Abstract
Summary: A genetic variant can be represented in the Variant Call Format (VCF) in multiple different ways. Inconsistent representation of variants between variant callers and analyses will magnify discrepancies between them and complicate variant filtering and duplicate removal. We present a software tool vt normalize that normalizes representation of genetic variants in the VCF. We formally define variant normalization as the consistent representation of genetic variants in an unambiguous and concise way and derive a simple general algorithm to enforce it. We demonstrate the inconsistent representation of variants across existing sequence analysis tools and show that our tool facilitates integration of diverse variant types and call sets.
Availability and implementation: The source code is available for download at http://github.com/atks/vt. More detailed documentation is available at http://genome.sph.umich.edu/wiki/Variant_Normalization.
Contact: hmkang@umich.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Methods for calling genetic variants from sequence data are rapidly evolving beyond single nucleotide polymorphisms (SNPs), to more complex variants such as short insertions and deletions (indels), short tandem repeats (STRs), multi-nucleotide polymorphisms (MNPs), structural variations (SVs) and others. These different classes of variants are typically represented in the Variant Call Format (VCF) (Danecek et al., 2011), which provides a format for storing variant calling results for diverse variant types generated by different tools.
Different sequence analysis software tools often represent the same sequence variant in different ways in a VCF file, making it non-trivial to integrate and compare variants across call sets. However, the impact of ambiguous variant representations on the analysis of sequence data is under-appreciated, and there is no standard guideline for consistent representation of variants.
Here we provide a formal definition and algorithm for variant normalization. Our definition and algorithm enable the representation of variants in an unambiguous, unique way. We show that existing variant calling software tools often do not consistently represent complex variants. Finally, we demonstrate how our normalization method helped integrate different variant call sets in the 1000 Genomes Project (1000 Genomes Project Consortium, 2012).
2 Variant normalization
2.1 Definitions
We define several terms related to variant normalization. A sequence is defined as a string of nucleotides. A reference sequence is a sequence representing the reference genome, and an alternate sequence is a sequence that differs from the reference sequence.
A variant is defined as a combination of a reference and at least one alternate sequence. A VCF entry is defined as a combination of (i) chromosome name, (ii) base position, (iii) reference allele and (iv) alternate alleles, where alleles are sequences of positive length. A VCF entry represents a variant, if—starting at the chromosome and base position indicated—its reference and alternate alleles exactly match the reference and alternate sequences of a variant while outside the portion represented by VCF is identical to the reference sequence. Figure 1 illustrates how multiple VCF entries can represent the same variant.
Fig. 1.
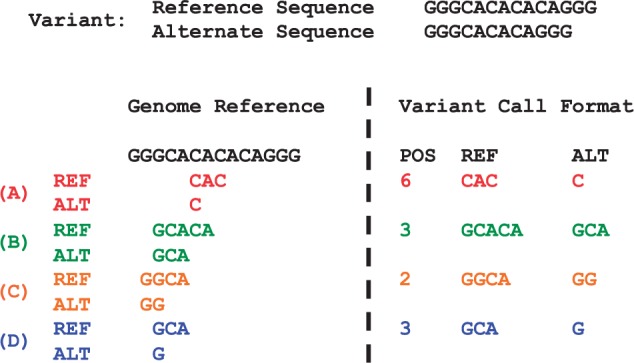
Example of VCF entries representing the same variant. Left panel aligns each allele to the reference genome, and the right panel represents the variant in VCF. (A) is not left-aligned (B) is neither left-aligned nor parsimonious, (C) is not parsimonious and (D) is normalized
A VCF entry is normalized if and only if it is left aligned and parsimonious. A VCF entry is left aligned if and only if its base position is smallest among all potential VCF entries having the same allele length and representing the same variant. A VCF entry is parsimonious if and only if the entry has the shortest allele length among all VCF entries representing the same variant. The concept of left alignment is used across different sequence analysis tools such as GATK (DePristo et al., 2011), but it has not been precisely defined. The left alignment and parsimony criteria ensure that a variant is unambiguously and concisely represented by a normalized VCF entry (see Lemma 1 in Supplementary material). Figure 1D is an example of normalized VCF entry.
2.2 Normalizing a VCF entry
While variant normalization is now clearly defined, verifying whether a VCF entry is normalized may appear challenging and even complicated.
We introduce a necessary and sufficient condition for a VCF entry to be normalized in a principled fashion:
The alleles end with at least two different nucleotides.
The alleles start with at least two different nucleotides, or the shortest allele has length 1.
The first condition ensures that the VCF entry is left aligned, and the second condition ensures that the VCF entry is parsimonious among all left aligned entries representing the same variant. Based on these simplified rules, a VCF entry can be normalized by the procedure described in Algorithm 1. Our algorithm has two parts. The first part focuses, counter-intuitively, on the rightmost base for each allele in bi-allelic or multi-allelic variant. Whenever this base is identical across all alleles, the variant start point can be shifted to the left. The second part simply trims redundant sequences at the beginning of each allele, ensuring that all alleles are represented uniquely and as tersely as possible (see Supplementary material for detailed proofs).
Algorithm 1 Normalize a VCF entry.
Input: A VCF entry and the reference genome sequence.
Output: A normalized VCF entry
1 : do
2 : if all alleles end with same nucleotide then
3 : truncate the rightmost nucleotide of each allele
4 : if any allele is length zero then
5 : extend all alleles by 1 nucleotide to the left
6 : while changes made in the VCF entry in the loop
7 : while all alleles start with same nucleotide and length ≥2 do
8 : truncate the leftmost nucleotide of each allele
9 : end while
10 : return the VCF entry
3 Results
3.1 Integration of 1000 Genomes Variant Calls
We applied our normalization method to the call sets contributed to the 1000 Genomes phase 3 consensus building process, excluding structural variants. The union of unfiltered call sets consists of 186 051 502 VCF entries. Among these, the majority represents SNPs, but 25 521 642 represent non-SNPs. We classified the non-SNP entries into MNPs, short insertions and deletions (indels) and the other complex variants (Table 1). 5 057 823 (19.8%) non-SNP entries needed normalization, resulting in 473 767 (1.9%) redundant entries. The leftAlignAndTrimVariants tool in GATK, which is designed to normalize simple bi-allelic indels, produces results that match our algorithm for 96.6% of the variants, but fails to normalize 615 515 variants, most (99.9%) of which are multi-allelic indels. All unnormalized variants were parsimonious but not left aligned.
Table 1.
Summary of unique non-SNP VCF entries across the 1000 Genomes phase 3 call sets, comparing before and after normalization
| Type | Counts/% of variants | MNP | Indel | Others |
|---|---|---|---|---|
| Bi-allelic | Total raw count | 1 471 391 | 12 885 278 | 744 473 |
| % Need normalization | 16.6 | 2.3 | 90.9 | |
| % Redundant | 5.7 | 1.4 | 9.1 | |
| After normalization | 1 387 196 | 12 710 933 | 676 395 | |
| Multi-allelic | Total raw count | 2 227 300 | 5 432 444 | 2 760 756 |
| % Need normalization | <0.01 | 37.2 | 65.8 | |
| % Redundant | <0.01 | 1.3 | 2.8 | |
| After normalization | 2 227 281 | 5 361 318 | 2 684 752 |
3.2 Existing variant resources including dbSNP
We also applied normalization on a published resource of array-based bi-allelic indels (Mills et al., 2011) and observed that 3994 of 9996 (40%) of VCF entries were not normalized, and 1092 (11%) were redundant. Next, we normalized 6 367 920 non-SNP variants deposited in dbSNP 141 (Sherry et al., 2001), and observed that 897 438(14.9%) entries were not normalized and 289 958 (4.6%) were redundant (Table 2). After normalization, the overlap with 1000 Genomes phase 3 increased from 49 and 26 to 81% and 28%, respectively, for the array-based indels list and for dbSNP, respectively. The overlap of the array-based indels list with dbSNP increased from 60 to 86% after normalizing both datasets. Our results highlight the impact of variant normalization on assessing the novelty and quality of non-SNP variants.
Table 2.
Summary of unique non-SNP VCF entries in bi-allelic indel array resources (Mills et al., 2011) and dbSNP 141, before/after normalization
| Counts/% of variants | Indel array | dbSNP 141 |
|---|---|---|
| Number before/after normalization | 9996/8904 | 6 367 920/6 077 962 |
| % 1000G overlaps (before/after) | 49%/81% | 26%/28% |
| % Need normalization | 40% | 14% |
| % Redundant | 11% | 4.6% |
4 Conclusion
Consistent representation of genetic variants is important in many contexts of sequence analysis, including evaluation of variant quality, integration across datasets and functional interpretation of variants. We demonstrate that a substantial fraction of existing tools and resources need to be normalized, and propose a formal and easy-to-implement standard to represent a variant in VCF, with publicly available implementation. We expect that our principled proposal for variant normalization will facilitate more accurate analysis and integration of genetic variants.
Supplementary Material
Acknowledgements
We thank 1000 Genomes analysis group for making individual call sets publicly available.
Funding
This study was funded through grants from the National Institutes of Health (NHGRI and NHLBI) (HG006513, HG007022, HL117626).
Conflict of Interest: none declared.
References
- 1000 Genomes Project Consortium (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., et al. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M.A., et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.E., et al. (2011) Natural genetic variation caused by small insertions and deletions in the human genome. Genome Res., 21, 830–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S.T., et al. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res., 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


