Abstract
There are two basic mechanisms that are associated with the maintenance of the telomere length, which endows cancer cells with unlimited proliferative potential. One mechanism, referred to as alternative lengthening of telomeres (ALT), accounts for approximately 10–15% of all human cancers. Tumours engaged in the ALT pathway are characterised by the presence of the single stranded 5′-C-rich telomeric overhang (C-overhang). This recently identified hallmark of ALT cancers distinguishes them from healthy tissues and renders the C-overhang as a clear target for anticancer therapy. We analysed structures of the 5′-C-rich and 3′-G-rich telomeric overhangs from human and Caenorhabditis elegans, the recently established multicellular in vivo model of ALT tumours. We show that the telomeric DNA from C. elegans and humans forms fundamentally different secondary structures. The unique structural characteristics of C. elegans telomeric DNA that are distinct not only from those of humans but also from those of other multicellular eukaryotes allowed us to identify evolutionarily conserved properties of telomeric DNA. Differences in structural organisation of the telomeric DNA between the C. elegans and human impose limitations on the use of the C. elegans as an ALT tumour model.
INTRODUCTION
Telomeres are specialised, functional DNA-protein structures found at the ends of all eukaryotic linear chromosomes. Telomeres help to protect the ends of chromosomes from being treated as damaged DNA in need of repair and also provide a means of complete replication of the chromosome. In most eukaryotes, telomeres comprise double-stranded DNA consisting of simple tandem repeat sequences (e.g. d(TTAGGG).(CCCTAA) in vertebrates) terminating in a single-stranded, 150- to 250-nt long G-rich 3′-overhang, herein referred to as the G-overhang (1,2). The G-overhang is essential for the formation of a protective higher-order structure referred to as the T-loop (3). Telomeric DNA in cultured cells has been shown to undergo progressive loss of 50−200 bases following each cell division due to incomplete replication of the lagging strand (4,5). This process ultimately leads to critically short telomeres, which triggers cell cycle arrest (senescence) or cell death (reviewed in 6,7). In most eukaryotic organisms, telomere maintenance relies on the activation of telomerase (8). Appropriate telomerase function in stem and germ cell populations counteracts replication-dependent telomere shortening (reviewed in 9). In contrast, repression of telomerase expression in most somatic tissues limits the proliferative potential of these cells (reviewed in 6,7). Thus, senescence, which is induced by telomere shortening and subsequent DNA damage signalling, is an essential tumour suppression mechanism, emphasised by the finding that repression of telomerase is lost from approximately 85–90% of cancer cells and primary tumours, endowing them with unlimited proliferative potential (10,11).
The inherent characteristic of the mammalian G-overhang is its ability to adopt a non-canonical tetraplex DNA structure, defined as the G-quadruplex, both in vitro (reviewed, e.g. in 12) and in vivo (13,14). The G-quadruplex structure relies on the formation of planar guanine tetrads marked by a Hoogsteen-type guanine-guanine base-pairing pattern. Folding of single-stranded, G-rich telomeric DNA, which serves as a substrate for telomerase, into a four-stranded G-quadruplex has been demonstrated to inhibit telomerase activity (15), as G-quadruplex formation ensures that the 3′-end is inaccessible to hybridisation with the telomerase RNA template, the first essential step in the telomerase catalytic cycle. Several small molecular weight ligands that stabilise telomeric G-quadruplex structures have displayed promising anticancer activity in tumour xenograft models, indicating that the stabilisation of telomeric G-quadruplexes might be applicable to the treatment of a wide range of human cancers (reviewed in, e.g. (16,12).
Although most human cancers rely on the activation of telomerase (8) to maintain the telomere length, a distinct telomerase-independent mechanism is active in a subset (∼10%) of tumours (17). In these tumours, telomeres are maintained using the homologous recombination (HR)-based alternative lengthening of telomeres (ALT) mechanism (17). Tumour cells engaged in this pathway display several distinctive characteristics (18,19) particularly the single–stranded, C-rich telomeric 5′-overhang, herein referred to as a C-overhang (20).
Analogous to mammalian G-rich telomeric DNA, the inherent property of the mammalian complementary C-rich telomeric DNA strand is its ability to form a non-canonical DNA tetraplex structure, defined as the i-motif (21,22). In contrast to the G-quadruplex, which requires the formation of Hoogsteen-type guanine-guanine base pairs, the i-motif structure is based on the formation of C+.C non-canonical base pairs, presuming the protonation of cytosine bases (23,24). Whereas G-quadruplex formation was demonstrated both in vitro and in vivo, i-motif formation from telomeric DNA has, thus far, only been observed in vitro either under acidic (21,22) or neutral pH conditions in the presence of molecular crowding mimics (25). Whether i-motif formation in mammalian telomeric C-rich DNA is only a peculiarity of its conformational space under specific environmental conditions or is associated with telomere function remains to be elucidated.
In this study, we present the results of comparative structural analyses of DNA constructs emulating both 3′-G-rich and 5′-C-rich single-stranded telomeric DNA corresponding to the G- and C-overhangs, respectively. Particularly, we focus on comparison of structural properties of telomeric DNAs from model nematode Caenorhabditis elegans and that of mammals. Thus far, C. elegans and mammals (humans and mice) provide the only reported cases of multicellular eukaryotes displaying linear chromosomes containing, next to its 3′-G-rich overhang, a 5′-C-rich single-stranded telomeric overhang (20,26). Importantly, while humans, similar to the majority of eukaryotic organisms, display a slightly alkaline intracellular pH (pHi) shift in the course of cell cycle progression (reviewed in 27), C. elegans is a rare example of a eukaryote that exhibits notable acidification (pHi ∼ 6.3) of the intracellular space during its development (28). Comparison of the unique structural characteristics of G- and C-rich telomeric DNA from the nematode C. elegans and those from other species allowed us identify the properties of telomeric DNA that are conserved during evolution. Our data suggest that the detected variability in the primary sequence of telomeric DNA repeats among eukaryotic species is not a consequence of a positive selection force during evolution but rather a manifestation of the conservation of telomeric DNA structural characteristics, which allow a limited set of permutations in the telomeric DNA primary sequence.
The implications of the C-overhang structure in the therapeutic intervention of tumours engaged in the ALT pathway and in the recently proposed use of C. elegans as a model for ALT (29,30) are discussed.
MATERIALS AND METHODS
DNA constructs
The unlabelled DNA oligonucleotides listed in Table 1 were purchased from IDT (USA). Sequences for the S. cerevisiae constructs (Table 1) were derived from study by Shampey et al. (31). Desalted and lyophilised oligonucleotides were dissolved in deionised water, and their concentrations were determined using an UNICAM 5625 UV/Vis spectrometer on the basis of UV absorption at 260nm measured at 90°C and molar absorption coefficients calculated according to Gray et al. (32). The 13C/15N fully isotopically labelled analogue of the CE20 sequence (see Table 1), which was extended at its 5′-end with a guanosine residue to restrict the terminal fraying effect at the bottom of the hairpin stem (oligonucleotide CE21 in Table 1), was prepared enzymatically (33) and was purified via ion exchange chromatography (Silantes, Germany). To avoid structural artefacts caused by the ion composition of the buffer (34), the oligonucleotide experiments were performed in buffer mimicking the ion composition of the intracellular environment (25 mM sodium phosphate, 110 mM KCl, 10 mM NaCl, 1 mM MgCl2, 130 nM CaCl2, 10% D2O (pH 7.5)), herein referred to as intracellular buffer, at a DNA strand concentration of approximately 0.3 mM (unlabelled oligonucleotides) or 0.5 mM (isotopically labelled oligonucleotide, CE21 - see Table 1). Notably, identical samples were employed for the acquisition of both the CD and NMR spectra as well as for native PAGE.
Table 1. List of telomeric DNA constructs employed.
| Organism | Telomeric repeat (5′ → 3′) | Construct | Name |
|---|---|---|---|
| H. sapiens | TAACCC | CCC(TAACCC)3 | HS21 |
| CCC(TAACCC)7 | HS45 | ||
| GGGTTA | (GGGTTA)3GGG | gHS21 | |
| (GGGTTA)7GGG | gHS45 | ||
| C. elegans | TAAGCC | CC(TAAGCC)3 | CE20 |
| GCC(TAAGCC)3 | CE21 | ||
| CC(TAAGCC)7 | CE44 | ||
| GGCTTA | (GGCTTA)3GG | gCE20 | |
| (GGCTTA)3GGC | gCE21 | ||
| (GGCTTA)7GG | gCE44 | ||
| (GGCTTA)7GGC | gCE45 | ||
| S. cerevisiae | C1–3A | CCACACCCACACCACACCC | SA-a19 |
| CCCAC(ACACCC)3 | SA-b23 | ||
| TG1—3 | GGGTGTGGTGTGGGTGTG | gSA-a19 | |
| (GGGTGT)3GTGGG | gSA-b23 | ||
| T. thermophyla | CCCCAA | CCCC(AACCCC)3 | TT22 |
CD spectroscopy
CD spectra were measured using the Jasco J815 spectrometer in 0.1- and 0.2-mm path length Hellma cells placed in a Peltier holder. CD signals were expressed as the difference in the molar absorption Δϵ of the left- and right-handed circularly polarised light. The molarity was related to nucleosides. Spectra were acquired at a rate of 100 nm/min and averaged from four measurements. The pH values of the samples were adjusted via the addition of HCl and measured using a Sentron Titan pH meter.
Native PAGE
Non-denaturing PAGE was performed in a temperature-controlled submersible apparatus (SE600; Hoefer Scientific) containing circulating buffer. The gels (16%, 29:1 acrylamide:bisacrylamide) were electrophoresed for 19 hours at 25 V and 20°C in the intracellular buffer. The gels were stained with Stains-All (Sigma). Densitometry was performed using a Personal Densitometer (SI, Model 375-A).
UV absorption melting and thermal difference spectra
Melting curves (5–90°C, 90–5°C and again 5–90°C) were measured using a Varian Cary 4000 spectrophotometer in 1-mm cells at a strand concentration of approximately 0.02 mM. The absorbance was recorded at 260 nm (foldback and i-motif) and at 297 nm (G-quadruplex), and it was considered as the absorbance of the ordered structure.
NMR spectroscopy
The 1D proton spectra of the unlabelled samples were measured at 700 MHz using a Bruker Avance III NMR spectrometer equipped with a triple resonance room temperature probe using the WATERGATE pulse sequence (35), including 1-ms rectangular selective pulses to suppress the water signal. All of the spectra were measured in water solution (90% H2O/10% D2O) at 20°C or 1°C unless stated otherwise. All of the NMR experiments of the 13C/15N-labelled samples were measured at 850 MHz using a Bruker Avance III spectrometer equipped with a triple-resonance cryogenic probe. All spectra of the labelled samples were measured in water solution (90% H2O/10% D2O) at 10°C or 1°C. The1H-13C 2D correlation spectrum was measured using base-optimised constant-time HSQC (36,37) with gradient selection. The constant time was set to 15 ms, and the proton-carbon evolution time 1/(2JHC) was set to 2.5 ms. The spectral widths were 9 ppm and 80 ppm in the 1H and 13C dimensions, respectively; the proton, carbon and nitrogen irradiation frequencies were set at 4.7 ppm, 120 ppm and 157.5 ppm, respectively. The 15N magnetisation was refocused during t1, and GARP decoupling was applied during acquisition using irradiation fields of 5.56 kHz and 1.47 kHz for 13C and 15N, respectively. All 180º carbon pulses were applied as 20% smoothed Chirp pulses (38) swept over 60 kHz. The NOESY spectrum, utilising a 250-ms mixing time and spectral widths of 21 ppm in both dimensions, were recorded using a 3–9–19 water suppression scheme and a 1-ms rectangular flip-back pulse (39,40) on each fully 13C/15N-labelled sample. In the directly and indirectly detected dimensions, 2048 and 1200 real points were recorded, respectively. The effect of heteronuclear coupling was removed using refocusing pulses in t1 evolution and decoupling during the detection period in the same manner as described for the HSQC experiment. The 2D 1H-1H version of the HCCH-TOCSY experiment to correlate the adenine H2 and H8 resonances (41,42) was measured using spectral widths of 10 ppm in both dimensions and 60 ms DIPSI-3 mixing using a 5.2-kHz irradiation field set at 150 ppm. GARP decoupling using a 5.56-kHz irradiation field was applied to carbon during acquisition. The HNN-COSY spectra (43,44) were measured with 21-ppm and 116-ppm spectral widths for 1H and 15N, respectively. The evolution times for the one-bond proton-nitrogen coupling 1/(2JHN) and the nitrogen-nitrogen coupling through hydrogen bond 1/(2JNN) were set to 5 ms and 30 ms, respectively. The WATERGATE scheme (35) consisting of 1-ms rectangular pulses was used to suppress the water signal, and GARP4 decoupling using a 1.47-kHz field was applied to nitrogen during acquisition.
Pull-down assay
Streptavidin magnetic beads (Promega; Streptavidine Magne-Sphere Paramagnetic Particles; 50 μg/rci) were washed twice with 0.5× SSC and once with 1× binding buffer (10 mM Tris, pH 6.7; 5 mM KCl; 10% glycerol; 1 mM DTT; 0.5 mM MgCl2; 1 mM EDTA; and 1 pill of protease inhibitor cocktail: complete, EDTA free, Roche). Biotinylated HS21 oligo (2.6 μM/rci) was bound to the beads in 1× binding buffer and placed in a roller at room temperature for 30 min. The non-bound DNA was removed by washing (3 times with 1× binding buffer at room temperature for 5 min). The magnetic beads coupled with biotinylated DNA oligos were separated into individual reactions, mixed with an increasing amount of TMPyP4 (Sigma Aldrich) (at a molar ratio of ∼ 0/1; 0.05/1; 1/1 or 20/1 TMPyP4/DNA) and incubated in a roller at 4°C for 1 hour. The oligonucleotide pull-down reactions were performed using recombinant human PCBP2 protein (OriGene Technologies). The mixtures of magnetic beads coupled with biotinylated DNA oligos in the presence of increasing concentrations of TMPyP4 and hPCBP2 (0.3 μg/rci) were incubated in the roller at 4°C for 30 minutes, followed by three washes with 1× binding buffer. The amount of bound protein was analysed via Western blot. The protein samples and molecular weight standards (Precision Plus Protein Standard, BioRad) were separated via SDS-PAGE in 12% resolving gels and were electro-transferred to PVDF membrane (Trans Blot Turbo mini size PVDF membrane, BioRad). The membrane was subjected to Western blot analysis using polyclonal rabbit antisera against human PCBP2 (OriGene Technologies) and goat anti-rabbit peroxidase-conjugated IgG (Jackson ImmunoResearch) as a secondary antibody. The immuno-reactive bands were detected using ECL detection reagents (Clarify Western ECL substrate, BioRad).
RESULTS
Vertebrate C-rich telomeric DNA folds into an i-motif structure in acidic pH
The structural properties of two DNA fragments based on four (HS21) and eight (HS45) repeats of human (vertebrate) telomeric C-rich DNA (see Table 1) were systematically investigated as a function of decreasing pH, which is used herein as a chemical perturbation that facilitates sampling of the DNA conformational space, via NMR, CD and absorption spectroscopy and native polyacrylamide gel electrophoresis (PAGE). As flanking nucleotides (generated during construct design) might influence the structural properties of four-repeat-based telomeric DNA constructs (45,46), the construct based on the eight-repeat sequence was used as an important control. Under slightly alkaline pH (pH ∼ 7.3), which corresponds to the physiological pH of metazoan cells, the CD spectra for HS21 and HS45 were virtually identical, and both sequences displayed spectral shape that is typical of unstructured C-rich DNA (Figure 1A). In agreement with the CD spectra, no imino signals were detected in the 1D 1H NMR spectra of HS21 or HS45, indicating the absence of hydrogen bonding-stabilised secondary structure in the investigated constructs (Figure 1B). Altogether, the NMR and CD spectra, along with the lack of cooperative transition upon regulated gradual thermal denaturation of both constructs (data not shown), clearly indicate that, under slightly alkaline conditions, both of the investigated constructs were predominantly unstructured. Upon slight acidification (pH ∼ 6.7) - i.e. to levels that mimic the response of the pHi of mammalian cells to mild hyperthermia - the 1D 1H NMR spectra of both constructs displayed a weak signal at approximately 15.3 ppm (data not shown) originating from the proton attached to nitrogen (N3) of cytosine, a hallmark of i-motif formation (47), while the CD spectra were virtually unchanged. Upon lowering pH to 5.5, the CD spectra of both constructs displayed positive bands at 285 nm and 220 nm and a negative peak at 250 nm. In agreement with the NMR data, this pattern was indicative of i-motif formation (48). Formation of the i-motif structure was also detected based on the UV absorption spectra as the pH-dependent absorbance decrease at 260 nm and absorbance increase at 290 nm (Figure 1A, inset). Whereas the decrease in the absorbance at 260 nm reflects formation of structured DNA, the increase in the absorbance at 290 nm is due to protonation of cytosine bases. The migration patterns of HS21 and HS45 via native PAGE at pH ∼ 5.5 demonstrated that the respective i-motifs are monomeric (Figure 1C, Supplementary Information - Figure S1). Our observation of intramolecular i-motif formation in the HS21 construct in response to acidic pH is in agreement with previous observation of intramolecular i-motif formation from four-repeat-based constructs of human C-rich telomeric DNA (21,47–49).
Figure 1.
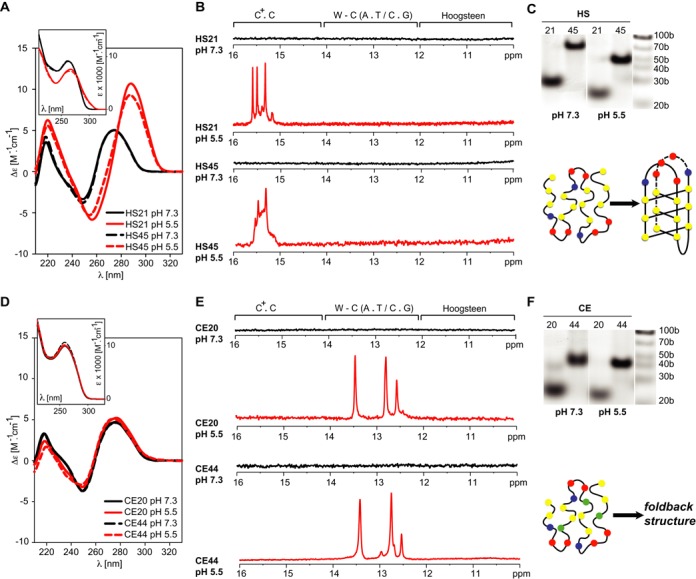
C-rich telomeric DNA from C. elegans and humans forms fundamentally different structures. LEFT: CD spectra of the DNA constructs based on human (A) and C. elegans (D) C-rich telomeric DNA repeats as a function of pH and the number of telomeric repeats. The spectra were acquired at 20°C. The insets show the pH-induced changes in the UV absorption spectra of the constructs. CENTRE: Imino regions of the 1D 1H NMR spectra of the DNA constructs based on human (B) and C. elegans (E) C-rich telomeric DNA repeats as a function of pH and the number of telomeric repeats. The spectra were acquired at 20°C. Regions of the spectra typical of imino protons involved in C.C+, Watson-Crick (W-C) and Hoogsteen base-pairing are indicated. RIGHT: Non-denaturing PAGE of the DNA constructs based on human (C) and C. elegans (F) C-rich telomeric DNA repeats as a function of pH and the number of telomeric repeats.
C. elegans C-rich telomeric DNA forms a foldback structure in acidic pH
Two DNA fragments based on four (CE20) and eight (CE44) C. elegans telomeric repeats (see Table 1) were used for structural investigations of C. elegans (nematode) telomeric C-rich DNA. Similar to the results of the human constructs at mildly alkaline pH (∼ 7.3), the CD spectra of the CE20 and CE44 fragments were virtually identical and their shape and non-cooperative melting corresponded to unstructured DNA (Figure 1D). In contrast to the human telomeric DNA, the CD and UV absorption spectra of both constructs did nearly not change with the gradual decrease in pH from 7.3 to 5.5 (Figure 1D). However, the NMR spectra displayed profound pH-dependent changes. In line with the CD results, the 1D 1H NMR spectra of both constructs were devoid of imino signals under mildly alkaline conditions (Figure 1E). However, weak signals in the imino region of the 1D 1H NMR spectrum between 12.5 and 13.5 ppm, clearly absent at pH ∼ 7.3, were detected upon mild acidification (pH ∼ 6.7) of the CE44 construct (data not shown). These signals became notably pronounced upon further acidification to pH ∼ 6.3 or pH ∼ 5.5. At these pH values, a similar pattern of imino signals was detected in the 1D 1H NMR spectra of both the CE20 and CE44 constructs (Figure 1E). The dispersions in the chemical shifts of the imino-protons in the 1D 1H NMR spectra of the CE20 and CE44 constructs (Figure 1E) suggested that both constructs adopted a similar secondary structure that was stabilised by canonical Watson-Crick (W-C) base pairs. The electrophoretic migration of both CE20 and CE44 indicated that the formed structures were monomeric (Figure 1F, Supplementary Information - Figure S1). Taken together, these data indicated that acidic pH promoted the formation of a foldback structure that is stabilised by W-C base pairs. Formation of the foldback structure provides an explanation for the virtual absence of changes in CD spectra of the CE20 and CE44 upon sample acidification (Figure 1D), since the B-type DNA structures generally displays only very small CD changes (50).
However, the data did not provide an explanation for the observed pH-dependency of the C. elegans C-rich telomeric DNA folding: no sign of cytosine(s) protonation, which is responsible for the pH dependence of i-motif formation in human C-rich telomeric DNA, was detected in the NMR, CD or absorption spectra. To identify the origin of the pH-dependent formation of the secondary structure of C. elegans C-rich telomeric DNA, we analysed the structural properties of a 13C/15N isotopically labelled construct (CE21) (Table 1) in detail using high-resolution NMR spectroscopy methods. (Note: The CE21 construct is an analogue of the CE20 sequence, which was extended at its 5′-end with a guanosine residue to restrict the terminal frying effect at the bottom of the hairpin stem. The response of the CE21 and CE20 to pH is identical (Supplementary Information - Figure S2)). In agreement with the properties of the imino region of the 1D 1H NMR spectrum, six pairs of cross peaks, two and four pairs at characteristic nitrogen frequencies for W-C A.T and W-C G.C base pairs, respectively, were detected in the 2D HNN-COSY spectra at pH 5.5 (Supplementary Information - Figure S3). The broad peak at ∼ 10.5 ppm, apparent in the 1D 1H NMR spectrum acquired at low temperature, did not display any correlation in the HNN-COSY spectrum (Supplementary Information - Figure S3). These findings indicated that the CE21 secondary structure was stabilised by two A.T and four G/C W-C base pairs and contained one unpaired, imino-proton bearing residue. This unpaired residue was designated as thymine based on the cross peak in the 2D 1H-1H NOESY spectrum between the imino proton resonating at ∼ 10.5 ppm and a signal from the proton resonating at ∼1.2 ppm, a frequency that is typical of a thymine methyl group (data not shown). The comparison of the 2D 1H-13C HSQC spectra acquired at alkaline (pH = 7.3) and acidic pH (5.5) provided an explanation of the pH-dependent formation of the foldback motif. At alkaline pH, the aromatic region of the HSQC spectrum was characterised by a low number of (mostly overlapped) cross peaks resonating at frequencies typical of unstructured DNA (Figure 2A). At acidic pH, the aromatic region of the HSQC spectrum displayed dispersed resonances, indicative of a well-defined secondary structure (Figure 2A). Notably, four anomaly-shifted cross peaks, two at ∼142/7.9 ppm and two at ∼150/6.7 ppm in the 13C/1H dimension, correlating to the four C2/H2 peaks of adenines, were detected (Figure 2A). These anomalous shifts in the C2/H2 peaks, two at ∼ 142/7.8 ppm and two at ∼ 150/6.7 ppm, are unique spectral markers of protonated adenine at N1 and the 5′-non-protonated adenine stacked on the protonated adenine, respectively (51,52). The network of inter- and intra-residue NOEs indicated that the adenines were stabilised in the protonated form by forming asymmetric A+.C wobble-type base pairs (52). Taken together, the NMR data at pH = 5.5 indicated that the formation of asymmetrical A+.C non-canonical wobble base-pair(s) is responsible for the pH-dependent formation of the foldback structure. The primary structural characteristics of the CE21 construct at acidic pH that are consistent with all spectral data are schematically represented in Figure 2B.
Figure 2.
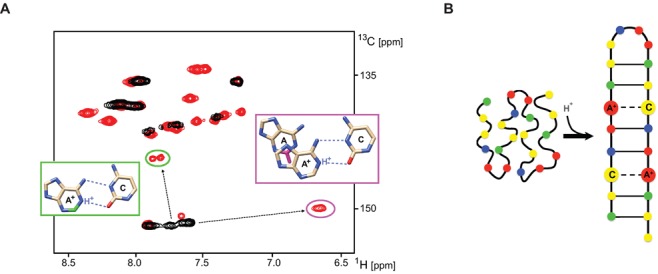
Formation of asymmetrical wobble A+.C base-pair is responsible for pH dependent formation of foldback motif within C. elegans C-rich telomeric DNA. (A) Aromatic regions of the 2D 1H-13C HSQC spectra of the CE21 construct recorded at 10°C and pH = 7.3 (black) and at pH = 5.5 (red). Anomalously shifted cross-peaks (C2/H2) from (two) protonated adenine bases are highlighted in green ellipse. Two anomalously shifted cross-peaks (C2/H2) due to 5′-stacking of non-protonated adenine to the protonated adenine base are highlighted in purple ellipse. Nuclei affected by the adenosine protonation are highlighted in the figure insets: C2-H2 adjoining the protonated N1 of adenosine (green) and C2-H2 exposed to the ring current the from 5′-protonated adenosine (purple). (B) Schematic representation of the low pH-induced foldback motif adopted by C-rich telomeric DNA from C. elegans.
The secondary structures within C-overhang appear to mirror the secondary structures within the G-overhang
Previous studies using short constructs based on four repeats of human telomeric DNA have demonstrated that both C- and G-rich strands can adopt intramolecular tetraplex structures (47). As shown by others for extended human telomeric G-rich sequences (53–56) (Figure 3A and B) and as demonstrated by our CD and NMR spectra (Figure 1), this property is preserved even for extended constructs of C-rich human telomeric DNA. In addition to vertebrate telomeric DNA, the simultaneous formation of tetraplex structures in both C- and G-rich strands was observed for telomeric DNA from evolutionarily distal eukaryotic species, such as plants, insects, or unicellular eukaryotes (22,57). However, whereas the C. elegans (nematode) G-rich telomeric DNA was previously shown to preferentially fold into a G-quadruplex structure (57), we find that the C-rich strand of the telomeric C. elegans DNA adopts duplex-based motif. While these observations on one hand support idea of the G-quadruplex in the G-rich strand as an evolutionarily conserved property of telomeric DNA, they argue against evolutionary conservation of the i-motif in the C-rich strand of telomeric DNA.
Figure 3.
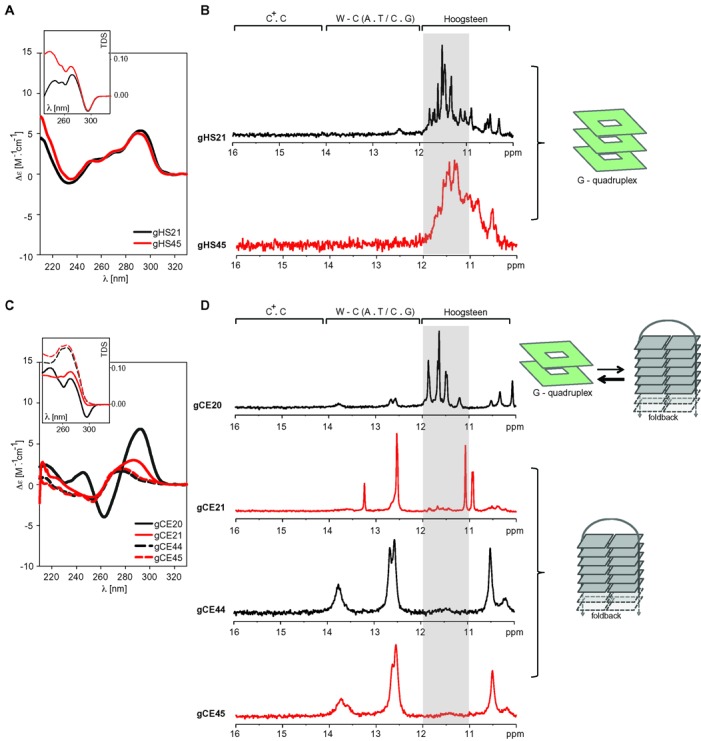
G-rich telomeric DNA from C. elegans and humans forms fundamentally different structures. LEFT: CD spectra of the DNA constructs based on human (A) and C. elegans (C) G-rich telomeric DNA repeats as a function of the number of telomeric repeats and/or the presence of the 3′-flanking residue. The insets display the thermal difference spectra (TDS) of each construct (58). CENTRE: Imino regions of the 1D 1H NMR spectra of the DNA constructs based on human (B) and C. elegans (D) G-rich telomeric DNA repeats showing the effect of the number of telomeric repeats and the presence of the 3′-flanking residue (C. elegans). RIGHT: Structural interpretation of the spectra.
Considering the importance of these implications and the fact that the flanking nucleotides (particularly those at the 3′-end) might substantially influence the structural properties of four-repeat-based telomeric DNA constructs (45–46,54), we re-examined the structural properties of G-rich telomeric DNA from C. elegans to exclude possible interpretational bias caused by the use of a short construct of nematode G-rich telomeric DNA in the study by Tran et al. (57). Four constructs were employed to re-investigate the structural properties of telomeric G-rich DNA from C. elegans: (i) the original four-repeat-based construct used in the study by Tran et al. (57), referred here to as the minimal construct (gCE20—Table 1), (ii) the minimal construct containing additional cytosine at the 3′-end to assess the influence of the 3′-end flanking nucleotide (gCE21—Table 1) and (iii) two extended telomeric DNA constructs based on eight-telomeric G-rich repeats with (gCE45) or without (gCE44) a 3′-terminal cytosine (Table 1). The CD and NMR spectra of all of these constructs are shown in Figure 3C and D, respectively. The gCE20 construct displayed CD spectrum typical of an anti-parallel G-quadruplex, including positive and negative bands at 295 nm and 265 nm, respectively (Figure 3C). This spectrum, as well as its interpretation, is fully consistent with those reported by Tran et al. (57). Also the absorption thermal difference (TD) spectrum of the gCE20 (Figure 3C, inset) evidenced the G-quadruplex formation (58). However, the CD and TD spectra of the gCE20 construct were notably distinct from the TD and CD spectra of all of the extended constructs (gCE21, gCE44 and gCE45). The shape of CD spectra for the gCE21, gCE44 and gCE45 corresponded to either unstructured DNA or the B-type structure, but not a G-quadruplex (Figure 3C). Additionally, the corresponding TD spectra lacked the 295 nm negative band characteristic of the G-quadruplex structure (Figure 3C, inset). In agreement with the interpretation of the CD/TD spectra, the imino region of the 1D 1H NMR spectrum of the gCE20 construct displayed characteristic signals in the ‘Hoogsteen’ region (11–12 ppm) (Figure 3D), indicating the formation of the G-quadruplex structure. In addition to the signals in the ‘Hoogsteen region’, weak signals in the ‘W-C’ region (12–14 ppm) were observed (Figure 3D). These signals can be attributed to minor population of ‘hairpin’ structure noted also in the original study by Tran et al. (57). However, the signals indicating Hoogsteen-type base pairing were clearly absent from the imino regions of the 1D 1H NMR spectra of all of the extended constructs (Figure 3D). Instead, the spectra of the extended constructs (gCE44, gCE45) displayed three dominant signals in the ‘W-C’ region between 12 and 14 ppm. The signal at ∼ 12.8 ppm was indicative of the G.C W-C base pair(s) formation and the signals between 13.2 and 13.8 ppm indicated the formation of A.T W-C base pairs. In conclusion, our data suggest that the G-quadruplex formation in nematode telomeric G-rich DNA observed by Tran et al. (57) is restricted to the gCE20 construct. Extension of the original C. elegans construct (gCE20) remodelled the conformation space of the C. elegans telomeric DNA towards formation of the foldback structure that was stabilised by W-C base pairs.
It needs to be emphasised that the telomeric DNA from both human and C. elegans is based on regular telomeric repeat sequences. However, telomeric DNA in many species, predominantly from the class of Saccharomycetes but including some plants, slime moulds and fungi, is characterised by irregular telomeric repeats (59,60). To investigate whether secondary structures within the C-rich telomeric DNA based on irregular repeats mirror the secondary structures within the corresponding G-rich telomeric DNA, we probed structural properties of four distinct constructs from the G- (gSA-a19 and gSA-b23) and C-rich (SA-a19 and SA-b23) telomeric DNA from the Saccharomyces cerevisiae (Table 1) using NMR spectroscopy. At mildly alkaline pH (pH ∼ 7.3), the imino regions of the 1D 1H NMR spectra of both gSA-a19 and gSA-b23 constructs displayed characteristic signals in the ‘Hoogsteen’ region (11–12 ppm) (Figure 4A), indicating the formation of the G-quadruplex structure. At this pH, no imino signals were detected in the 1D 1H NMR spectra of both SA-a19 and SA-b23, indicating the absence of hydrogen bonding-stabilised secondary structure in the investigated constructs (Figure 4B). Yet, upon sample acidification (pH ∼ 5.5), the 1D 1H NMR spectra of both constructs displayed characteristic signals in the ‘i-motif’ region (15–16 ppm). Altogether, our data indicate that the telomeric DNA from S. cerevisiae can form stable secondary structures of the same type (tetraplex) in the G- as well as in the C-rich strand. Similar to the results for the human and the C. elegans C-rich constructs, formation of the secondary structure within the S. cerevisie C-rich strand is pH dependent.
Figure 4.
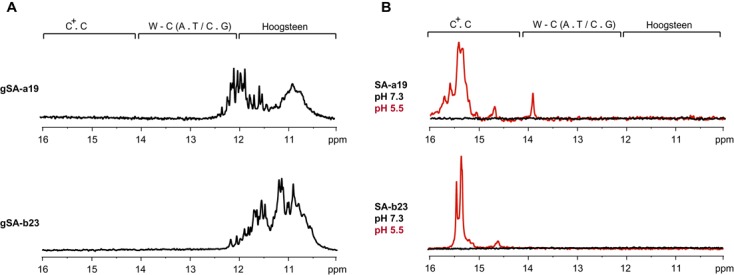
(A) Imino regions of the 1D 1H NMR spectra of the DNA constructs based on S. cerevisiae G-rich telomeric DNA repeats (gSA-a19 and gSA-b23) at pH 7.3 (B) Imino regions of the 1D 1H NMR spectra of the DNA constructs based on S. cerevisiae C-rich telomeric DNA repeats (SA-a19 and SA-b23) at pH 7.3 (black) and pH 5.5 (red). The spectra were acquired at 700 MHz at 20°C.
The UV melting experiments
The stabilities of the CE21 and CE45 were assessed via thermal denaturation followed by monitoring of absorbance changes at 260 nm at pH ∼ 5.5. The thermodynamic stabilities of corresponding G-rich constructs, namely gCE21 and gCE45, were assessed via thermal denaturation followed by monitoring of absorbance changes at 260 nm at pH ∼ 7.3 (Figure 5). The melting temperatures for individual constructs estimated from thermal denaturation experiments were 43.7°C, 45.6°C, 50.3°C and 52.2°C for the CE21, CE45, gCE21 and gCE45, respectively.
Figure 5.
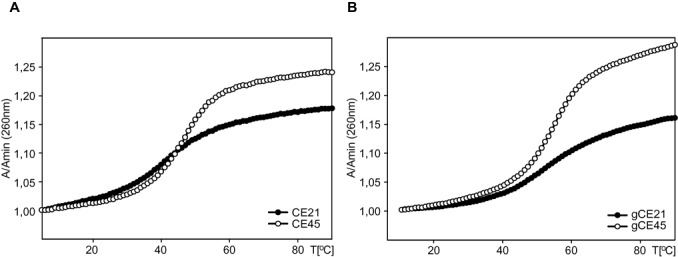
Thermal melting curves recorded at 260 nm and at 0.02 mM oligonucleotide strand concentration: (A) CE21 and CE45; (B) gCE21 and gCE45. Thermal melting curves for the C- and G-rich sequences were acquired at pH = 5.5 and pH = 7.3, respectively. The individual constructs are labelled according Table 1.
DISCUSSION
Identified structural properties of both C. elegans and human telomeric DNA are summarised in Figure 6. Their comparison with the properties of telomeric DNA from other eukaryotic species (Figure 7) provides interesting and important insights into the evolution of telomeric DNA.
Figure 6.
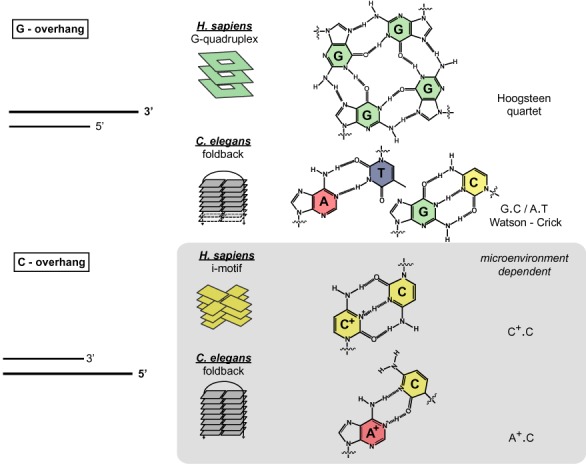
Schematic representations of the structural differences of C. elegans and human 5′- and 3′- telomeric C- and G-overhangs. C. elegans telomeric repeats from C- and G-rich strands form simple foldback motifs. Whereas the foldback motif formation in the G-rich strand relies on the formation of canonical Watson-Crick (G.C and A.T) base pairs, the formation of the foldback structure in the C-rich strand relies on the environmentally induced formation of non-canonical wobble A+.C pair(s). Human telomeric repeats in the G-rich strand form a G-quadruplex structure relying on the formation of so-called G-quartet stabilised by a Hoogsteen-type network of hydrogen bonds. Human C-rich telomeric DNA forms an intercalated tetraplex (i-motif), which is stabilised by non-canonical wobble C.C+ base pairs.
Figure 7.
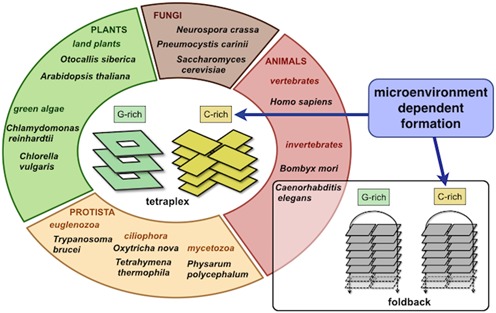
The structural characteristics of the C. elegans telomeric DNA are distinct from those of other eukaryotes. Changes in the telomeric repeat primary sequence (Supplementary Information—Table S1) appear to be ‘neutral’ with respect to the specific structural properties of both the G- and C-rich strand, namely: i) ability to form stable secondary structures of the same type in the G- as well as in the C-rich strand and ii) microenvironment dependent formation of secondary structure within the C-rich strand.
In the past two decades, it has become apparent that the primary sequence of the minisatellite repeat at the functional telomere does not define the major phylogenetic groups (61). On the one hand, the human type of telomeric repeat (GGGTTA.TAACCC) is found to be scattered across all major phyla, ranging from primitive moulds to invertebrates and mammals. On the other hand, many closely related species carry distinct primary telomeric repeat sequences, and several examples of species carrying mixed arrays of telomeric repeats exist (62,63). Although it is clear that more than one type of minisatellite repeat might fulfil the function of telomeric DNA, the factors that control and drive the variability of telomeric sequences during evolution are unknown (64). Thus far, cumulative evidence has established that the primary telomeric sequence is not the primary determinant of telomere function; however, there must be some universal epigenetic hallmarks of telomeric DNA.
Previous findings regarding the formation of the i-motif and the G-quadruplex in the C- and G-rich strands from a broad range of phylogenetically distal multicellular eukaryotes have implicated the i-motif and G-quadruplex as evolutionarily conserved characteristics of telomeric DNA (65). However, the phylogenetic conservation of the i-motif, implying its functional importance, has been controversial. One argument states that acidic pH required for the i-motif formation in vitro appears generally incompatible with the pH of intracellular environment. However, it needs to be considered that other general physical mechanisms such as molecular crowding or confinement, both related to chromatine compaction state (66,67), can induce the intramolecular i-motif structure at physiological pH (68). Another (phylogenetic) argument states that the apparent conservation of the i-motif is a coincidental characteristic of the conformational space of C-rich DNA that plays no functional role and is only a consequence of the propagation of sequence information from the complementary evolutionarily conserved G-rich (and G-quadruplex-forming) strand.
Our findings of the foldback motif formation within the C. elegans telomeric C-rich DNA clearly argue against the phylogenetic conservation of the i-motif. However, on the other hand, our findings identify a microenvironment-dependent secondary structure formation, regardless of an actual structural type, as a plausible conserved property of the telomeric C-rich strand indicating its functional role independent of the G-rich counterpart.
Analogously, the detection of the foldback motif in the G-rich strand of C. elegans telomeric DNA refute the G-quadruplex formation as an evolutionarily conserved property of the G-rich strand. Our data rather suggest that the ability to form thermodynamically stable secondary structure, regardless of an actual structural type, is the property of the G-rich strand of telomeric DNA that is conserved in the course of evolution.
Noteworthy, we observe that the melting temperatures of the short, CE21 (43.7°C) and gCE21 (50.3°C), and the corresponding extended, CE44 (45.6°C) and gCE44 (52.2°C), constructs from C. elegans telomeric DNA are remarkably similar with differences being generally less than 2°C. (Figure 5). This similarity indicates that both G- and C-rich telomeric DNA might consist of four-repeat based foldback motifs organised in ‘bead-on-a-string’ like fashion, i.e. in arrangement analogous to that observed in extended human telomeric G-rich DNA (54–55,69), rather than forming long continuous hairpins.
Altogether, our data are indicative that the properties of the telomeric DNA are evolutionarily conserved but not at the primary sequence level. Changes in the telomeric repeat sequence are clearly tolerated during evolution; however, these changes appear to be ‘neutral’ (70) with respect to the specific structural properties of both the G- and C-rich strand (Figure 7), namely: i) ability to form stable secondary structures of the same type in the G- as well as in the C-rich strand and ii) microenvironment dependent formation of secondary structure within the C-rich strand.
Importantly, the conservation of a secondary structure formation within the C-rich strand of human telomeric DNA implies the i-motif importance for the telomere function. In this respect, a logical question arises—can targeting of the 5′ C-overhang, a characteristic of ALT tumours, be employed for the selective elimination of tumour cells engaged in the ALT pathway? Although addressing this question is beyond the scope of this study, several lines of indirect evidence provide support for the concept of the i-motif as a negative regulator of telomere maintenance and, thus, the potential for the human telomeric i-motif as a therapeutic target. Recently, Chen et al. (29) showed that treatment of K562 and HeLa cells with single walled carbon nanotube (SWCN), a selective stabiliser of human telomeric i-motif, leads to telomere dysfunction via telomere uncapping marked by the displacement of several telomere binding proteins including the poly-C binding protein (PCBP) that has been identified as a specific telomere C-rich strand binder (71,72). Along the same line, Fujimori et al. (73) showed that treatment of telomerase-negative, ALT-positive U2OS cells with the a small molecular weight cationic porphyrin TMPyP4, known stabiliser of both human telomeric i-motif (74) and human (telomeric) DNA G-quadruplex (75,76), resulted in inhibition of cell growth and elevated levels of apoptosis supposedly due to telomere dysfunction. Unfortunately, complexity of the reported cellular phenotypes did not allow identification of primary events responsible for the telomere dysfunction. We observed that the TMPyP4 could directly interfere with the binding of the PCBP to human telomeric C-rich DNA in in-vitro assay (Supplementary Information—Figure S4). This observation provides support for explanation by Chen et al. (29) that displacement of PCBP binding/shielding protein might indeed be due to the SWCN induced i-motif formation within the telomeric DNA. However, it needs to be stressed that all available evidence is only indirect, and detailed studies will be needed to validate this concept.
Finally, C. elegans mutants that can survive in the absence of a functional telomerase pathway and that fully rely on the ALT mechanism were recently proposed as a multi-cellular model organism for ALT (30,77–78). The telomeric DNA in this model organism appears to display all of the important characteristics underlying ALT-maintained telomeres, including the presence of the 5′-C-rich overhang. Although this organism appears to represent an outstanding tool for elucidating the mechanisms underlying the ALT pathway, our structural data implicating distinct types of secondary structures in both 3′-G-rich and 5′-C-rich overhangs between C. elegans and humans emphasises the important limitation of C. elegans as an ALT model. This model organism should be used with caution in situations involving the use of ligands targeting telomeric DNA.
It is important to stress that our conclusions presented here were drawn based on the properties of model oligonucleotides in vitro. The conformational properties of telomeric repeats in the context of genomic DNA in vivo might differ from those of isolated oligonucleotides employed in our study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Czech Science Foundation [13–28310S to L.T. and M.V., P205/12/0466 to M.V.]; CEITEC [CZ.1.05/1.1.00/02.0068]. L.T. was supported by the R&D grant from Instruct, career development grant from the European Organization for Molecular Biology [IG2535] and by the Marie-Curie Re-integration grant. S.F.T. was supported from the SoMoPro II Programme, co-financed by the European Union and the South Moravian Region (Czech Republic). We thank Michaela Krafcikova for help with NMR spectra acquisition. Funding for open access charge: Czech Science Foundation [13-28310S].
Author Contributions: P.S. and K.B. conducted the CD/UV and native PAGE experiments; L.T., R.F. and S.F.T. conducted the NMR experiments; S.F.T. conducted the pull-down assays; P.S., K.B., R.F., S.F.T., L.T. and M.V. designed the experiments; and M.V. and L.T. wrote the manuscript.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wright W.E., Tesmer V.M., Huffman K.E., Levene S.D., Shay J.W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makarov V.L., Hirose Y., Langmore J.P. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 3.Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 4.Allsopp R.C., Vaziri H., Patterson C., Goldstein S., Younglai E.V., Futcher A.B., Greider C.W., Harley C.B. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelhardt M., Martens U.M. The implication of telomerase activity and telomere stability for replicative aging and cellular immortality (Review) Oncol. Rep. 1998;5:1043–1052. doi: 10.3892/or.5.5.1043. [DOI] [PubMed] [Google Scholar]
- 6.Aubert G., Lansdorp P.M. Telomeres and aging. Physiol. Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- 7.Shore D., Bianchi A. Telomere length regulation: coupling DNA end processing to feedback regulation of telomerase. EMBO J. 2009;28:2309–2322. doi: 10.1038/emboj.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greider C.W., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 9.Hiyama E., Hiyama K. Telomere and telomerase in stem cells. Br. J. Cancer. 2007;96:1020–1024. doi: 10.1038/sj.bjc.6603671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 11.Lipps H.J., Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell. Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Phan A.T. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 13.Hansel R., Foldynova-Trantirkova S., Lohr F., Buck J., Bongartz E., Bamberg E., Schwalbe H., Dotsch V., Trantirek L. Evaluation of parameters critical for observing nucleic acids inside living Xenopus laevis oocytes by in-cell NMR spectroscopy. J. Am. Chem. Soc. 2009;131:15761–15768. doi: 10.1021/ja9052027. [DOI] [PubMed] [Google Scholar]
- 14.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zahler A.M., Williamson J.R., Cech T.R., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 16.De Cian A., Lacroix L., Douarre C., Temime-Smaali N., Trentesaux C., Riou J.F., Mergny J.L. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Bryan T.M., Englezou A., Dalla-Pozza L., Dunham M.A., Reddel R.R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 18.Ogino H., Nakabayashi K., Suzuki M., Takahashi E., Fujii M., Suzuki T., Ayusawa D. Release of telomeric DNA from chromosomes in immortal human cells lacking telomerase activity. Biochem. Biophys. Res. Commun. 1998;248:223–227. doi: 10.1006/bbrc.1998.8875. [DOI] [PubMed] [Google Scholar]
- 19.Cesare A.J., Reddel R.R. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 20.Oganesian L., Karlseder J. Mammalian 5′ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol. Cell. 2011;42:224–236. doi: 10.1016/j.molcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed S., Kintanar A., Henderson E. Human telomeric C-strand tetraplexes. Nat. Struct. Biol. 1994;1:83–88. doi: 10.1038/nsb0294-83. [DOI] [PubMed] [Google Scholar]
- 22.Leroy J.L., Gueron M., Mergny J.L., Helene C. Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif. Nucleic Acids Res. 1994;22:1600–1606. doi: 10.1093/nar/22.9.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehring K., Leroy J.L., Gueron M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 24.Lieblein A.L., Kramer M., Dreuw A., Furtig B., Schwalbe H. The nature of hydrogen bonds in cytidine…H+…cytidine DNA base pairs. Angew. Chem. 2012;51:4067–4070. doi: 10.1002/anie.201200549. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J., Wei C., Jia G., Wang X., Feng Z., Li C. Formation of i-motif structure at neutral and slightly alkaline pH. Mol. Biosyst. 2010;6:580–586. doi: 10.1039/b919600e. [DOI] [PubMed] [Google Scholar]
- 26.Raices M., Verdun R.E., Compton S.A., Haggblom C.I., Griffith J.D., Dillin A., Karlseder J. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell. 2008;132:745–757. doi: 10.1016/j.cell.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 27.Madshus I.H. Regulation of intracellular pH in eukaryotic cells. Biochem. J. 1988;250:1–8. doi: 10.1042/bj2500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wadsworth W.G., Riddle D.L. Acidic intracellular pH shift during Caenorhabditis elegans larval development. Proc. Natl. Acad. Sci. U.S.A. 1988;85:8435–8438. doi: 10.1073/pnas.85.22.8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y., Qu K., Zhao C., Wu L., Ren J., Wang J., Qu X. Insights into the biomedical effects of carboxylated single-wall carbon nanotubes on telomerase and telomeres. Nat. Commun. 2012;3:1074–1088. doi: 10.1038/ncomms2091. [DOI] [PubMed] [Google Scholar]
- 30.Lackner D.H., Karlseder J. C. elegans survivors without telomerase. Worm. 2013;2:e21073. doi: 10.4161/worm.21073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shampay J., Szostak J.W., Blackburn E.H. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- 32.Gray D.M., Hung S.H., Johnson K.H. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 33.Masse J.E., Bortmann P., Dieckmann T., Feigon J. Simple, efficient protocol for enzymatic synthesis of uniformly 13C, 15N-labeled DNA for heteronuclear NMR studies. Nucleic Acids Res. 1998;26:2618–2624. doi: 10.1093/nar/26.11.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fiala R., Spackova N., Foldynova-Trantirkova S., Sponer J., Sklenar V., Trantirek L. NMR cross-correlated relaxation rates reveal ion coordination sites in DNA. J. Am. Chem. Soc. 2011;133:13790–13793. doi: 10.1021/ja202397p. [DOI] [PubMed] [Google Scholar]
- 35.Piotto M., Saudek V., Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 36.Vuister G.W., Bax A. Measurement of two-bond JCOH alpha coupling constants in proteins uniformly enriched with 13C. J. Biomol. NMR. 1992;2:401–405. doi: 10.1007/BF01874818. [DOI] [PubMed] [Google Scholar]
- 37.Santoro J., King G.C. A constant-time 2D overbodenhausen experiment for inverse correlation of isotopically enriched species. J. Mag. Reson. 1992;97:202–207. [Google Scholar]
- 38.Bohlen J.M., Bodenhausen G. Experimental aspects of Chirp NMR spectroscopy. J. Magn. Reson. 1993;102:293–301. [Google Scholar]
- 39.Sklenar V., Peterson R.D., Rejante M.R., Feigon J. Two- and three-dimensional HCN experiments for correlating base and sugar resonances in 15N,13C-labeled RNA oligonucleotides. J. Biomol. NMR. 1993;3:721–727. doi: 10.1007/BF00198375. [DOI] [PubMed] [Google Scholar]
- 40.Lippens G., Dhalluin C., Wieruszeski J.M. Use of a water flip-back pulse in the homonuclear NOESY experiment. J. Biomol. NMR. 1995;5:327–331. doi: 10.1007/BF00211762. [DOI] [PubMed] [Google Scholar]
- 41.Legault P., Farmer B.T., Mueller L., Pardi A. Through-bond correlation of adenine protons in a C-13-labeled ribozyme. J. Am. Chem. Soc. 1994;116:2203–2204. [Google Scholar]
- 42.Marino J.P., Prestegard J.H., Crothers D.M. Correlation of adenine H2/H8 resonances in uniformly C-13 labeled RNAs by 2D HCCH-TOCSY - A new tool for H-1 assignment. J. Am. Chem. Soc. 1994;116:2205–2206. [Google Scholar]
- 43.Dingley A.J., Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide (2)J(NN) couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 44.Pervushin K., Ono A., Fernandez C., Szyperski T., Kainosho M., Wuthrich K. NMR scalar couplings across Watson-Crick base pair hydrogen bonds in DNA observed by transverse relaxation-optimized spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14147–14151. doi: 10.1073/pnas.95.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai J., Carver M., Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vorlickova M., Tomasko M., Sagi A.J., Bednarova K., Sagi J. 8-oxoguanine in a quadruplex of the human telomere DNA sequence. FEBS J. 2012;279:29–39. doi: 10.1111/j.1742-4658.2011.08396.x. [DOI] [PubMed] [Google Scholar]
- 47.Phan A.T., Mergny J.L. Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simonsson T., Pribylova M., Vorlickova M. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem. Biophys. Res. Commun. 2000;278:158–166. doi: 10.1006/bbrc.2000.3783. [DOI] [PubMed] [Google Scholar]
- 49.Phan A.T., Leroy J.L. Intramolecular i-Motif Structures of Telomeric DNA. J. Biomol. Struct. Dyn. 2000;17(Suppl. 1):245–251. doi: 10.1080/07391102.2000.10506628. [DOI] [PubMed] [Google Scholar]
- 50.Kypr J., Kejnovska I., Renciuk D., Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Legault P., Pardi A. In-situ probing of adenine protonation in RNA by C-13 NMR. J. Am. Chem. Soc. 1994;116:8390–8391. [Google Scholar]
- 52.Stefl R., Oberstrass F.C., Hood J.L., Jourdan M., Zimmermann M., Skrisovska L., Maris C., Peng L., Hofr C., Emeson R.B., et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Azarkh M., Singh V., Okle O., Dietrich D.R., Hartig J.S., Drescher M. Intracellular conformations of human telomeric quadruplexes studied by electron paramagnetic resonance spectroscopy. ChemPhysChem. 2012;13:1444–1447. doi: 10.1002/cphc.201100980. [DOI] [PubMed] [Google Scholar]
- 54.Hansel R., Lohr F., Trantirek L., Dotsch V. High-resolution insight into G-overhang architecture. J. Am. Chem. Soc. 2013;135:2816–2824. doi: 10.1021/ja312403b. [DOI] [PubMed] [Google Scholar]
- 55.Vorlickova M., Chladkova J., Kejnovska I., Fialova M., Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petraccone L., Spink C., Trent J.O., Garbett N.C., Mekmaysy C.S., Giancola C., Chaires J.B. Structure and stability of higher-order human telomeric quadruplexes. J. Am. Chem. Soc. 2011;133:20951–20961. doi: 10.1021/ja209192a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran P.L., Mergny J.L., Alberti P. Stability of telomeric G-quadruplexes. Nucleic Acids Res. 2011;39:3282–3294. doi: 10.1093/nar/gkq1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mergny J.L., De Cian A., Ghelab A., Sacca B., Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohn M. Molecular Diversity of Telomeric Sequences. In: Nosek J, Tomaska L, editors. Origin and Evolution of Telomeres. Landes Bioscience; 2008. pp. 70–82. [Google Scholar]
- 60.Gunisova S., Elboher E., Nosek J., Gorkovoy V., Brown Y., Lucier J.F., Laterreur N., Wellinger R.J., Tzfati Y., Tomaska L. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA. 2009;15:546–559. doi: 10.1261/rna.1194009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fajkus J., Sykorova E., Leitch A.R. Telomeres in evolution and evolution of telomeres. Chromosome Res. 2005;13:469–479. doi: 10.1007/s10577-005-0997-2. [DOI] [PubMed] [Google Scholar]
- 62.Sykorova E., Lim K.Y., Kunicka Z., Chase M.W., Bennett M.D., Fajkus J., Leitch A.R. Telomere variability in the monocotyledonous plant order Asparagales. Proc. Biol. Sci. 2003;270:1893–1904. doi: 10.1098/rspb.2003.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rotkova G., Sklenickova M., Dvorackova M., Sykorova E., Leitch A.R., Fajkus J. An evolutionary change in telomere sequence motif within the plant section Asparagales had significance for telomere nucleoprotein complexes. Cytogenet Genome Res. 2004;107:132–138. doi: 10.1159/000079584. [DOI] [PubMed] [Google Scholar]
- 64.Tomaska L., Nosek J. Telomere heterogeneity: taking advantage of stochastic events. FEBS Lett. 2009;583:1067–1071. doi: 10.1016/j.febslet.2009.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garavis M., Gonzalez C., Villasante A. On the origin of the eukaryotic chromosome: the role of noncanonical DNA structures in telomere evolution. Genome Biol. Evol. 2013;5:1142–1150. doi: 10.1093/gbe/evt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walter A., Chapuis C., Huet S., Ellenberg J. Crowded chromatin is not sufficient for heterochromatin formation and not required for its maintenance. J. Struct. Biol. 2013;184:445–453. doi: 10.1016/j.jsb.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 67.Bancaud A., Huet S., Daigle N., Mozziconacci J., Beaudouin J., Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009;28:3785–3798. doi: 10.1038/emboj.2009.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rajendran A., Nakano S., Sugimoto N. Molecular crowding of the cosolutes induces an intramolecular i-motif structure of triplet repeat DNA oligomers at neutral pH. Chem. Commun. (Camb.) 2010;46:1299–1301. doi: 10.1039/b922050j. [DOI] [PubMed] [Google Scholar]
- 69.Yu H.Q., Miyoshi D., Sugimoto N. Characterization of structure and stability of long telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2006;128:15461–15468. doi: 10.1021/ja064536h. [DOI] [PubMed] [Google Scholar]
- 70.Gray M.W., Lukes J., Archibald J.M., Keeling P.J., Doolittle W.F. Cell biology. Irremediable complexity. Science. 2010;330:920–921. doi: 10.1126/science.1198594. [DOI] [PubMed] [Google Scholar]
- 71.Bandiera A., Tell G., Marsich E., Scaloni A., Pocsfalvi G., Akintunde Akindahunsi A., Cesaratto L., Manzini G. Cytosine-block telomeric type DNA-binding activity of hnRNP proteins from human cell lines. Arch. Biochem. Biophys. 2003;409:305–314. doi: 10.1016/s0003-9861(02)00413-7. [DOI] [PubMed] [Google Scholar]
- 72.Fenn S., Du Z., Lee J.K., Tjhen R., Stroud R.M., James T.L. Crystal structure of the third KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.6 A resolution. Nucleic Acids Res. 2007;35:2651–2660. doi: 10.1093/nar/gkm139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fujimori J., Matsuo T., Shimose S., Kubo T., Ishikawa M., Yasunaga Y., Ochi M. Antitumor effects of telomerase inhibitor TMPyP4 in osteosarcoma cell lines. J. Orthop. Res. 2011;29:1707–1711. doi: 10.1002/jor.21451. [DOI] [PubMed] [Google Scholar]
- 74.Fedoroff O.Y., Rangan A., Chemeris V.V., Hurley L.H. Cationic porphyrins promote the formation of i-motif DNA and bind peripherally by a nonintercalative mechanism. Biochemistry. 2000;39:15083–15090. doi: 10.1021/bi001528j. [DOI] [PubMed] [Google Scholar]
- 75.Qin Y., Rezler E.M., Gokhale V., Sun D., Hurley L.H. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wheelhouse R.T., Sun D.K., Han H.Y., Han F.X.G., Hurley L.H. Cationic porphyrins as telomerase inhibitors: the interaction of tetra-(N-methyl-4-pyridyl)porphine with quadruplex DNA. J. Am. Chem. Soc. 1998;120:3261–3262. [Google Scholar]
- 77.Lackner D.H., Raices M., Maruyama H., Haggblom C., Karlseder J. Organismal propagation in the absence of a functional telomerase pathway in Caenorhabditis elegans. EMBO J. 2012;31:2024–2033. doi: 10.1038/emboj.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng C., Shtessel L., Brady M.M., Ahmed S. Caenorhabditis elegans POT-2 telomere protein represses a mode of alternative lengthening of telomeres with normal telomere lengths. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7805–7810. doi: 10.1073/pnas.1119191109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


