will be those with the least correlation with the already selected fragments. The best possible set of fragments would be a set of N independent fragments, such that , and denotes the number of structures with the same precursor. Finally, when quantifying the uncertainty about the structure identities in experiments using just a single fragment (as in precursor and neutral loss scanning, and some MRM experiments), the independence assumption is not necessary.
2.2 Estimating likelihoods of ionization and common fragment production
We use logistic regressions to model likelihoods of a structure ionizing and producing fragmentation products common in METLIN. To limit over-fitting and eliminate uninformative predictors, we fit the logistic regressions with an L1-regularized objective:
where yi indicates that metabolite i ionizes and produces a common fragmentation product given the ionization mode and collision energy, si is a vector representation of the chemical structure, θ is a vector of the fitted weights, α is a regularization constant, and wi is the importance weight of each metabolite, of which there is a total of M. The metabolite importance weights were used to offset the effect of having unbalanced class sizes by setting wi = 1 for , and for y = 1. We identified the optimal regularization parameters for each model using 10-fold cross-validation. All models were fit using LIBLINEAR (Fan et al., 2008).
2.3 Representing chemical structures
We represent each structure in METLIN in two ways. First, we represent the presence and absence of substructures with a binary vector by using several chemical fingerprints. We used the Extended Connectivity Fingerprint and Chemical Hashed Fingerprint from JChem version 6.0.0 (2013, http://www.chemaxon.com) and FP2, FP3, FP4 and MACCS fingerprints from the OpenBabel chemical toolbox (OBoyle et al., 2011).
Second, we represent each METLIN structure in terms of its similarity with each training set structure. We do this using the Tanimoto similarity coefficient, which has been successfully applied as a kernel in a variety of chemical classification and regression problems (Girschik et al., 2012; Swamidass et al., 2005). The Tanimoto coefficient is defined:
for two bit vectors si and sj. We used the Extended Connectivity Fingerprints to compute the similarities between structures with fragmentation data and all structures in METLIN.
2.4 Defining common fragmentation products
When a molecule fragments, some pieces of the molecule may be charged while other pieces may be neutral. Pieces of the molecule that are charged are defined as fragments and can be detected directly by MS. Pieces of the molecule that are neutral cannot be detected directly, but can be calculated indirectly by the difference between precursor and fragment mass-to-charge values. The latter are defined as neutral losses. We consider both fragments and neutral losses to be types of fragmentation products. When considering fragments and neutral losses for multiple compounds, fragments and neutral losses represent the same fragmentation event only when the precursor mass is the same. However, no fragment/neutral-loss pair is exclusive to a single precursor mass.
Our ability to detect a fragmentation product depends on the ionization mode of the instrument, the mass being targeted, the collision energy during fragmentation and the relative abundance of the produced ion. We therefore define four sets of common fragmentation products representing different minimal relative signal intensities, and define each product by its nominal mass, collision energy and instrument polarity. We consider a fragmentation product common if it occurs in at least 50 structures, which we assume to be the fewest number of structures that we could use to estimate likelihoods of fragment production.
2.5 Evaluating fragmentation product predictions
We are interested in quantifying the extent to which we assign higher likelihoods to structures that ionize and produce common fragmentation products over structures that do not ionize or produce them. To do this, we use the area under the receiver operating characteristic curve (AUC), which tracks the trade-off between true- and false-positive rates over a range of decision boundary values. An AUC of 1 represents perfect classification, and an AUC of 0.5 represents random classification.
Additionally, we compare performance conditioned on precursor structures having the same mass. We do this because every QqQ experiment contains masses of both the fragment and the precursor ions, and this can be interpreted as a more realistic estimate of classification performance once a precursor and product ion are observed. To evaluate AUCs conditioned on precursor mass, we group the validation data by nominal precursor mass and report the average of the AUCs in each precursor group that contains structures of both classes.
2.6 Substructure search baseline
One type of QqQ experiment scans all metabolites for a characteristic fragment or neutral loss (Han et al., 2012). The assumption in these experiments is that the targeted fragmentation product is a specific substructure representative of a class of metabolites. We test how well this assumption holds-up for known fragmentation products, as well as how well it generalizes to other substructures. To do this, we use the MetFrag annotations of the fragments present in the METLIN database and perform a chemical substructure search on the intact precursor structures. This is done using the substructure search function in the OpenBabel chemical toolbox (OBoyle et al., 2011). The result is a classifier that assigns a positive result to all structures that contain the substructure, and a negative otherwise, and reflects the conventional thinking in designing certain QqQ experiments.
2.7 Selecting ensembles of fragmentation products
We select ensembles of up to 12 fragmentation products for MRM experiments. To do this, for each precursor mass (rounded to the nearest 0.1 Da) in METLIN, we first select the fragment with the smallest resulting conditional entropy, and then greedily add each additional fragment. We do this using two sets of structures, just those in KEGG, or all of METLIN.
2.8 Evaluating ensembles of fragmentation products
We use two metrics to evaluate the quality of the selected fragments: (i) the portion of metabolites that would be detected given the selected fragments, and (ii) the ranks of posterior probabilities for true metabolite identities given that we selected at least one fragment that they produce. To do this, we retrain fragment likelihoods using 10-fold cross-validation, with the previously determined regularization parameters. This time, the held-out structures are used to evaluate our two metrics instead of fragment production likelihoods. To evaluate which structures would be detected, we evaluate how many of the held-out structures produce at least one of the selected fragments. To evaluate the rank of the posterior likelihoods of the true metabolite identities, we use the fragmentation pattern of the selected metabolite and compute the posterior as defined in Section 2.1 for every structure with that precursor mass.
3 Results and discussion
3.1 Description of METLIN
The METLIN database (Tautenhahn et al., 2013) is currently the largest repository of structures and collision cell induced fragmentation patterns of known metabolites. Each metabolite standard was analyzed in positive and negative ionization modes, and detected fragmentation spectra are obtained at four collision energies on a high-resolution QTOF mass spectrometer. All spectra are annotated using MetFrag (Wolf et al., 2010), and a summary of the available data are presented in Table 1. Our work uses METLIN as of October 22, 2013.
Table 1.
Database summary statistics
| Field | Count |
|---|---|
| METLIN structures | 82 696 |
| METLIN structures with MS/MS data | 11 676 |
| KEGG structures | 17 252 |
| KEGG structures with MS/MS data | 2200 |
| Structures ionized in positive mode | 10 056 |
| Structures ionized in negative mode | 3796 |
| Total fragments | 1 877 378 |
| Unique fragment annotations | 133 898 |
| Unannotated fragments | 384 202 |
| Unique neutral loss annotations | 50 231 |
| Unannotated neutral losses | 617 225 |
In developing a method for metabolite identification using a QqQ instrument, we first identify common fragmentation products that will be detected given a sufficiently abundant precursor ion. Assuming that fragment abundances sum to the precursor abundance, we define four limit of detection thresholds relative to the precursor abundance. Considering the nominal masses of all fragments, we then identify the common fragmentation products for each intensity threshold. This results in a total of 6683 fragments summarized in Table 2.
Table 2.
Summary of selected common fragments
| Signal threshold | Fragment type | Number of fragments | Metabolites per fragment | Annotations per fragment | Fragments per metabolite |
|---|---|---|---|---|---|
| 0.01 | Frag | 1963 | 131 (51–3033) | 66 (1–575) | 38 (0–155) |
| 0.01 | NL | 1817 | 156 (51–10 051) | 5 (0–80) | 39 (2–155) |
| 0.05 | Frag | 912 | 84.5 (51–2079) | 39 (1–214) | 11 (0–33) |
| 0.05 | NL | 809 | 93 (51–10 036) | 3 (0–30) | 12 (0–33) |
| 0.10 | Frag | 466 | 76 (51–1658) | 29 (1–125) | 5 (0–15) |
| 0.10 | NL | 382 | 83 (51–9966) | 2 (0–21) | 6 (0–18) |
| 0.20 | Frag | 189 | 66 (51–1046) | 19 (1–73) | 2 (0–8) |
| 0.20 | NL | 145 | 89 (51–9775) | 2 (0–16) | 3 (0–10) |
Signal threshold represents the minimal abundance of a fragment normalized by the total intensity of all fragments. NL denotes neutral loss. The last three columns are formatted median (min-max).
3.2 Prediction of common-fragment production likelihoods
To obtain likelihoods of observing common fragments given precursor structures, we must predict whether the structures ionize given the ionization mode, and whether they produce a common fragmentation product given that they ionized and were exposed to a specified collision energy. The data in METLIN can be used to obtain these likelihoods independently, first considering all structures with mass spectrometry data to predict ionization, and then using only the structures that ionize to predict fragmentation. This results in average AUCs of 0.94 and 0.96 when predicting ionization for positive and negative modes, respectively. In an effort to avoid propagating errors between the models, we instead jointly predict ionization and fragment production using all 11 676 structures in a single model for each fragmentation product.
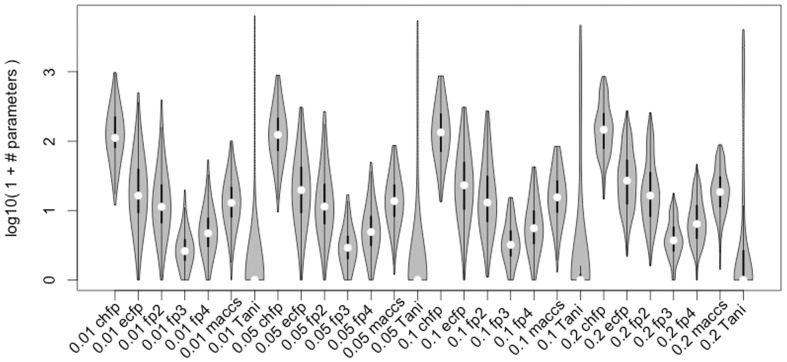
We use global and local descriptors to represent the structures in METLIN. Locally, we use chemical fingerprints to represent each structure in terms of its constituent substructures. Globally, we represent the similarities between structures using Tanimoto coefficients between training and test structures. To limit over-fitting and select only the most informative predictors, we fit out our logistic regressions with an L1 regularization penalty. This results in a median of 299 predictors per fragmentation product, distributed as shown in Figure 2. Interestingly, global similarities of structures become more important as minimal signal intensities increase, suggesting that molecular class is important for the abundance of the detected fragment.
Fig. 2.
Distribution of the number of fitted parameters to estimate fragment production likelihoods by predictor type and signal intensity. The x-axis is formatted minimal signal, predictor type. The black line and white dots are box and whisker plots, the gray area is the kernel density
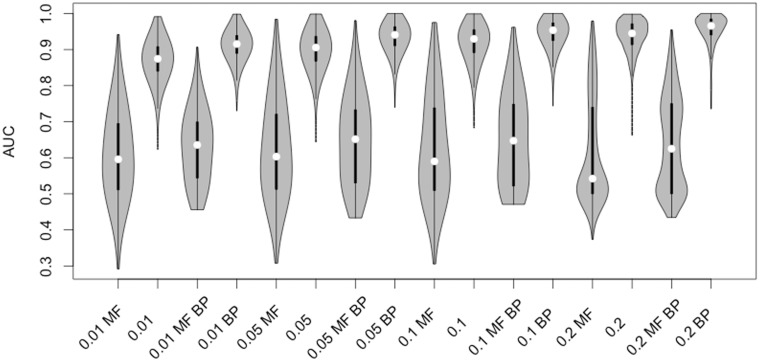
To evaluate the quality of the assigned fragment production likelihoods, we use AUC, which approximates the probability that an occurring fragmentation event is assigned a higher likelihood than a fragmentation event that does not occur. Considering the performance of just the structure representations, we obtain a median AUC of 0.935. However, because QqQ experiments always provide precursor masses coupled with fragment masses, we could limit our analysis to only those validation set structures that share precursor masses. This reduces the validation sets from 1176 structures to a median of 62 structures. Within these validation sets, our median AUC improves to 0.957. The distribution of AUCs by fragmentation product is shown in Figure 3.
Fig. 3.
Violin plots of fragment prediction AUCs at four minimal signal thresholds. MF denotes the MetFrag substructure search baseline, BP denotes that structures were grouped by precursor. The black line and white dots are box and whisker plots, the gray area is the kernel density
To compare our likelihoods against a baseline method, we emulated the conventional approach taken by QqQ mass spectrometrists, namely a substructure search. When used in previous QqQ work, such as lipidomics (Han et al., 2012) or non-targeted CoA profiling (Zimmerman et al., 2013), a specific fragment representative of a class of metabolites is selected, and precursors containing the functional group that produces the fragment are considered to be likely matches. We generalized this approach to work with every annotated common fragment in METLIN. Although the substructure search method has worked well for characteristic fragments in past research, our results show that it does not extend well to arbitrary annotated fragments, and that our likelihood estimates are significantly better at prioritizing fragment-producing precursors.
3.3 Selecting ensembles of fragmentation products for QqQ experiments
Having shown that METLIN contains many commonly detectable fragmentation products and that we are able to model their likelihoods, we now aim to identify subsets of those fragmentation products for designing tractable QqQ experiments. Depending on the type of biological sample and on the chromatography conditions of the QqQ experiment, there is a varying capacity for profiling precursor-product transitions. Because our goal is to discriminate between a median of 62 structures, we need to select a median of at least six fragments. We anticipate that not all selected fragments will be independent, and therefore select up to 12 fragments, which verges on the technical limitations of high-throughput QqQ experiments.
We assess the performance of our method in 16 conditions, when using four signal to noise thresholds, two ionization modes, and two databases. To compare the performance of the method, we use two different metrics. First, we emulate the conventional mode of evaluating MRM methods, by simply requiring that a targeted fragment be detected. To do this, for every structure that produces at least one detectable fragment under the specified conditions, we ask: what portion of the metabolites in the database produce at least one of the selected fragments? To go further than the conventionally designed MRM experiments, we then assess how well we are able to rank detected structures given the selected fragments.
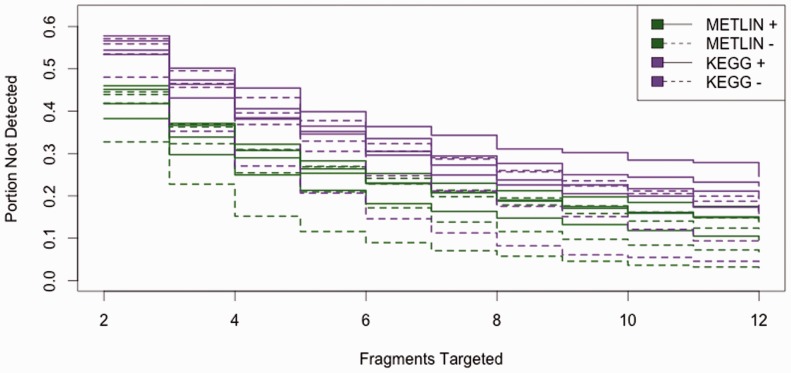
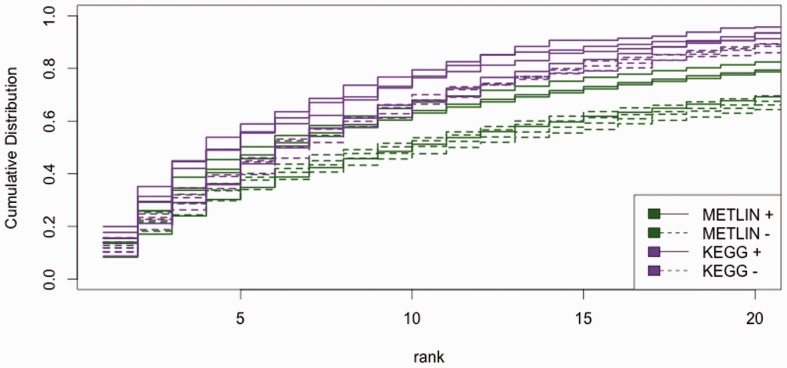
The complete results are available in Supplementary Figure S1 and Supplementary Table S1. Figure 4 shows that by selecting 12 fragmentation products, we detect as many as 0.973 of METLIN structures ionizing in negative mode and 0.904 of METLIN structures ionizing in positive mode. In comparison, we detect as many as 0.970 of KEGG structures ionizing in negative mode and 0.842 of KEGG structures ionizing in positive mode. Using 12 fragments, Figure 5 shows that 0.825 of METLIN structures are returned in the top 20 matches for positive-mode data and 0.700 of METLIN structures are returned in the top 20 matches for negative-mode data. Comparatively, 0.961 of KEGG structures are returned in the top 20 matches for positive-mode data, while 0.893 KEGG structures are returned in the top 20 matches for negative-mode data. This performance can be valuable for some experiments, but our primary intent is to provide a benchmark for future work improving on our results.
Fig. 4.
Portion of metabolites not detected given the number of selected fragments
Fig. 5.
Ranks of detected metabolites using 12 fragments per precursor
3.4 Applications
We have demonstrated our ability to prioritize precursor structures that produce common fragmentation products and to select sets of fragmentation products that prioritize true metabolite identifications. The number of structures in METLIN and KEGG, however, exceeds the number of MRM experiments that QqQ instruments can perform in a single analytical run. This can be overcome by either performing multiple analytical runs, or by limiting analysis to a portion of all METLIN or KEGG structures, for instance to those detected at sufficient abundance using MS1 profiling.
Applications of our database-driven design of MRM experiments will be improved by incorporating two additional sources of information about structural identities. First, after initial MS1 profiling of the sample, the prior probability of an ion identity given the precursor mass can be obtained by modeling the expected retention times of each structure (Creek et al., 2011; Hall et al., 2012; Stanstrup et al., 2013). Second, while our work selected all fragments simultaneously, if multiple experiments are used to profile a sample, fragments can be selected based on obtained experimental results, restricting the likely structure candidates for each experiment. We therefore anticipate that performance will improve as our approach is applied to experimental data.
4 Conclusion
Although conventional large-scale metabolite profiling detects thousands of ions with high-resolution mass spectrometers, the analysis of these datasets and subsequent structural identification of metabolites has proven challenging. In contrast, profiling and structural identification of a targeted number of metabolites by using QqQ-based MRM experiments is robust and efficient once experimental methods are established. Here, we have explored the design of QqQ-based MRM experiments toward profiling of each metabolite in the METLIN and KEGG databases. To overcome the lack of fragmentation data for a majority of the structures in these databases, we modeled the likelihoods that these structures produce common fragmentation products. We demonstrated that ensembles of our predicted fragmentation products can be used to effectively prioritize METLIN and KEGG structures with the same precursor mass. Although this is a first step toward addressing the challenge of developing MRM experiments for tens of thousands of metabolites, future research is needed to improve upon the specificity and reduce the number of MRMs suggested here by using additional experimental parameters such as retention time.
Supplementary Material
Funding
This work was supported by the National Institutes of Health Grants R01 ES022181 (GJP) and L30 AG0 038036 (GJP).
Conflict of Interest: none declared.
References
- Allen F., et al. (2015) Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics , 11, 98–110. [Google Scholar]
- Bailey T.L., et al. (1995) Unsupervised learning of multiple motifs in biopolymers using EM. Mach. Learn., 21, 51–80. [Google Scholar]
- Bajad S.U., et al. (2006) Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A , 1125, 76–88. [DOI] [PubMed] [Google Scholar]
- Creek D., et al. (2011) Toward global metabolomics analysis with hydrophilic interaction liquid chromatography mass spectrometry: improved metabolite identification by retention time prediction. Anal. Chem. , 83, 8703–8710. [DOI] [PubMed] [Google Scholar]
- Fan R.-E., et al. (2008) LIBLINEAR: a library for large scale linear classification. J. Mach. Learn. Res. , 9, 1871–1874. [Google Scholar]
- Girschick T., et al. (2012) Similarity boosted quantitative structure-activity relationship—a systematic study of enhancing structural descriptors by molecular similarity. J. Chem. Inf. Model. , 53, 1017–1025. [DOI] [PubMed] [Google Scholar]
- Hall L.M., et al. (2012) Development of Ecom50 and retention index models for nontargeted metabolomics: identification of 1,3-dicyclohexylurea in human serum by HPLC/mass spectrometry. J. Chem. Inf. Model. , 52, 1222–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., et al. (2012) Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analysis. Mass Spectrom. Rev. , 31, 134–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen M., et al. (2012) Metabolite identification and molecular fingerprint prediction through machine learning. Bioinformatics , 28, 2333–2341. [DOI] [PubMed] [Google Scholar]
- Jain M., et al. (2012) Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science , 336, 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangas L.J., et al. (2012) In silico identification software (ISIS): a machine learning approach to tandem mass spectral identification of lipids. Bioinformatics , 28, 1705–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T., et al. (2013) LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods , 10, 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng A.Y., Jordan M.I. (2002) On Discriminative vs. Generative Classifiers: A comparison of Logistic Regression and Naive Bayes, Neural Information Processing Systems. Vol. 14. [Google Scholar]
- OBoyle N.M., et al. (2011) OpenBabel: an open chemical toolbox. J. Cheminform. , 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti G.J., et al. (2012) Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol., 13, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., et al. (2014) Metabolite identification through multiple kernel learning on fragmentation trees. Bioinformatics, 30, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanstrup J., et al. (2013) Metabolite profiling and beyond: approaches for the rapid processing and annotation of human blood serum mass spectrometry data. Anal. Bioanal. Chem. , 405, 5037–5048. [DOI] [PubMed] [Google Scholar]
- Swamidass S.J., et al. (2005) Kernels for small molecules and the prediction of muta-genicity, toxicity, and anti-cancer activity. Bioinformatics , 21, 1359–1368. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R., et al. (2013) An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. , 30, 826–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., et al. (2010) In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinformatics , 11, 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., et al. (2013) Nontargeted profiling of coenzyme A thioesters in biological samples by tandem mass spectrometry. Anal. Chem. , 85, 8284–8290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.