Abstract
Objective
To conduct an epigenome-wide analysis of DNA methylation and obesity traits.
Design and Methods
We quantified DNA methylation in CD4+ T-cells using the Illumina Infinium Human Methylation450 array in 991 participants of the Genetics of Lipid Lowering Drugs and Diet Network. We modeled methylation at individual cytosine-phosphate-guanine (CpG) sites as a function of body mass index (BMI) and waist circumference (WC), adjusting for age, gender, study site, T-cell purity, smoking, and family structure.
Results
We found epigenome-wide significant associations between eight CpG sites and BMI and five CpG sites and WC, successfully replicating the top hits in whole blood samples from the Framingham Heart Study (n=2,377) and the Atherosclerosis Risk in Communities study (n=2,105). Top findings were in CPT1A (meta-analysis P= 3.5×10−37 for BMI and P=2.2×10−16 for WC), PHGDH (meta-analysis P= 4.7×10−15 for BMI and 2.2×10−8 for WC), CD38 (meta-analysis P= 3.7×10−11 for BMI and 6.1×10−13 for WC) and long intergenic non-coding RNA 00263 (meta-analysis P= 1.2×10−13 for BMI and 5.8×10−10 for WC), regions with biologically plausible relationships to adiposity.
Conclusions
This large-scale epigenome-wide study discovered and replicated robust associations between DNA methylation at CpG loci and obesity indices, laying the groundwork for future diagnostic and/or therapeutic applications.
Keywords: body mass index, waist circumference, obesity, epigenetics, genomics, CpG methylation
Introduction
Body mass index (BMI) and waist circumference (WC) are quantitative traits characterized by multifactorial etiology, relevance to disease risk, and high heritability (1). However, known genetic polymorphisms explain only a modest fraction of inherited variation in both obesity-related traits (2). Emerging evidence suggests that epigenetic changes play a key role in both heritable and environmental influences on obesity (3). Consistent with that paradigm, recent studies identified DNA methylation patterns in several biologically relevant regions as important correlates of BMI (4, 5, 6).
Epigenetic processes such as DNA methylation underlie the gene-environment interactions in complex traits by changing gene expression (7). This relationship between DNA methylation and gene expression is further complicated by DNA sequence variation, which contributes to both processes through expression- (eQTL) and methylation quantitative trait loci (meQTL). In the pathogenesis of obesity, changes in DNA methylation may represent both the cause and the consequence, acting through inflammation, oxidative stress, hypoxia, or other biological pathways (4, 8, 9). Beyond changes in mean DNA methylation levels, obesity is also associated with increased variance in DNA methylation, possibly reflecting adaptation to changing environmental conditions (5).
Because most population studies to date have pursued the candidate gene approach, many potentially novel associations between DNA methylation patterns and obesity remain unexplored. Large-scale studies with well-characterized methylome data are poised to fill this gap. To that end, we quantified methylation levels at ~470,000 cytosine-phosphate-guanine (CpG) sites in CD4+ T-cells from 991 participants of the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) study (https://dsgweb.wustl.edu/goldn/). Our choice of cell type was informed by evidence of association between obesity-related immune dysfunction and DNA methylation (6) as well as relative abundance of CD4+ T-cells. Subsequently, we performed an epigenome-wide association study of BMI and WC and replicated several of our top findings in blood leukocyte samples from the Framingham Heart Study (FHS) and the Atherosclerosis Risk in Communities (ARIC) study.
Methods
Study subjects
The design of the family-based GOLDN study is described in detail in prior publications (10). Briefly, we recruited families with at least two siblings from the participants of the National Heart, Lung, and Blood Institute Family Heart Study at the genetically homogeneous sites of Minneapolis and Salt Lake City. All participants (n=1327) self-identified as European Americans. The goal of the trial was to identify genetic and epigenetic factors that mediated response to acute lipid-raising (i.e. postprandial lipemia challenge) or lipid-lowering (3 week fenofibrate therapy) interventions. Before each study visit, participants abstained from eating for at least 8 hours, drinking alcohol for 24 hours, and using lipid-lowering medications for 4 weeks. We collected DNA samples and BMI/WC measurements for this epigenome-wide study at the baseline visit. The descriptions of the FHS and ARIC populations, as well as other methods pertaining to replication analyses, are summarized in Supporting Information. GOLDN, FHS, and ARIC study protocols were approved by Institutional Review Boards at each participating university and all participants provided written informed consent.
Measurement of obesity traits and covariates
On the day of anthropometric data collection, GOLDN participants wore light clothes and no shoes. Weight was measured by a beam balance and height was ascertained by a stadiometer. BMI was calculated as weight in kilograms divided by height in meters squared. WC was measured over the unclothed abdomen at the umbilicus at the end of a normal expiration (11). Smoking (current vs. not) and demographic covariates were self-reported via questionnaire.
DNA isolation and bisulfite conversion
In the GOLDN study, DNA was isolated from CD4+ T-cells harvested from frozen buffy coat samples from peripheral blood using positive selection by antigen-specific magnetic beads (Invitrogen, Carlsbad, CA, USA). We chose CD4+ T-cells for reasons of both biological plausibility, as many metabolism-related genes are expressed in immune cells (12), as well as logistics, as they are the most abundant lymphocyte in humans. We lysed cells captured on the beads and extracted DNA using DNeasy kits (Qiagen, Venlo, Netherlands). Bisulfite conversion was performed on 500 ng of genomic DNA using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA) prior to quantifying epigenome-wide methylation (13).
Epigenome-wide association study
All three studies used the Infinium Human Methylation450 array (Illumina, San Diego, CA) to quantify genome-wide DNA methylation. The GOLDN analysis is described in extensive detail in a manuscript from our group (13). After the standard steps of whole genome amplification, hybridization, and imaging, GOLDN, ARIC, and FHS investigators used Illumina GenomeStudio software to estimate β scores, defined as the proportion of total signal from the methylation-specific probe or color channel, and detection P-values, defined as the probability that the total intensity for a given probe falls within the background signal intensity.
Normalization and quality control
During the quality control stage in GOLDN, we removed any β scores with an associated detection P-value greater than 0.01 and samples with more than 1.5% missing data points, as well as CpGs where the probe sequence mapped either to a location that did not match the annotation file, or to more than one locus (13). Additionally, we excluded any CpG probes where more than 10% of samples failed to yield adequate intensity. A total of 991 samples and 461,281 CpGs were used in the analysis. The resulting β scores were normalized using the ComBat package (http://www.bu.edu/jlab/wp-assets/ComBat/Download.html) for R software (14). A detailed published description of our normalization methods and the performance of ComBat is available (13). Briefly, we performed the normalization on random subsets of 20,000 CpGs per run, with each array of 12 samples used as a “batch,”adjusting for both plate and position on the plate. We separately normalized probes from the Infinium I and II chemistries and subsequently adjusted the β scores for Infinium II probes as described earlier (13). To adjust for cell purity, we generated principal components (PCs) based on the β scores of all autosomal CpGs that passed quality control. Additionally, we estimated predicted CD4+ T-cell purity using a linear model based deconvolution method derived from the methods described by Abbas et al. (15) and Houseman et al (16). To estimate the percentage of CD4+ T-cells versus other cells in each sample, we used a reference data set of CD4+ T-cells and granulocytes extracted from fresh blood sample. Predicted CD4+ T-cell purity (%) was robustly associated with the first PC (r2=0.85) but not other PCs (17).
Statistical analysis
In the GOLDN study, we modeled associations between methylation scores at each CpG site and obesity traits using linear mixed models, adjusted for age, sex, study site, current smoking, the first 4 CD4+ T-cell purity PCs, and pedigree as a random effect using the lmekin function of the kinship package (http://cran.r-project.org/src/contrib/Archive/kinship/) in R (18). Previously, we tested genetic ancestry as a potential confounder and did not find associations with the outcome, likely due to the homogeneity of our study population. We implemented a stringent Bonferroni correction to address the multiple testing problem, setting the statistical significance level at 0.05/470,000= 1.1×10−7. We constructed a Manhattan plot to visualize the results.
CpG sites that reached a P-value < 1.0 ×10−7 (8 for BMI and 5 for WC) were tested in the replication stage. Consequently, the Bonferroni-corrected P-value in the replication stage was 0.006 for BMI and 0.01 for WC. BMI replication analyses were performed in both FHS and ARIC, while WC data were only available in ARIC. Further details on statistical methods in the replication stage are available in Supporting Information. We meta-analyzed P-values from GOLDN, 2 batches of FHS, and ARIC. Because of the different magnitude of effects between FHS and the other two cohorts at the CPT1A locus, the meta-analysis P-value calculation for cg00574958 used the Chernoff bound of the chi-squared cumulative distribution function. At all other loci, we calculated the summary P-values exactly.
Results
Table 1 summarizes the general characteristics of the GOLDN, FHS, and ARIC populations. Approximately half of GOLDN participants were recruited at each of the Minnesota and Salt Lake City study sites, while 93% of the ARIC cohort came from the Mississippi site, with only 7% recruited at the North Carolina Site; all FHS participants were recruited in Framingham, Massachusetts. The discovery and replication cohorts differ most strikingly on the selection of samples for methylation analysis, with the former restricted to CD4+ T-cells and the latter both using whole blood. FHS participants were considerably older than either GOLDN or ARIC participants. With the exception of the sex distribution, the two FHS laboratory batches were demographically and anthropometrically similar. The main distinction between GOLDN/FHS and ARIC is the racial composition of the cohorts; while GOLDN and FHS are entirely comprised of European Americans, the methylation data in ARIC was only available on African Americans. Compared to GOLDN, ARIC participants were on average older, more likely to be female, and more likely to report current smoking. The mean values for BMI and WC in all three cohorts slightly exceeded the clinical guidelines for healthy weight (19, 20).
Table 1.
Demographic and anthropometric characteristics of the study populations.
| GOLDN (n=991) | ARIC (n=2,106) | FHS Case-Control (n=1,935) | FHS Random Sample (n=442) | |
|---|---|---|---|---|
| Age, yearsa | 49 ± 16 | 56 ± 6 | 65 ± 9 | 71 ± 8 |
| Sex, % female | 52 | 63 | 61 | 30 |
| Race, % | ||||
| European American, % | 100 | -- | 100 | 100 |
| African American, % | -- | 100 | -- | -- |
| Current smokers, % | 7 | 24 | 9 | 6 |
| Body mass index, kg/m | 28 ± 6 | 30 ± 6 | 28 ± 6 | 29 ± 5 |
| Waist circumference, cm | 97 ± 16 | 101 ± 15 | -- | -- |
Values are shown as mean ± SD or %
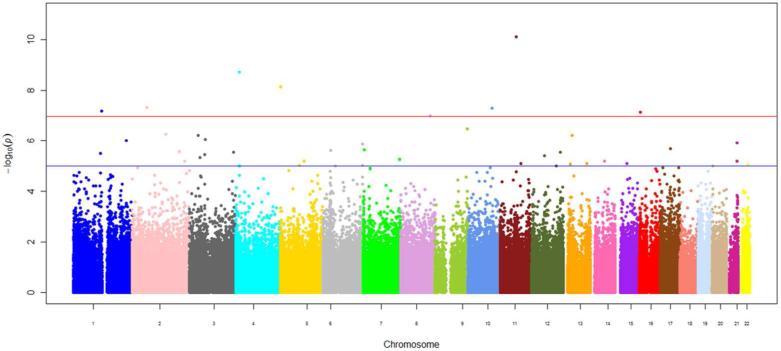
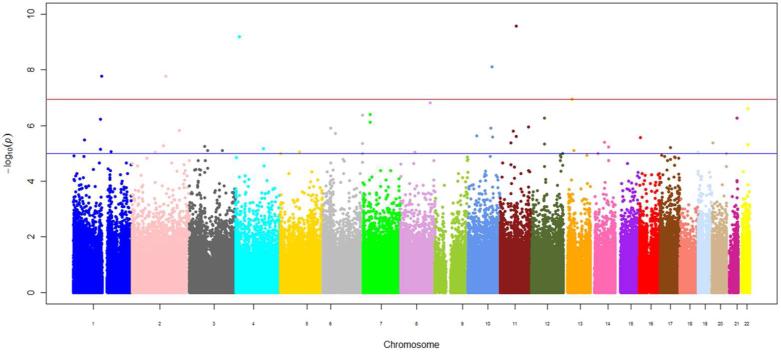
Using the Bonferroni-corrected statistical significance level of 1.1×10−7, we identified eight loci where methylation status was associated with BMI (Table 2, Figure 1) and five loci for WC (Table 3, Figure 2). In the MI case-control batch from FHS, we successfully replicated significant associations with four out of the 8 BMI loci, located in CPT1A, PHGDH, CD38, and a site on chromosome 10 located in the promoter of a long non-coding intergenic RNA (lincRNA). Of those, associations with CpG sites in CPT1A and PHGDH also replicated in the larger FHS random sample. In ARIC, we replicated associations between BMI with CpG sites in CPT1A and CD38, as well as the association of WC with the CpG site in the lincRNA on chromosome 10. The directions of the associations were also confirmed: at a CpG site in the first intron of CPT1A, the β score was inversely associated with BMI and WC, while the methylation status of a CpG site adjacent to the transcriptional start site of CD38 was positively associated with both phenotypes in all three studies. The replicated association between the methylation status of the lincRNA CpG site and obesity traits was also uniformly positive.
Table 2.
Top CpG methylation sites associated with body mass index in GOLDN (n=991), FHS (n=1,935 for batch 1 and 442 for batch 2), and ARIC (n=2,105) at the level of genome-wide significance (P<1.1×10−7) in the discovery phase.
| Marker | Chr | Gene | GOLDN | FHS Case-Control | FHS Random Sample | ARIC | Meta P | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| βa ± SE | P | β ± SE | P | β ± SE | P | β ± SE | P | ||||
| cg00574958 | 11 | CPT1A | −0.0009 ± 0.0001 | 7.6×10−11 | −0.04 ± 0.004 | 2.4×10−24 | −0.04 ± 0.01 | 1.2×10−5 | −0.003 ± 0.0007 | 7.2×10−6 | 3.5×10−37 |
| cg04332373 | 4 | CD38 | 0.0013 ± 0.0002 | 1.9×10−9 | 0.01 ± 0.004 | 0.004 | 0.002 ± 0.01 | 0.84 | 0.003 ± 0.0009 | 0.0009 | 3.7×10−11 |
| cg17287155 | 5 | AHRR | 0.0015 ± 0.0003 | 7.5×10−9 | 0.005 ± 0.004 | 0.22 | −0.006 ± 0.009 | 0.50 | 0.001 ± 0.0008 | 0.13 | 2.5×10−7 |
| cg26164488 | 2 | NA | 0.0012 ± 0.0002 | 4.9×10−8 | 0.005 ± 0.004 | 0.19 | −0.006 ± 0.009 | 0.55 | 0.005 ± 0.002 | 0.02 | 2.4×10−7 |
| cg07504977 | 10 | NA | 0.0011 ± 0.0002 | 5.0×10−8 | 0.02 ± 0.004 | 1.9×10−7 | 0.02 ± 0.01 | 0.04 | 0.004 ± 0.002 | 0.03 | 1.2×10−13 |
| cg14476101 | 1 | PHGDH | −0.0015 ± 0.0003 | 6.6×10−8 | −0.02 ± 0.004 | 1.7×10−8 | −0.03 ± 0.01 | 0.006 | −0.003 ± 0.001 | 0.05 | 4.7×10−15 |
| cg26680760 | 16 | NA | 0.0013 ± 0.0002 | 7.4×10−8 | 0.008 ± 0.004 | 0.06 | 0.008 ± 0.01 | 0.42 | −0.0006 ± 0.0009 | 0.51 | 1.7×10−6 |
| cg26140475 | 8 | NA | 0.001 ± 0.0002 | 1.0×10−7 | 0.003 ± 0.004 | 0.41 | −0.006 ± 0.009 | 0.51 | 0.002 ± 0.001 | 0.05 | 1.8×10−6 |
The β symbols in this table denote regression coefficients, not methylation scores
Figure 1.
Manhattan plot of epigenome-wide results of testing for association between epigenome-wide methylation and body mass index. The X-axes display the chromosome on which the CpG site is located, the Y-axes display –log10(P-value). The red horizontal line indicates the threshold for epigenome-wide statistical significance after a Bonferroni correction.
Table 3.
Top CpG methylation sites associated with waist circumference in GOLDN (n=991) and ARIC (n=2,106) at the level of genome-wide significance (P<1.1×10−7 ) in the discovery phase.
| Marker | Chr | Gene | Discovery (GOLDN) | Replication (ARIC) | Meta P | ||||
|---|---|---|---|---|---|---|---|---|---|
| P | SE | P | P | SE | P | ||||
| cg00574958 | 11 | CPTIA | −0.0003 | 0.00005 | 2.6×10−10 | −0.004 | 0.0007 | 2.1×10−8 | 2.2×10−16 |
| cg04332373 | 4 | CD38 | 0.0005 | 0.00008 | 6.2×10−10 | 0.004 | 0.0009 | 0.00003 | 6.1×10−13 |
| cg07504977 | 10 | NA | 0.0004 | 0.00007 | 7.6×10−9 | 0.005 | 0.002 | 0.003 | 5.8×10−10 |
| cg25349939 | 2 | GTDC1 | 0.0005 | 0.00008 | 1.6×10−8 | 0.0009 | 0.0008 | 0.26 | 8.4×10−8 |
| cg14476101 | 1 | PHGDH | −0.0006 | 0.0001 | 1.7×10−8 | −0.003 | 0.002 | 0.06 | 2.2×10−8 |
Figure 2.
Manhattan plot of epigenome-wide results of testing for association between epigenome-wide methylation and waist circumference. The X-axes display the chromosome on which the CpG site is located, the Y-axes display –log10(P-value). The red horizontal line indicates the threshold for epigenome-wide statistical significance after a Bonferroni correction.
Using information provided by the Illumina array manufacturer, we functionally annotated the top hits and found that four of them (cg26164488, cg07504977, cg26140475, and cg25349939) represent enhancer elements (bioinformatically determined), one (cg14476101) is located within a reprogramming-specific differentially methylated region, and one (cg26140475) is located within a general differentially methylated region. The cg26140475 hit is also located at a DNAse hypersensitive site. Finally, Illumina has bioinformatically determined that the hits in CD38, AHRR, and the intergenic region on chromosome 16 are implicated in promoter activity.
Discussion
Our study contributes to the growing body of evidence in support of epigenetic correlates of complex disease phenotypes. Specifically, we have discovered associations between DNA methylation measured in immune cells at several CpG sites and two obesity-related traits, BMI and WC, in a cohort of 991 healthy adults of European American descent. The top associations were replicated in whole blood samples taken from two independent study samples including both European Americans and African Americans, indicating the robustness of our findings.
The biological significance of the CPT1A locus is hard to overstate. Using data from GOLDN and FHS, we recently identified a CpG site in CPT1A as an important epigenetic determinant of fasting triglyceride and very low-density lipoprotein cholesterol levels (17). Interestingly, the observed associations with BMI and WC did not persist upon adjustment for triglycerides (data not shown). CPT1A encodes carnitine palmitoyltransferase 1A, the rate-limiting enzyme for mitochondrial fatty acid oxidation. Public databases show that CPT1A is expressed in CD4+ T-cells (21) and is implicated in several metabolic processes. Prior studies have linked genetic variation in CPT1A and indices of obesity. For example, a large-scale candidate gene study found that the L479 allele is associated with reduced adiposity in Yup'ik individuals (22). Another population-based study conducted in French Canadians found that CPT1A genotype modified the association between dietary fat intake and adiposity variables (BMI, weight, and WC) (23). In light of these studies and the well-established connection between sequence variation and epigenetic changes (24), we have previously conducted a genome-wide study to search for CPT1A methylation quantitative trait loci; however, no evidence of the latter was found (17).
The Infinium Human Methylation450 array measurement of CPT1A methylation in GOLDN was validated using bisulfite sequencing and found to be inversely correlated with CPT1A expression in buffy coats (17). Based on previously published reports and our findings, the following paradigm emerges: higher methylation status of CPT1A results in decreased expression of the gene, which in turn is negatively correlated with BMI and WC. Because this is a cross-sectional study, we cannot infer temporality or causality; future studies should consider repeated measurement analysis to establish temporality of the association. Additionally, as dietary factors such as intake of long-chain monounsaturated fatty acids have also been shown to regulate CPT1A expression (25) as well as DNA methylation patterns (26), future investigations would benefit from incorporating dietary information to provide further insight into these findings.
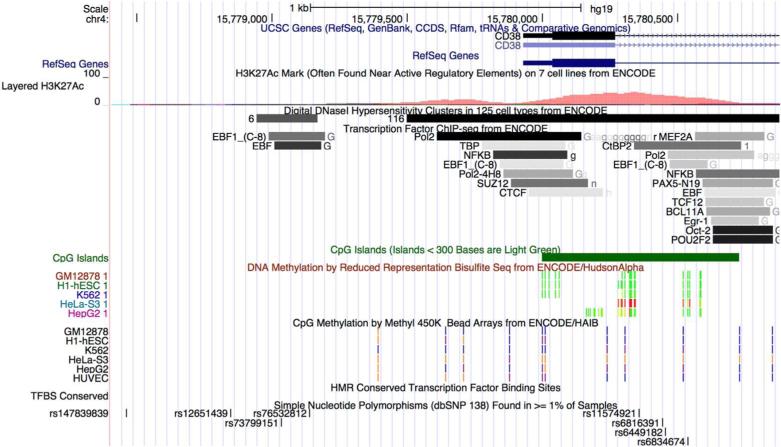
The second CpG methylation locus that achieved epigenome-wide significance and was replicated in ARIC (as well as in the case-control study from FHS) is located 289 bp upstream of the transcription start site of CD38, an immunologically relevant gene expressed in CD4+ T-cells. It falls in the upstream shore of a CpG island, has a strong polymerase II (Pol II) ChIP-seq signal and a minor peak of the histone mark H3K27Ac in immune cells, and is differentially methylated by cell type (Figure 3). These bioinformatics data likely indicate that methylation at this locus is trailing Pol II activity and may be correlated with CD38 expression in CD4+ T-cells. Several lines of evidence link the gene product of CD38 with metabolic traits. First, CD38-knockout mice are resistant to developing diet-induced obesity, liver steatosis, and glucose intolerance via a mechanism that involves an increase in intracellular NAD(+) levels and decreased protein acetylation through sirtuin activation (27, 28). In human macrophages and adipocytes, quercetin, which is a CD38 inhibitor, attenuates inflammation and insulin resistance (29). In our study, we observed a positive relationship between CD38 methylation and adiposity traits. Because we did not measure CD38 RNA directly, we cannot fully describe the mechanisms linking changes in DNA methylation and adiposity traits, especially in light of recent evidence suggesting that DNA methylation may be correlated with both positive and negative changes in gene expression (24). Because of the immunologic relevance of CD38, it is likely that the difference between the magnitude of estimates in the discovery and replication cohorts (as well as non-replication in the random sample from FHS) is due to using CD4+ T-cells vs. whole blood samples, in which differential immune cell composition could dilute the association. Our findings emphasize the relevance of CD38 to human metabolic traits and may lay the groundwork for understanding the epigenetic mechanisms of emerging therapies for obesity and inflammation.
Figure 3.
ENCODE annotation of the genomic region containing the transcription start site of CD38.
Another promising finding emerged from an intergenic region on chromosome 10. The cg07504977 locus is located on the north shelf of a CpG island and maps to the promoter of a lincRNA, LINC00263 (Figure 4). The methylation status of this locus is positively associated with BMI in both GOLDN and FHS (although not in ARIC), as well as with WC in both GOLDN and ARIC. Although the specific function of LINC00263 is unknown, evidence points to the functional importance of long non-coding RNAs in adipogenesis (30) and obesity-related syndromes (31). In particular, a recent study demonstrated that 10% of all transcriptionally active regions in mature adipocytes map to chromosome 10, with many of those regions clustering proximally to LINC00263 (32). As shown in Figure 4, that region also contains H3K27Ac marks, providing further evidence for its regulatory relevance.
Figure 4.
ENCODE annotation of the genomic region containing the long intergenic non-coding RNA on chromosome 10.
We identified robust associations between the methylation status of a CpG locus in PHGDH and BMI in samples from GOLDN and FHS, but not ARIC. PHGDH encodes the phosphoglycerate dehydrogenase enzyme, which catalyzes the first step in the phosphorylation pathway of serine biosynthesis. Previously, the methylation of the same locus has been linked to blood concentrations of 4-androsten-3beta,17beta-diol, a steroid hormone upregulated in obesity (33, 34). The metabolic implications of PHGDH are further underscored by evidence of its overexpression in approximately 70% of estrogen receptor-negative breast cancers, as it drives metabolic changes leading to rapid cellular growth (35). While the effects of phosphoglycerate dehydrogenase on oncogenesis are well-characterized, our findings warrant further examination of this enzyme in other metabolic and obesity-related phenotypes. Interestingly, the PHGDH locus and the LINC00263 locus described above replicated in FHS but not ARIC; the reasons for non-replication could include demographic variability, differences in covariate adjustment, and chance. Moreover, many environmental differences between the European Americans in FHS and GOLDN and the African Americans in ARIC accumulate across the entire life course and even across generations, starting with pregnancy and affecting both DNA methylation and fat accumulation from the early years (36).
Interestingly, we did not observe statistically significant associations between BMI and the methylation status of three sites in HIF3A, which was recently identified and validated as a novel epigenetic obesity determinant in a genome-wide analysis (4). Specifically, in the analysis of the GOLDN data the P-values for β scores at cg22891070, cg27146050, and cg16672562 were 0.13, 0.35, and 0.16 respectively. Differences could be due to cohort makeup (healthy individuals in GOLDN vs. both healthy blood donors and MI patients in the Cardiogenics Consortium or venous thrombosis patients in one of the two replication cohorts), choice of tissue (CD4+ T-cells vs. whole blood samples), or chance. Although findings in our study as well as in Cardiogenics were independently replicated, these discrepancies highlight the need for future functional studies as follow-up to large-scale methylation assays.
Our study documents novel associations between CD4+ T-cell DNA methylation in several biologically plausible genomic regions and human obesity traits, i.e. BMI and WC. The associations with the CPT1A locus were robust in all replication analyses despite the differences in sample collection types (isolated CD4+ T-cells in GOLDN vs. whole blood in FHS and ARIC) and racial composition. The sample size, high resolution of epigenetic phenotyping, and successful replication represent distinguishing strengths of our study. However, the results presented here need to be interpreted in light of several potential limitations. First, because DNA methylation can both precede phenotypic changes as well as result from environmental influences such as obesity (37), follow-up analyses of repeated measurements are required to further causal understanding of the observed associations. Second, the effect of our findings on gene expression has been previously established for CPT1A but not for other genes that contain the top differentially methylated CpG sites in this study. Third, because both DNA methylation and adiposity traits are associated with a range of environmental inputs, residual confounding remains a possible explanation for our findings despite covariate adjustment. For example, the third top hit in our BMI analysis is a CpG site in AHRR, the methylation status of which was compellingly linked to smoking (38, 39). When we restricted the GOLDN analyses to never smokers (n=703, data not shown), the P-values for AHRR increased from 7.5×10−9 to 2.2×10−7, beyond the threshold for epigenome-wide significance. The same attenuation did not take place for other top hits, namely in CPT1A and CD38, suggesting that those associations are less likely to be a mere artifact of smoking. Fourth, the CpG sites in CPT1A, CD38, and AHRR contain known sequence variants (rs78442314, rs77882313, and rs6899226 respectively), which could influence the binding potential of the probe and affect methylation measurements. However, prior genome-wide analyses did not yield evidence of association between any sequence polymorphism and the methylation score at the top CPT1A locus (17). Fifth, the between-cohort differences in statistical methods, especially in normalization approaches, may contribute to non-replication; while that is unlikely to impact our top findings, it may present problems for loci with more modest signals (40). Finally, the methylation patterns in CD4+ T-cells used in the study may have been altered during the freezing and thawing process and may not generalize to other cell types. The differences between CD4+ T-cells and other cell types present in whole blood samples are particularly relevant as such heterogeneity can confound epigenomic association results, mirroring statistical issues created by population stratification. Despite that concern, we are reassured by successful replication of our top findings in whole blood samples from ARIC and FHS.
On balance, our study highlights the importance of DNA methylation in complex human traits and identifies several candidate regions as promising epigenetic correlates of obesity. Further investigations of these regions will further characterize the heritable components of BMI and WC beyond sequence variants, potentially leading to novel therapeutic and preventative approaches to the challenge posed by the global obesity epidemic.
Supplementary Material
What is already known about this subject
Body mass index (BMI) and waist circumference (WC) are quantitative traits characterized by multifactorial etiology, relevance to disease risk, and high heritability.
Known genetic polymorphisms explain only a modest fraction of inherited variation in obesity-related traits.
Emerging evidence suggests that epigenetic changes such as DNA methylation are a key influence on obesity.
What this study adds
We found epigenome-wide significant associations between the methylation status of eight genomic sites and BMI, as well as that of five sites and WC, successfully replicating our top findings in ethnically diverse populations.
Top findings were in the genes encoding biologically relevant proteins such as carnitine palmitoyltransferase 1A (CPT1A), phosphoglycerate dehydrogenase, and CD38 molecule.
Acknowledgments
The authors thank the staff and participants of ARIC, FHS, and GOLDN for their important contributions.
Footnotes
Author contributions: Substantial contributions to conception and design: DMA, DKA, SA, EB, JB, EWD, MRI, DL, JMO, HKT; Acquisition of data: DMA, DKA, EB, IBB, JB, EWD, MF, MLG, PNH, DL, JMO, JSP, KST, HKT, MYT; Analysis: DMA, DKA, SA, EWD, WG, LL, LL, CL, MM, JS, KST, DZ; Interpretation of data: DMA, DKA, SA, EWD, WG, BH, LL, LL, CL, MM, JSP, JS, KST, DZ; Drafting of article: SA. All authors were involved in revising the manuscript critically for important intellectual content, and all granted final approval of the submitted and published versions.
Conflicts of interest
The authors have no conflicts of interest to declare.
Contributor Information
Stella Aslibekyan, Department of Epidemiology, School of Public Health, University of Alabama at Birmingham, Birmingham, AL, 35294, USA.
Ellen W. Demerath, Division of Epidemiology and Community Health, School of Public Health, University of Minnesota, Minneapolis, MN, 55455, USA
Michael Mendelson, Population Sciences Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, 20824, USA; and Framingham Heart Study, Framingham, MA, 01702, USA; and Department of Cardiology, Boston Children's Hospital, Boston, MA, 02215, USA.
Degui Zhi, Department of Biostatistics, School of Public Health, University of Alabama at Birmingham, Birmingham, AL, 35294, USA.
Weihua Guan, Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, MN, 55455, USA.
Liming Liang, Population Sciences Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, 20824, USA; and Framingham Heart Study, Framingham, MA, 01702, USA; and Departments of Epidemiology and Biostatistics, School of Public Health, Harvard University, Boston, MA, 02115, USA.
Jin Sha, Department of Epidemiology, School of Public Health, University of Alabama at Birmingham, Birmingham, AL, 35294, USA.
James S. Pankow, Division of Epidemiology and Community Health, School of Public Health, University of Minnesota, Minneapolis, MN, 55455, USA
Chunyu Liu, Framingham Heart Study, Framingham, MA, 01702, USA; and Department of Biostatistics, Boston University, Boston, MA, 02118, USA.
Marguerite R. Irvin, Department of Epidemiology, School of Public Health, University of Alabama at Birmingham, Birmingham, AL, 35294, USA
Myriam Fornage, Brown Foundation Institute of Molecular Medicine, The University of Texas Health Science Center, Houston, TX 77030, USA; and Human Genetics Center, School of Public Health, The University of Texas Health Science Center at Houston, TX 77030, USA.
Bertha Hidalgo, Department of Epidemiology, School of Public Health, University of Alabama at Birmingham, Birmingham, AL, 35294, USA.
Li-An Lin, Human Genetics Center, School of Public Health, The University of Texas Health Science Center at Houston, TX 77030, USA.
Krista Stanton Thibeault, HudsonAlpha Institute for Biotechnology, Huntsville, AL, 34806, USA.
Jan Bressler, Human Genetics Center, School of Public Health, The University of Texas Health Science Center at Houston, TX 77030, USA.
Michael Y. Tsai, Division of Laboratory Medicine and Pathology, University of Minnesota, Minneapolis, MN, 55455, USA
Megan L. Grove, Human Genetics Center, School of Public Health, The University of Texas Health Science Center at Houston, TX 77030, USA
Paul N. Hopkins, School of Medicine, University of Utah, Salt Lake City, UT, 84108, USA
Eric Boerwinkle, Brown Foundation Institute of Molecular Medicine, The University of Texas Health Science Center, Houston, TX 77030, USA; and Human Genetics Center, School of Public Health, The University of Texas Health Science Center at Houston, TX 77030, USA.
Ingrid B. Borecki, Division of Statistical Genomics, Department of Genetics, Washington University in St Louis, St Louis, MO, 63108, USA
Jose M. Ordovas, Department of Epidemiology, Atherothrombosis and Imaging, Centro Nacional de Investigaciones Cardiovasculares, Madrid, Spain and Instituto Madrileño de Estudios Avanzados Alimentacion, Madrid, Spain; and Jean Mayer USDA Human Nutrition Research Center on Aging, Tufts University, Boston, MA, 02115, USA
Daniel Levy, Population Sciences Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, 20824, USA; and Framingham Heart Study, Framingham, MA, 01702, USA.
Hemant K. Tiwari, Department of Biostatistics, School of Public Health, University of Alabama at Birmingham, Birmingham, AL, 35294, USA
Devin M. Absher, HudsonAlpha Institute for Biotechnology, Huntsville, AL, 34806, USA.
Donna K. Arnett, Department of Epidemiology, School of Public Health, University of Alabama at Birmingham, Birmingham, AL, 35294, USA.
References
- 1.Hemani G, Yang J, Vinkhuyzen A, Powell JE, Willemsen G, Hottenga JJ, et al. Inference of the genetic architecture underlying BMI and height with the use of 20,240 sibling pairs. Am J Hum Genet. 2013;93:865–875. doi: 10.1016/j.ajhg.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vattikuti S, Guo J, Chow CC. Heritability and genetic correlations explained by common SNPs for metabolic syndrome traits. PLoS Genet. 2012;8:e1002637. doi: 10.1371/journal.pgen.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwenk RW, Vogel H, Schurmann A. Genetic and epigenetic control of metabolic health. Molecular metabolism. 2013;2:337–347. doi: 10.1016/j.molmet.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Su S, Barnes VA, De Miguel C, Pollock J, Ownby D, et al. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics. 2013;8:522–533. doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Zhu H, Snieder H, Su S, Munn D, Harshfield G, et al. Obesity related methylation changes in DNA of peripheral blood leukocytes. BMC Med. 2010;8:87. doi: 10.1186/1741-7015-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franks PW, Ling C. Epigenetics and obesity: the devil is in the details. BMC Med. 2010;8:88. doi: 10.1186/1741-7015-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milagro FI, Campion J, Cordero P, Goyenechea E, Gomez-Uriz AM, Abete I, et al. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. Faseb j. 2011;25:1378–1389. doi: 10.1096/fj.10-170365. [DOI] [PubMed] [Google Scholar]
- 9.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annual review of immunology. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 10.Corella D, Arnett DK, Tsai MY, Kabagambe EK, Peacock JM, Hixson JE, et al. The - 256T>C polymorphism in the apolipoprotein A-II gene promoter is associated with body mass index and food intake in the genetics of lipid lowering drugs and diet network study. Clin Chem. 2007;53:1144–1152. doi: 10.1373/clinchem.2006.084863. [DOI] [PubMed] [Google Scholar]
- 11.Warodomwichit D, Shen J, Arnett DK, Tsai MY, Kabagambe EK, Peacock JM, et al. ADIPOQ polymorphisms, monounsaturated fatty acids, and obesity risk: the GOLDN study. Obesity (Silver Spring) 2009;17:510–517. doi: 10.1038/oby.2008.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouwens M, Afman LA, Muller M. Activation of peroxisome proliferator-activated receptor alpha in human peripheral blood mononuclear cells reveals an individual gene expression profile response. BMC Genomics. 2008;9:262. doi: 10.1186/1471-2164-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, et al. genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 2013;9:e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 15.Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One. 2009;4:e6098. doi: 10.1371/journal.pone.0006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irvin MR, Zhi D, Joehanes R, Mendelson M, Aslibekyan S, Claas SA, et al. Epigenome-Wide Association Study of Fasting Blood Lipids in the Genetics of Lipid Lowering Drugs and Diet Network Study. Circulation. 2014;130:565–572. doi: 10.1161/CIRCULATIONAHA.114.009158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atkinson B, Therneau T. kinship: mixed-effects Cox models, sparse matrices, and modeling data from large pedigrees. 2007.
- 19.Clinical Guidelines on the Identification, Evaluation, and Treatment of Overweight and Obesity in Adults--The Evidence Report. National Institutes of Health. Obes Res. 1998;6(Suppl 2):51s–209s. [PubMed] [Google Scholar]
- 20.Lean ME, Han TS, Morrison CE. Waist circumference as a measure for indicating need for weight management. Bmj. 1995;311:158–161. doi: 10.1136/bmj.311.6998.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemas DJ, Wiener HW, O'Brien DM, Hopkins S, Stanhope KL, Havel PJ, et al. Genetic polymorphisms in carnitine palmitoyltransferase 1A gene are associated with variation in body composition and fasting lipid traits in Yup'ik Eskimos. J Lipid Res. 2012;53:175–184. doi: 10.1194/jlr.P018952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robitaille J, Houde A, Lemieux S, Perusse L, Gaudet D, Vohl MC. Variants within the muscle and liver isoforms of the carnitine palmitoyltransferase I (CPT1) gene interact with fat intake to modulate indices of obesity in French-Canadians. Journal of molecular medicine (Berlin, Germany) 2007;85:129–137. doi: 10.1007/s00109-006-0116-7. [DOI] [PubMed] [Google Scholar]
- 24.Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang ZH, Miyahara H, Iwasaki Y, Takeo J, Katayama M. Dietary supplementation with long-chain monounsaturated fatty acids attenuates obesity-related metabolic dysfunction and increases expression of PPAR gamma in adipose tissue in type 2 diabetic KK-Ay mice. Nutr Metab (Lond) 2013;10:16. doi: 10.1186/1743-7075-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang FF, Morabia A, Carroll J, Gonzalez K, Fulda K, Kaur M, et al. Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population. J Nutr. 2011;141:1165–1171. doi: 10.3945/jn.110.134536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbosa MT, Soares SM, Novak CM, Sinclair D, Levine JA, Aksoy P, et al. The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. Faseb j. 2007;21:3629–3639. doi: 10.1096/fj.07-8290com. [DOI] [PubMed] [Google Scholar]
- 28.Escande C, Nin V, Price NL, Capellini V, Gomes AP, Barbosa MT, et al. Flavonoid apigenin is an inhibitor of the NAD+ ase CD38: implications for cellular NAD+ metabolism, protein acetylation, and treatment of metabolic syndrome. Diabetes. 2013;62:1084–1093. doi: 10.2337/db12-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overman A, Chuang CC, McIntosh M. Quercetin attenuates inflammation in human macrophages and adipocytes exposed to macrophage-conditioned media. Int J Obes (Lond) 2011;35:1165–1172. doi: 10.1038/ijo.2010.272. [DOI] [PubMed] [Google Scholar]
- 30.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powell WT, Coulson RL, Crary FK, Wong SS, Ach RA, Tsang P, et al. A Prader-Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum Mol Genet. 2013;22:4318–4328. doi: 10.1093/hmg/ddt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi F, Yang F, Liu X, Chen H, Ji T, Jiang L, et al. RNA-seq identified a superAlong intergenic transcript functioning in adipogenesis. RNA biology. 2013;10:991–1001. doi: 10.4161/rna.24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen AK, Zeilinger S, Kastenmuller G, Romisch-Margl W, Brugger M, Peters A, et al. Epigenetics meets metabolomics: an epigenome-wide association study with blood serum metabolic traits. Hum Mol Genet. 2014;23:534–545. doi: 10.1093/hmg/ddt430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azziz R, Potter HD, Bradley EL, Jr., Boots LR. delta 5-Androstene-3 beta,17 betaAdiol in healthy eumenorrheic women: relationship to body mass and hormonal profile. Fertil Steril. 1994;62:321–326. doi: 10.1016/s0015-0282(16)56885-x. [DOI] [PubMed] [Google Scholar]
- 35.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuzawa CW, Sweet E. Epigenetics and the embodiment of race: developmental origins of US racial disparities in cardiovascular health. Am J Hum Biol. 2009;21:2–15. doi: 10.1002/ajhb.20822. [DOI] [PubMed] [Google Scholar]
- 37.Ge ZJ, Luo SM, Lin F, Liang QX, Huang L, Wei YC, et al. DNA methylation in oocytes and liver of female mice and their offspring: effects of highAfatAdietAinduced obesity. Environ Health Perspect. 2014;122:159–164. doi: 10.1289/ehp.1307047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, et al. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPICATurin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22:843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 39.Zeilinger S, Kuhnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu MC, Joubert BR, Kuan PF, Haberg SE, Nystad W, Peddada SD, et al. A systematic assessment of normalization approaches for the Infinium 450K methylation platform. Epigenetics. 2014;9:318–329. doi: 10.4161/epi.27119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.