Abstract
In vitro construction of Escherichia coli ribosomes could elucidate a deeper understanding of these complex molecular machines and make possible the production of synthetic variants with new functions. Toward this goal, we recently developed an integrated synthesis, assembly and translation (iSAT) system that allows for co-activation of ribosomal RNA (rRNA) transcription and ribosome assembly, mRNA transcription and protein translation without intact cells. Here, we discovered that macromolecular crowding and reducing agents increase overall iSAT protein synthesis; the combination of 6% w/v Ficoll 400 and 2 mM DTBA yielded approximately a five-fold increase in overall iSAT protein synthesis activity. By utilizing a fluorescent RNA aptamer, fluorescent reporter proteins and ribosome sedimentation analysis, we showed that crowding agents increase iSAT yields by enhancing translation while reducing agents increase rRNA transcription and ribosome assembly. Finally, we showed that iSAT ribosomes possess ∼70% of the protein synthesis activity of in vivo-assembled E. coli ribosomes. This work improves iSAT protein synthesis through the addition of crowding and reducing agents, provides a thorough understanding of the effect of these additives within the iSAT system and demonstrates how iSAT allows for manipulation and analysis of ribosome biogenesis in the context of an in vitro transcription-translation system.
INTRODUCTION
Ribosome biogenesis is an intricate and highly coordinated process that occurs in all living cells. The process requires the synthesis and ordered arrangement of ribosomal RNA (rRNA) and ribosomal proteins (r-proteins), resulting in one of nature's more sophisticated and complex macromolecular machines. The Escherichia coli ribosome, for example, is a 2.5 MDa machine that consists of a large 50S subunit and a small 30S subunit. It carries out sequence-defined polymerization of amino acids, decoding messenger RNA (mRNA) templates into polypeptides at an astounding rate of up to 21 amino acids per second with only one error per ∼104 amino acids (1,2).
Freed from cell viability constraints, extensive in vitro studies of E. coli ribosomes have led to a deep understanding of ribosome structure and function (3–5). For example, such studies have unraveled a number of important features of ribosomes including assembly mechanisms (6–8), rRNA modifications (9,10), rRNA-protein interactions (11,12) and r-protein assembly maps (13–15). However, these studies reconstitute E. coli ribosomes using native rRNA and r-proteins in an approach that does not mimic co-transcription of rRNA and ribosome assembly as it happens in cells, and, for the large subunit, require non-physiological conditions utilizing temperature and magnesium shifts to overcome kinetic traps. Further, classic reconstitution procedures largely fail to incorporate synthetic 23S rRNA of the large subunit into highly active particles (16,17).
To address these limitations, we developed an integrated synthesis, assembly and translation (iSAT) technology for the in vitro construction of ribosomes from template-derived rRNA in a ribosome-free S150 crude extract (18) and improved the system through transcriptional tuning (19) and by alleviating substrate limitations (20) to yield highly active ribosomes. The value of the iSAT system lies in the ability to co-activate the processes of rRNA synthesis and processing, ribosome assembly and translation as they occur in vivo. The iSAT system therefore provides a unique and powerful approach for the interrogation and manipulation of E. coli ribosome biogenesis in the context of a transcription and translation environment. In addition, the iSAT system is well poised to contribute toward emerging efforts to build minimal cells (21–26) and engineer ribosomal variants (27,28).
Unfortunately, the translational activity of ribosomes purified from iSAT reactions was only about 20% the activity of native ribosomes purified directly from E. coli cells (19). Therefore, we explored the use of additives that we hypothesized could improve overall iSAT protein synthesis activity. First, we sought to assess the impact of macromolecular crowding agents on the iSAT system. The iSAT reaction environment is inherently different than the intracellular environment, as E. coli cells contain ∼200–320 mg/ml protein (29) while iSAT reactions contain about 2 mg/ml total E. coli protein (19). We therefore expected that dilution could limit iSAT efficiency since multiple macromolecules with weak affinity for each other must interact for ribosome assembly and translation to occur. This hypothesis is supported in previous studies of other in vitro transcription and translation systems that have used macromolecular crowding agents to increase effective molecule concentration by either reducing the volume of available solvent or by preventing components from diffusing away before recycling (i.e. a cage effect). For example, macromolecular crowding agents have been shown to increase transcription in a cell-free protein synthesis (CFPS) system (30), improve RNA folding (31) and alter protein–protein interactions (32–34). For these reasons, we investigated the effect of macromolecular crowding conditions on the iSAT system using the polymers polyethylene glycol (PEG) and Ficoll. Second, we sought to assess the impact of various reducing agents on iSAT in an attempt to mimic the highly reduced cytoplasmic environment of E. coli (35,36). While dithiothreitol (DTT) and β-mercaptoethanol (BME) are commonly used in biological applications, Tris(2-carboxyethyl) phosphine (TCEP) and dithiobutylamine (DTBA) have been developed as alternative reducing agents with different activities and stabilities in various biochemical applications (37–39). We were interested in studying the effects of these reducing agents on iSAT activity.
Here we report on addition of crowding and reducing agents to the iSAT system. Using a multiple assay strategy, we thoroughly explored the effects of the additives on ribosome biogenesis (specifically rRNA synthesis and ribosome assembly), reporter mRNA transcription, and reporter protein translation (Figure 1). First, traditional iSAT batch reactions were performed with crowding and reducing agents to identify the additives and concentrations that maximized iSAT protein synthesis activity. To isolate the processes of transcription and translation, the same additive effects were assessed for purified E. coli 70S ribosomes in a transcription-translation (TX-TL) reaction. Next, effects of crowding and reducing agents on protein synthesis in iSAT or 70S TX-TL reactions were separated into transcriptional and translational effects through the use of an mRFP1-spinach aptamer reporter construct, which allows for simultaneous monitoring of mRNA and reporter protein concentrations over time. Additionally, the effects of additives on iSAT ribosome biogenesis were visualized through sucrose gradient sedimentation analysis of translating and non-translating iSAT reactions. Finally, we combined translation and ribosome profile data to determine the translation elongation rate of iSAT ribosomes and to compare the activity of iSAT ribosomes with in vivo-assembled E. coli ribosomes. Such an investigation is unique to the iSAT system and can provide biological insight into additive effects on ribosomal component synthesis and ribosome assembly.
Figure 1.
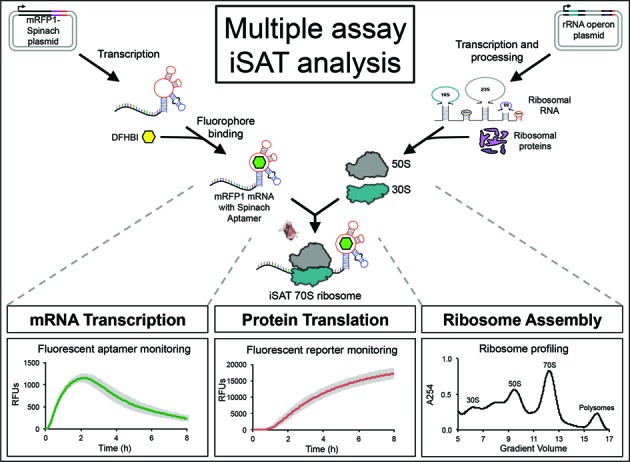
A three-pronged iSAT approach for studying in vitro ribosome construction, where iSAT is an integrated method for the assembly of ribosomes from in vitro transcribed rRNA and subsequent translation by these ribosomes in the same compartment. Analyses to study these co-activated processes include measurements of mRNA transcription through use of a fluorescent RNA aptamer, measurements of protein translation through use of a fluorescent reporter protein and assessment of ribosome assembly through ribosome profiling.
MATERIALS AND METHODS
Plasmid construction
DNA plasmids encoding mRFP1 and spinach aptamer were constructed from gBlocks (IDT) using previously reported sequences (40) with 5′ and 3′ additions to allow for digestion-ligation construction (Supplementary Table S1). The plasmid pY71sfGFP (41) was used as a source of the pY71 vector. For pY71mRFP1 construction, pY71sfGFP and mRFP1 were digested with NdeI and SalI, purified, ligated and transformed into C2987 competent E. coli cells. A similar procedure was used for constructing pY71mRFP1-SpA by digesting pY71mRFP1 and spinach aptamer (SpA) with AflII and SalI. The constructs pT7AM552A, pY71sfGFP and pK7Luc have been previously reported (18,19,41).
Component preparation
S150 extract, E. coli 70S ribosomes, total protein of 70S ribosomes (TP70) and T7 RNA polymerase (RNAP) were prepared as previously reported (19,42) except the dialysis and storage buffers were altered to use only iSAT salt components; iSAT buffer consists of 50 mM HEPES-KOH (pH 7.6), 10 mM MgGlu, 200 mM KGlu, 2 mM DTT, 1 mM spermidine and 1 mM putrescine.
iSAT reactions
iSAT reactions of 15 μl were set-up as previously described (19) with component concentrations shown in Supplementary Table S2. Reactions were prepared in polymerase chain reaction tubes with optically clear flat caps and incubated at 37°C in a CFX96™ real-time thermal cycler (Bio-Rad). iSAT reactions contained reporter protein plasmids encoding mRFP1 (pY71mRFP1), mRFP1-spinach aptamer (pY71mRFP1-SpA), superfolder GFP (sfGFP) (pY71sfGFP) or luciferase (pK7Luc). Red fluorescence of mRFP1 was monitored using the CFX96™ real-time thermal cycler as previously described for sfGFP (19) (excitation: 560–590 nm, emission: 610–650 nm). For reactions with mRFP1-spinach aptamer, the fluorophore DFHBI (Lucerna, Inc.) was included in the reactions at 60 μM. Green fluorescence of the spinach aptamer with DFHBI was simultaneously monitored (excitation: 450–490 nm, emission: 510–530 nm). Reactions without T7 RNAP or TP70 served as transcriptional or translational background controls, respectively. Additives were included at the described final concentrations. Crowding agents were prepared in 60% w/v stock solutions in nuclease-free water, while reducing agents were prepared in 1 M stock solutions in nuclease-free water. Crowding and reducing agents were purchased from Sigma-Aldrich, except for Ficoll 70, which was purchased from Santa Cruz Biotechnology.
iSAT ribosome purification
Several 15 μl iSAT reactions with 6% Ficoll 400 and 2 mM DTBA were prepared and incubated for 2 h at 37°C, then pooled together. Purified 70S E. coli ribosomes were recovered as previously described (19), with pelleted iSAT ribosomes resuspended in iSAT buffer, aliquoted and flash-frozen.
70S transcription and translation (TX-TL) reactions
70S TX-TL reactions were prepared in the same manner as iSAT reactions (Supplementary Table S2). Purified E. coli 70S ribosomes, purified iSAT 70S ribosomes or commercial E. coli ribosomes, (New England Biolabs Inc., P0763S) were included in the 70S TX-TL reactions, and pT7AM552A and TP70 were excluded. Reactions without ribosomes were used as negative controls for residual S150 extract activity.
Ribosome profiling through sedimentation analysis
Sucrose gradients were prepared from Buffer C (10 mM Tris–OAc (pH = 7.5 at 4°C), 60 mM NH4Cl, 7.5 mM Mg(OAc)2, 0.5 mM ethylenediaminetetraacetic acid, 2 mM DTT) with 10 and 40% sucrose in SW32.1 polycarbonate tubes using a Biocomp Gradient Master. Gradients were placed in SW32.1 buckets and chilled to 4°C. Meanwhile, several 15 μl iSAT reactions with and without additives were prepared and incubated at 37°C for 1 or 2 h. Reactions were pooled and 50 or 200 μl of iSAT reactions were carefully loaded onto chilled gradients. The gradients were ultra-centrifuged to 22 500 rpm for 17 h at 4°C, using an Optima L-80 XP ultracentrifuge (Beckman-Coulter) at medium acceleration and braking (‘5’ setting for each). Gradients were analyzed with a BR-188 Density Gradient Fractionation System (Brandel) by pushing 60% sucrose into the gradient at 0.75 ml/min. Traces of A254 readings versus elution volumes were obtained for each gradient, with readings adjusted to match baselines based on blank sucrose readings. iSAT reactions without the operon plasmid were performed to establish a background reading that was subtracted from experimental traces. Gradient fractions were collected and analyzed for rRNA content by gel electrophoresis in 1% agarose and imaged in a GelDoc Imager (Bio-Rad). Ribosome profile peaks were identified based on the rRNA content as representing 30S or 50S subunits, 70S ribosomes, or polysomes of two 70S ribosomes on a single mRNA. No peaks were observed for larger polysomes.
RESULTS
Crowding and reducing agents improve iSAT protein synthesis activity
To determine the effect of macromolecular crowding and reducing conditions on in vitro ribosome construction, we initially explored different concentrations of crowding and reducing agents on iSAT protein synthesis of the monomeric form of red fluorescent protein, mRFP1. mRFP1 was chosen since we intended to couple mRFP1 with a green fluorescent RNA aptamer in later experiments. As previously described, iSAT reactions consist of transcription of rRNA from an rRNA operon, assembly of rRNA with purified total ribosomal proteins of the 70S ribosome (TP70) and measurement of ribosome activity by transcription and translation of a reporter protein (in this case mRFP1) (18,19). These processes occur simultaneously at 37°C in 15 μl reactions. The reactions consist of crude ribosome-free S150 E. coli extract containing cytoplasmic translation and assembly factors, and salts, buffers and substrates necessary for transcription and translation.
For crowding agents, we tested molecular weight variants of PEG and Ficoll at concentrations ranging from 0 to 6% w/v. Both Ficoll and PEG more than doubled the active mRFP1 yield at particular concentrations (Figure 2A). Molecular weight variants of PEG or Ficoll behaved similarly, suggesting that polymer lengths within the tested ranges were not as important as volume exclusion. Addition of PEG resulted in the highest activity at 2 or 4% w/v, with iSAT protein synthesis activity decreasing at higher concentrations. Our results are consistent with previous works in S30 crude extract-based transcription and translation systems that have shown inhibitory effects at increasing PEG concentrations, which is attributed to the fact that PEG is able to dehydrate protein surfaces to cause protein precipitation and translation inhibition (30,43–45). Addition of Ficoll had the greatest positive effect on iSAT activity at 6% w/v, which was the highest tested concentration. Increasing Ficoll beyond 6% w/v was not feasible due to volume restrictions and difficulties in accurately pipetting highly viscous solutions. Previously, Ficoll has been shown to result in less protein precipitation than PEG (30).
Figure 2.
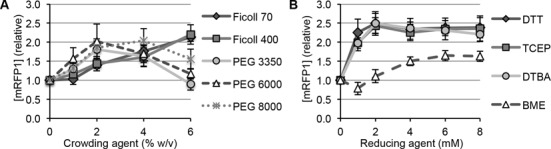
iSAT protein synthesis with the inclusion of macromolecular crowding and reducing agents. (A) Molecular weight variants of the crowding agents PEG and Ficoll were included in iSAT reactions at concentrations ranging from 0 to 6% w/v and (B) reducing agents DTT, TCEP, DTBA and BME were included in iSAT reactions at concentrations ranging from 0 to 8 mM. mRFP1 production after 18 h is relative to reactions with no additives. Values represent averages (n ≥ 3) and error bars represent 1 standard deviation (s.d.).
We subsequently tested the addition of reducing agents DTT, BME, TCEP and DTBA on iSAT protein synthesis activity at concentrations of 0 to 8 mM (Figure 2B). Even though iSAT reactions initially contain ∼1.0 mM DTT from the storage buffers of S150 extract, TP70 and T7 RNAP, all four reducing agents enhanced iSAT protein synthesis of mRFP1, with 2–8 mM DTT, TCEP or DTBA more than doubling protein synthesis. The reducing agents, excluding BME, showed very similar effects at the same concentrations. This suggests that the mechanism(s) of action for these reducing agents in iSAT reactions are similar, despite reported differences in other applications (37–39).
Next, we tested combinations of crowding and reducing agents at maximum effective concentrations (Table 1). The various combinations resulted in increases of iSAT protein synthesis ranging from 2.7- to 4.3-fold as compared to reactions without crowding and reducing additives. Subsequent analysis of the iSAT system used 6% Ficoll 400 and 2 mM DTBA because this combination of additives resulted in the highest relative mRFP1 production (Table 1). We note, however, that the requirements of particular assays or the synthesis of different proteins may favor the use of different combinations of crowding and reducing agents.
Table 1. iSAT protein synthesis of reactions containing combinations of crowding and reducing agents.
| Relative mRFP1 production | No crowding agent | Ficoll 70 (6% w/v) | Ficoll 400 (6% w/v) | PEG 6000 (2% w/v) | PEG 8000 (4% w/v) |
|---|---|---|---|---|---|
| No reducing agent | 1.0 | 2.1 | 2.2 | 2.0 | 2.0 |
| DTT (2mM) | 2.5 | 3.6 | 4.0 | 3.2 | 3.3 |
| DTBA (2mM) | 2.6 | 3.9 | 4.3 | 3.5 | 3.4 |
| TCEP (2mM) | 2.5 | 3.8 | 4.0 | 3.2 | 2.7 |
Crowding and reducing agents were included in iSAT reactions in combinations of concentrations with the greatest improvement on iSAT protein synthesis (Figure 2). Reactions were incubated at 37°C for 18 h and synthesis of mRFP1 was measured. mRFP1 production is shown relative to reactions with no additional crowding or reducing agents. Values represent averages (n ≥ 3) with 1 s.d.
Since the aforementioned results relied on the proper folding and fluorescence of the reporter protein mRFP1, we next sought to determine if the observed improvements were related to a general improvement in protein synthesis or simply protein folding and activation. Therefore, Ficoll 400 and DTBA were tested for their effects, individually and in combination, on the synthesis of various reporter proteins in the iSAT system as measured by 14C-leucine radioactive incorporation (Supplementary Methods and Supplementary Figure S1). For mRFP1, superfolder GFP (sfGFP), and luciferase, each additive resulted in a significant increase in iSAT protein synthesis, with the combination of additives providing further improvement ranging from 3.0- to 6.5-fold. This result suggests an increase in overall protein synthesis and not merely improved protein folding. Additionally, sfGFP production from batch iSAT reactions with Ficoll 400 and DTBA was 10.1 ± 0.6 μM, which exceeds our highest previously reported iSAT protein synthesis for sfGFP, which occurred in a semi-continuous reaction (20). This improvement is important for technological applications (e.g. constructing ribosomal variants with novel functionalities) and also for efforts to achieve the break-even milestone of ribosomes that are capable of constructing ribosomes (7434 peptide bonds are needed to make a complete set of r-proteins) (18).
Separation of biological process effects
Since crowding agents were expected to better mimic the high concentrations of molecules in cells and reducing agents were expected to ensure cytoplasmic redox mimicry, we hypothesized that the additives impacted iSAT reactions in different ways. To test this hypothesis, we asked how crowding and reducing agents affected each of the three primary processes that occur simultaneously in iSAT reactions: rRNA synthesis and ribosome assembly (e.g. ribosome biogenesis), reporter mRNA transcription and reporter protein translation. We explored the impact of additives on transcription and translation by removing the process of ribosome assembly. To do this, we performed cell-free transcription and translation (TX-TL) reactions in S150 extracts with purified E. coli 70S ribosomes in place of rRNA operons and purified r-proteins. The resulting reactions, which we term 70S TX-TL reactions, represent the reconstruction of processes occurring in traditional cell-free protein synthesis reactions utilizing S12 or S30 crude cell lysates (46–49), with the key difference being that S150 extracts and purified 70S ribosomes are used in place of S12 or S30 extracts. Expression of mRFP1 in 70S TX-TL reactions shows that Ficoll 400 provides a 63 ± 13% increase in protein yield, but additional reducing agent, DTBA, provides no significant increase (Figure 3). This result suggests that crowding agents affect iSAT reactions through the processes of mRNA transcription or translation, while additional reducing agents affect iSAT reactions through other processes, such as rRNA synthesis or ribosome assembly.
Figure 3.
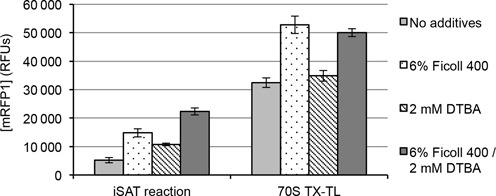
Comparison of iSAT and 70S TX-TL protein synthesis under crowding and reducing conditions. A total of 6% w/v Ficoll 400 and 2 mM DTBA were included in iSAT and 70S TX-TL reactions, individually or in combination. Reactions were incubated at 37°C for 18 h and synthesis of mRFP1 was measured in relative fluorescent units (RFUs). Values represent averages (n ≥ 4) and error bars represent 1 s.d.
In order to better understand the effect of crowding and reducing agents, we increased the resolution at which we studied the in vitro system by monitoring the processes of mRNA transcription and reporter protein translation. Previous studies have examined in vitro transcription and translation systems using either radioactive nucleotide incorporation (50), molecular beacons (51), binary FRET probes (52,53) or fluorescent RNA aptamers (40,54) in tandem with reporter proteins. We chose to use the previously reported mRFP1-spinach aptamer gene construct to allow for simultaneous real-time monitoring of transcription and translation (40). The spinach aptamer is an RNA structure that displays green fluorescence when bound to the fluorophore 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI) (Figure 1) (55).
Using the mRFP1-spinach aptamer gene construct, we carried out 15 μL batch iSAT reactions and also 70S TX-TL reactions for 8 h at 37°C. Measurements were taken every 5 min to profile mRNA and protein production with and without DTBA and Ficoll 400 (Figure 4). The effect of DTBA and Ficoll 400 on translation in iSAT and 70S TX-TL reactions is consistent with our observations reported in Figure 3. Furthermore, crowding and reducing agents do not appear to affect reaction duration in either case, suggesting that changes to substrate stability are not responsible for increases in protein synthesis.
Figure 4.
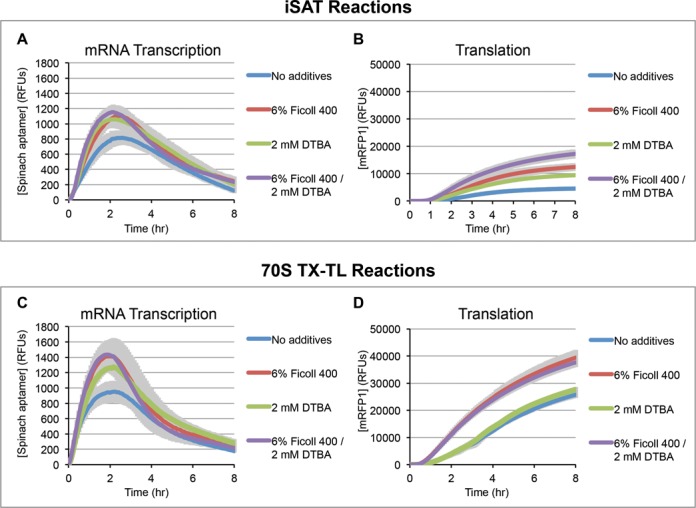
Comparison of transcription and translation under crowding and reducing conditions for (A) mRNA transcription and (B) translation in iSAT reactions and (C) mRNA transcription and (D) translation in 70S TX-TL reactions. Reactions were performed with the mRFP1-spinach construct and the fluorophore DFHBI. mRNA transcription (A and C) was measured by green fluorescence of the spinach aptamer of the mRNA bound to DFHBI. Translation (B and D) was measured by the red fluorescence of mRFP1. Readings were taken every 5 min. Lines represent smoothed averages (n ≥ 3) and shading represents smoothed error bars (1 s.d.).
For iSAT reactions, the additional transcriptional data from the spinach aptamer shows increased maximum mRNA production rates in the presence of crowding and reducing agents (Figure 4A). For example, the initial mRNA transcription rate over the first 30 min increases 35 ± 19% and 38 ± 21% when reactions are supplemented with Ficoll 400 and DTBA, respectively (Figure 4A). The combination of Ficoll 400 and DTBA results in increases of 71 ± 18% in initial transcription rate and 42 ± 20% in maximum mRNA concentration. In 70S TX-TL reactions, the addition of Ficoll 400 and DTBA individually and in combination showed significant improvement in maximum mRNA concentration: 64 ± 34% for Ficoll 400 and DTBA together (t = 4.52, d.f. = 4, P = 0.01, where t represents an unpaired t-test value, d.f. stands for degrees of freedom and P represents the two-tailed P-value), 51 ± 33% for Ficoll 400 alone (t = 3.62, d.f. = 4, P = 0.02) and 34 ± 20% for DTBA alone (t = 4.62, d.f. = 4, P = 0.01) (Figure 4C). Interestingly, addition of DTBA alone did not have a significant effect on 70S TX-TL translation of the reporter protein (Figure 4D). The fact that DTBA increases mRNA transcription but not mRFP1 translation suggests that mRNA transcription is not limiting for the 70S TX-TL reactions. It also suggests that an increase in transcription, if relevant to iSAT activity, may lie in the effect on rRNA transcription.
The observations that reducing agents increase iSAT protein synthesis but not 70S TX-TL protein synthesis suggest that ribosome biogenesis may be more sensitive to redox environment than are transcription and translation. The E. coli cytoplasm is highly reduced, so we hypothesized that iSAT activity would be decreased in an oxidized environment. Therefore, we tested the effect of the oxidant iodoacetamide (IAM) on iSAT and 70S TX-TL protein synthesis (Supplementary Figure S2) (56,57). At 200 μM IAM, iSAT reactions retain only 10 ± 3% protein synthesis activity, while 70S TX-TL reactions retain 50 ± 9% activity. This result supports our hypothesis that redox environment affects ribosome biogenesis more than it affects combined mRNA transcription and translation. A decade ago, Yin and Swartz showed the ability to carry out cell-free protein synthesis in an oxidizing environment (57), which was in many ways surprising given that the cytoplasm is reduced. Our data imply that the reducing environment is important for ribosome biogenesis.
Ribosome profiling of iSAT reactions shows that reducing agents increase ribosome biogenesis
Based on the previous data, we hypothesized that reducing agents in the iSAT system likely function to improve ribosome biogenesis (which in iSAT is composed of rRNA transcription and ribosome assembly) since they improve iSAT protein synthesis but not 70S TX-TL protein synthesis. To assess ribosome biogenesis directly, we utilized sucrose gradient sedimentation analysis (7,8,48,58). Several iSAT reactions, with or without additives, were incubated at 37°C for 2 h and then loaded onto 10–40% sucrose gradients. After ultracentrifugation, sucrose gradients were fractionated and analyzed by spectrophotometry. Profile peaks of spectrophotometric traces were identified by gel electrophoresis of gradient fractions to determine the presence of 16S and 23S rRNA (Supplementary Figures S3 and S4). While Ficoll 400 was used in previous experiments, its large molecular weight (∼400 kDa) was found to interfere with the ribosome profiles, particularly the small 30S subunit (∼900 kDa). Therefore, we used 2% PEG 6000 in place of 6% Ficoll 400 for ribosome profiling, as their effects on iSAT activity were similar (Figure 2A and Table 1). For the reducing agent, we continued to use 2 mM DTBA.
To isolate ribosome biogenesis, we performed ribosome profiling of iSAT reactions without reporter plasmid, thus preventing translation initiation, which requires mRNA templates. Without additives, ribosome profiling showed clear 30S and 50S peaks, with additional peaks likely representing partially formed ribosomal particles (Figure 5A, Supplementary Figure S3). Beyond the 50S peak, a small peak indicates formation of 70S ribosomes either on residual mRNA in the extract or through loose association of subunits, as iSAT reactions contain 7.5 mM Mg2+.
Figure 5.
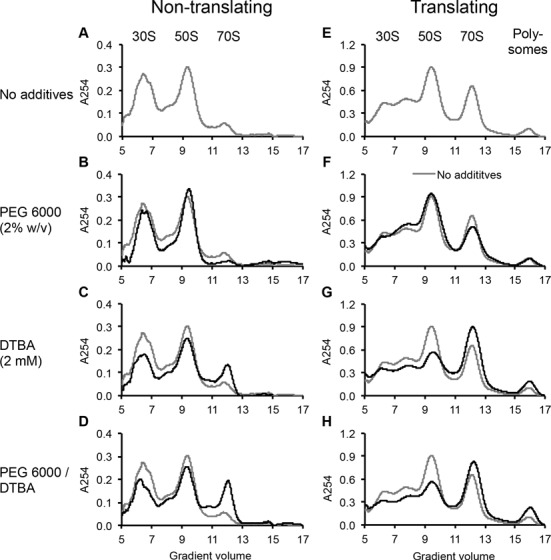
Ribosome profiling of iSAT reactions with and without crowding and reducing agents. Profiles represent A254 readings as relative signals versus gradient volume, in ml, measured from the top of 10–40% sucrose gradients, with peak identities labeled. For non-translating iSAT reactions (A–D), no reporter plasmid was added to 50 μl iSAT reactions including (A) no additives (gray, repeated in B–D), (B) 2% PEG 6000, (C) 2 mM DTBA or (D) 2% PEG 6000 and 2 mM DTBA. For translating iSAT reactions (E–H), pY71mRFP1-SpA was included as a reporter plasmid in 200 μl iSAT reactions including (E) no additives (gray, repeated in F–H), (F) 2% PEG 6000, (G) 2 mM DTBA or (H) 2% PEG 6000 and 2 mM DTBA.
Addition of PEG 6000 to non-translating iSAT reactions had little to no effect on the ribosome profile (Figure 5B). In contrast, the addition of DTBA dramatically altered the ribosome profile, most notably in the 2.4-fold increase of the 70S peak (Figure 5C). This correlates with a decrease in the 30S and 50S peaks, suggesting that the reducing agent influences subunit synthesis and assembly or subunit association, even without the presence of reporter mRNA. Even though we observe a 70S peak in non-translating iSAT reactions, we hypothesize that these may be capable of translation. Previous work by Underwood et al. in an E. coli S30 crude extract-based transcription and translation system showed that stably associated 70S ribosomes can transition to actively translating particles (48). The effect of DTBA on the ribosome profiles is retained in the presence of PEG 6000 (Figure 5D).
Ribosome profiles were also analyzed for complete iSAT reactions with reporter protein transcription and translation (Figure 5E–H, Table 2). The inclusion of the reporter protein plasmid dramatically altered iSAT ribosome profiles. For translating iSAT reactions without additives, the 70S peak is much larger in the presence of mRNA, and polysomes are now observed. Polysomes represent two or more ribosomes translating from the same mRNA (Figure 5E, Supplementary Figure S4, Table 2).
Table 2. Approximate concentrations of ribosomal components in translating iSAT reactions.
| Approximately concentration of each component in iSAT reaction (nM) | ||||
|---|---|---|---|---|
| No additives | 2% PEG 2000 | 2mM DTBA | 2%PEG 6000/2mM DTBA | |
| 30S + 50S | 192 | 212 | 153 | 139 |
| 70S | 65 | 62 | 102 | 98 |
| Polysomes | 7 | 7 | 13 | 8 |
| Total | 264 | 281 | 268 | 253 |
Ribosome profiles of translating iSAT reactions (Figure 5E–H) were analyzed from 5 to 17 ml elution volumes, with the separation of 50S and 70S peaks and 70S and polysome peaks determined by drop-down approximation from the lowest value between the peaks (∼11 and 15 ml, respectively). Areas under peaks were determined and converted to concentrations using the areas under peaks of known quantities of purified 70S ribosomes on sucrose gradients.
Addition of PEG 6000 to translating iSAT reactions had little effect on ribosome profiles (Figure 5F), which was similar to non-translating iSAT reactions. However, previous results show that PEG 6000 increases protein synthesis (Figure 2). Therefore, with this information and previous evidence that mRNA transcription is not a limiting factor for protein synthesis, we conclude that crowding agents increase protein synthesis by increasing the rate of translation while not impacting ribosome biogenesis. However, it is still possible that crowding agents improve the quality of the assembled ribosomes, which ribosome sedimentation analysis cannot assess. Advanced studies of the assembled ribosomes are required to make this distinction and are the basis of future works. Meanwhile, DTBA supplementation to iSAT reactions decreased subunit peaks and increased 70S and polysome peaks by 60% (Figure 5G, Table 2). Increased concentrations of 70S and polysome peaks are maintained in the presence of both DTBA and PEG 6000 (Figure 5H, Table 2). This result combined with the previous 70S TX-TL reactions showing no effect on protein synthesis from DTBA addition suggests that reducing agents improve ribosome biogenesis in iSAT by enabling assembly of more ribosomes and/or the construction of more active ribosomal particles and this improvement likely accounts for the increase in protein synthesis in iSAT reactions containing additional reducing agents.
Protein synthesis activity of iSAT-assembled ribosomes is similar to that of purified E. coli ribosomes
Protein synthesis and sucrose gradient ribosome profiling data were combined to calculate the translation elongation rate of ribosomes, as previously detailed for ribosomes within an E. coli cell-free protein synthesis reaction (48). For this calculation, iSAT and 70S TX-TL reactions with 2% PEG 6000 and 2 mM DTBA were performed in triplicate and analyzed for bulk translation rate of sfGFP and 70S monosome and polysome formation during the linear rate of protein synthesis (Supplementary Figure S5). By dividing bulk translation rate by the observed concentration of 70S ribosomes and polysomes, translation elongation rates were determined for iSAT ribosomes and E. coli ribosomes in our crude S150 extract. We determined iSAT ribosomes to have an elongation rate of 0.9 ± 0.1 amino acids per second (AA/s), while E. coli ribosomes in the 70S TX-TL reactions have an elongation rate of 1.6 ± 0.1 AA/s (Supplementary Table S3). A previous study of E. coli ribosomes in a crude S30 cell-free system determined an elongation rate of 1.5 ± 0.2 AA/s, though this included a correction for the observation that only 72% of ribosomes were actively translating (48). Such a correction is not possible with our current data, but the issue of non-translating 70S ribosomes may account for some of the difference between elongation rates for iSAT and E. coli ribosomes. However, even this current estimation highlights that the iSAT-assembled ribosomes have similar activity to those purified from E. coli cells.
To enable direct comparison of iSAT ribosome activity to activity of purified E. coli ribosomes, 70S iSAT ribosomes were collected from ribosome profile fractions by ultracentrifugation and re-suspended in iSAT buffer. We then carried out 70S TX-TL reactions with either 300 nM purified iSAT ribosomes or 300 nM E. coli 70S ribosomes. In TX-TL reactions, purified iSAT ribosomes show 71 ± 10% and 73 ± 9% of the activity of E. coli ribosomes at 1 and 18 h, respectively (Supplementary Figure S6). This comparison of iSAT and E. coli ribosome activity shows a dramatic improvement from our previous reporting of 20% activity at 6 h (19). Additionally, samples of pelleted iSAT and E. coli 70S ribosomes were submitted for proteomic analysis (Supplementary Methods and Supplementary Figure S7). From this analysis, we observed that all r-proteins were identified in the purified iSAT ribosomes, suggesting that they are all capable of assembling with in vitro transcribed rRNA. It is unclear if the lower-represented proteins, such as S1, L33, L36, are indicative of incomplete ribosome assembly or merely an artifact of the steps involved in purifying iSAT assembled ribosomes resulting in loss of some r-proteins. Overall, these data show high similarity between iSAT and E. coli ribosomes, both in activity and r-protein content, though further work is required to identify the remaining points of difference, which could include incomplete post-transcriptional modification of synthetic rRNA. Further, our results demonstrate that sedimentation analysis is a valuable tool to characterize iSAT reactions and that a multiple assay approach is a useful platform for making novel observations about ribosome synthesis and assembly.
DISCUSSION
We discovered that the inclusion of crowding and reducing agents in iSAT reactions dramatically increases protein synthesis activity. We then used several different assays to tease apart the functions of crowding and reducing agents on the individual co-activated processes of ribosome biogenesis, transcription, and translation in iSAT reactions. Crowding agents, which increase the effective molecular concentration in the otherwise dilute iSAT reaction, were found to increase mRNA transcription and reporter protein translation in both iSAT and 70S TX-TL reactions (Figure 4), but did not affect sedimentation profiles of translating iSAT ribosomes (Figure 5). Reducing agents, which mimic the cytoplasmic environment, were also found to increase mRNA transcription levels in both iSAT and 70S TX-TL reactions (Figure 4). However, reducing agents did not increase translation of 70S TX-TL reactions, suggesting that mRNA is synthesized in excess in these reactions. Additionally, reducing agents were found to increase the number of translating iSAT monosomes and polysomes (Figure 5), likely leading to the observed increase in translation activity for iSAT reactions that is not observed in 70S TX-TL reactions. Therefore, for the iSAT system, we attribute the primary effect of crowding agents on iSAT yields to improved protein translation, and the primary effect of reducing agents on iSAT yields to improved ribosome biogenesis, noting that the effects of crowding and reducing agents on other processes of the iSAT system may overlap. Given previous results in S30 extract-based CFPS showing high activity in oxidizing conditions, the impact of reducing agents was especially noteworthy, suggesting a potentially important role of reducing agents in ribosome synthesis and assembly that would not have been possible to reveal without the iSAT system.
We also observed that the effects of the individual crowding and reducing agents on iSAT protein synthesis activity are not fully additive when used in various combinations. For example, 6% Ficoll 400 yields a 2.2-fold improvement in relative mRFP1 production and 2 mM DTBA yields a 2.5-fold improvement in relative mRFP1 production, but combined they yield only a 4.3-fold improvement, noting that this observation is more pronounced for other combinations (Figure 2, Table 1). The non-additive effects are likely a consequence of the fact that crowding and reducing agents both enhance transcription (Figure 4; 6% Ficoll 400 and 2mM DTBA). Transcription is required for both ribosome synthesis and assembly (rRNA transcription) and protein synthesis (mRNA transcription) and this overlap may lead to interdependencies.
The technological improvements arising from the addition of crowding and reducing agents are an important step toward complete cell-free synthesis of ribosomes and the ability to construct and evolve ribosome variants. One important benchmark for synthetic ribosome activity is the ability to synthesize enough peptide bonds for translation of a complete set of ribosomal proteins (7434 peptide bonds) (18). With the addition of reducing and crowding agents to iSAT reactions, we observe synthesis of ∼1.0 to 2.4 mM peptide bonds based on 14C-leucine incorporation for translation of various reporter proteins (Supplementary Figure S1). Since our sedimentation analysis indicates that iSAT reactions contain approximately 110 nM ribosomes in 70S and polysome forms (Supplementary Figure S5, Table 2), our results suggest that iSAT ribosomes are capable of translating >8000 peptide bonds per ribosome. While our data do not allow us to calculate the exact number of peptide bonds synthesized per ribosome, as we cannot yet distinguish the possibilities of an active subpopulation of ribosomes versus a homogenous pool of partially active ribosomes, the use of crowding and reducing agents has brought iSAT protein synthesis activity past the break-even milestone for ribosomal protein synthesis. Even so, there are opportunities to understand a finer resolution of details about the iSAT system, such as (i) the differences in iSAT-assembled ribosomes and purified E. coli ribosomes that may impact activity and (ii) whether or not iSAT rRNA is post-transcriptionally modified. Both are key areas for future work.
The research reported here highlights the great potential of the iSAT system to better elucidate the systems biology of ribosome biogenesis and protein synthesis. For example, similar studies to those performed here could be carried out to understand the effects of temperature, pH, ribosomal mutations that are not viable in vivo or strain mutations that change the protein composition of S150 extract. This depth of understanding is not achievable in cellular studies or other crude lysate systems. With the iSAT system, we expect that ribosome biogenesis can be probed and dissected in novel and insightful ways that will provide new understanding of this complicated process. We also anticipate that advances reported here will contribute meaningfully toward efforts to build minimal cells and construct synthetic ribosomes with novel and useful properties.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors would like to acknowledge Dr Ioanna Ntai of the Proteomics Center of Excellence at Northwestern University for analysis of r-protein samples and Jennifer Kay for thoughtful review of the manuscript.
Author Contributions: B.R.F., O.K.J. and M.C.J. conceived the study and wrote the manuscript. O.K.J. prepared DNA constructs. B.R.F. and O.K.J. designed experiments and B.R.F. performed experiments. M.C.J. provided a supervisory role.
FUNDING
Office of Naval Research [N00014-11-1-0363]; Army Research Office [W911NF-11-1-0445]; NAKFI Keck Futures Award [NAKFI SB5]; David and Lucile Packard Foundation [2011-37152]. Funding for open access charge: Office of Naval Research [N00014-11-1-0363]; Army Research Office [W911NF-11-1-0445].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bremer H., Dennis P.P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd edn. Washington, DC.: American Society for Microbiology; 1996. pp. 1553–1569. [Google Scholar]
- 2.Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaczanowska M., Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shajani Z., Sykes M.T., Williamson J.R. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 5.Ogle J.M., Carter A.P., Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 6.Nierhaus K.H. Reconstitution of ribosomes. In: Spedding G, editor. Ribosomes and Protein Synthesis: A Practical Approach. Oxford: IRL Press; 1990. pp. 161–189. [Google Scholar]
- 7.Nierhaus K.H., Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1974;71:4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Traub P., Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. U.S.A. 1968;59:777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham P.R., Richard R.B., Weitzmann C.J., Nurse K., Ofengand J. The absence of modified nucleotides affects both in vitro assembly and in vitro function of the 30S ribosomal subunit of Escherichia coli. Biochimie. 1991;73:789–796. doi: 10.1016/0300-9084(91)90058-9. [DOI] [PubMed] [Google Scholar]
- 10.Green R., Noller H.F. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA. 1996;2:1011–1021. [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H., Abeysirigunawarden S.C., Chen K., Mayerle M., Ragunathan K., Luthey-Schulten Z., Ha T., Woodson S.A. Protein-guided RNA dynamics during early ribosome assembly. Nature. 2014;506:334–338. doi: 10.1038/nature13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agalarov S.C., Williamson J.R. A hierarchy of RNA subdomains in assembly of the central domain of the 30 S ribosomal subunit. RNA. 2000;6:402–408. doi: 10.1017/s1355838200991945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herold M., Nierhaus K.H. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J. Biol. Chem. 1987;262:8826–8833. [PubMed] [Google Scholar]
- 14.Mulder A.M., Yoshioka C., Beck A.H., Bunner A.E., Milligan R.A., Potter C.S., Carragher B., Williamson J.R. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talkington M.W., Siuzdak G., Williamson J.R. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green R., Noller H.F. Reconstitution of functional 50S ribosomes from in vitro transcripts of Bacillus stearothermophilus 23S rRNA. Biochemistry. 1999;38:1772–1779. doi: 10.1021/bi982246a. [DOI] [PubMed] [Google Scholar]
- 17.Semrad K., Green R. Osmolytes stimulate the reconstitution of functional 50S ribosomes from in vitro transcripts of Escherichia coli 23S rRNA. RNA. 2002;8:401–411. doi: 10.1017/s1355838202029722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jewett M.C., Fritz B.R., Timmerman L.E., Church G.M. In vitro integration of ribosomal RNA synthesis, ribosome assembly, and translation. Mol. Syst. Biol. 2013;9:678. doi: 10.1038/msb.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritz B.R., Jewett M.C. The impact of transcriptional tuning on in vitro integrated rRNA transcription and ribosome construction. Nucleic Acids Res. 2014;42:6774–6785. doi: 10.1093/nar/gku307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Fritz B.R., Anderson M.J., Schoborg J.A., Jewett M.C. Characterizing and alleviating substrate limitations for improved in vitro ribosome construction. ACS Synth. Biol. 2014 doi: 10.1021/sb5002467. doi:10.1021/sb5002467. [DOI] [PubMed] [Google Scholar]
- 21.Forster A.C., Church G.M. Towards synthesis of a minimal cell. Mol. Syst. Biol. 2006;2:45. doi: 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glass J.I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M.R., Maruf M., Hutchison C.A., 3rd, Smith H.O., Venter J.C. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U.S.A. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jewett M.C., Forster A.C. Update on designing and building minimal cells. Curr. Opin. Biotechnol. 2010;21:697–703. doi: 10.1016/j.copbio.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juhas M., Eberl L., Glass J.I. Essence of life: essential genes of minimal genomes. Trends Cell Biol. 2011;21:562–568. doi: 10.1016/j.tcb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Stano P., Luisi P.L. Semi-synthetic minimal cells: origin and recent developments. Curr. Opin. Biotechnol. 2013;24:633–638. doi: 10.1016/j.copbio.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Li J., Gu L., Aach J., Church G.M. Improved cell-free RNA and protein synthesis system. PLoS One. 2014;9:e106232. doi: 10.1371/journal.pone.0106232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann H., Wang K., Davis L., Garcia-Alai M., Chin J.W. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 28.Wang K., Neumann H., Peak-Chew S.Y., Chin J.W. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 29.Cayley S., Lewis B.A., Guttman H.J., Record M.T., Jr Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J. Mol. Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- 30.Ge X., Luo D., Xu J. Cell-free protein expression under macromolecular crowding conditions. PLoS One. 2011;6:e28707. doi: 10.1371/journal.pone.0028707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilburn D., Roh J.H., Guo L., Briber R.M., Woodson S.A. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J. Am. Chem. Soc. 2010;132:8690–8696. doi: 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillip Y., Sherman E., Haran G., Schreiber G. Common crowding agents have only a small effect on protein-protein interactions. Biophys. J. 2009;97:875–885. doi: 10.1016/j.bpj.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batra J., Xu K., Qin S., Zhou H.X. Effect of macromolecular crowding on protein binding stability: modest stabilization and significant biological consequences. Biophys. J. 2009;97:906–911. doi: 10.1016/j.bpj.2009.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou H.X. Influence of crowded cellular environments on protein folding, binding, and oligomerization: biological consequences and potentials of atomistic modeling. FEBS Lett. 2013;587:1053–1061. doi: 10.1016/j.febslet.2013.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang C., Sinskey A.J., Lodish H.F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 36.Derman A.I., Prinz W.A., Belin D., Beckwith J. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 37.Burns J.A., Butler J.C., Moran J., Whitesides G.M. Selective reduction of disulfides by tris (2-carboxyethyl) phosphine. J. Org. Chem. 1991;56:2648–2650. [Google Scholar]
- 38.Getz E.B., Xiao M., Chakrabarty T., Cooke R., Selvin P.R. A comparison between the sulfhydryl reductants tris(2-carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. Anal. Biochem. 1999;273:73–80. doi: 10.1006/abio.1999.4203. [DOI] [PubMed] [Google Scholar]
- 39.Lukesh J.C., 3rd, Palte M.J., Raines R.T. A potent, versatile disulfide-reducing agent from aspartic acid. J. Am. Chem. Soc. 2012;134:4057–4059. doi: 10.1021/ja211931f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chizzolini F., Forlin M., Cecchi D., Mansy S.S. Gene position more strongly influences cell-free protein expression from operons than T7 transcriptional promoter strength. ACS Synth. Biol. 2014;3:363–371. doi: 10.1021/sb4000977. [DOI] [PubMed] [Google Scholar]
- 41.Bundy B.C., Swartz J.R. Site-specific incorporation of p-propargyloxyphenylalanine in a cell-free environment for direct protein-protein click conjugation. Bioconjug. Chem. 2010;21:255–263. doi: 10.1021/bc9002844. [DOI] [PubMed] [Google Scholar]
- 42.Swartz J.R., Jewett M.C., Woodrow K.A. Cell-free protein synthesis with prokaryotic combined transcription-translation. Methods Mol. Biol. 2004;267:169–182. doi: 10.1385/1-59259-774-2:169. [DOI] [PubMed] [Google Scholar]
- 43.Bakke C.K., Jungbauer L.M., Cavagnero S. In vitro expression and characterization of native apomyoglobin under low molecular crowding conditions. Protein Expr. Purif. 2006;45:381–392. doi: 10.1016/j.pep.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Nakano H., Tanaka T., Kawarasaki Y., Yamane T. Highly productive cell-free protein synthesis system using condensed wheat-germ extract. J. Biotechnol. 1996;46:275–282. [Google Scholar]
- 45.Nakano H., Tanaka T., Kawarasaki Y., Yamane T. An increased rate of cell-free protein synthesis by condensing wheat-germ extract with ultrafiltration membranes. Biosci. Biotechnol. Biochem. 1994;58:631–634. doi: 10.1271/bbb.58.631. [DOI] [PubMed] [Google Scholar]
- 46.Sun Z.Z., Hayes C.A., Shin J., Caschera F., Murray R.M., Noireaux V. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J. Vis. Exp. 2013:e50762. doi: 10.3791/50762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jewett M.C., Swartz J.R. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 2004;86:19–26. doi: 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
- 48.Underwood K.A., Swartz J.R., Puglisi J.D. Quantitative polysome analysis identifies limitations in bacterial cell-free protein synthesis. Biotechnol. Bioeng. 2005;91:425–435. doi: 10.1002/bit.20529. [DOI] [PubMed] [Google Scholar]
- 49.Kim D.M., Kigawa T., Choi C.Y., Yokoyama S. A highly efficient cell-free protein synthesis system from Escherichia coli. Eur. J. Biochem. 1996;239:881–886. doi: 10.1111/j.1432-1033.1996.0881u.x. [DOI] [PubMed] [Google Scholar]
- 50.Karzbrun E., Shin J., Bar-Ziv R.H., Noireaux V. Coarse-grained dynamics of protein synthesis in a cell-free system. Phys. Rev. Lett. 2011;106:048104. doi: 10.1103/PhysRevLett.106.048104. [DOI] [PubMed] [Google Scholar]
- 51.Stogbauer T., Windhager L., Zimmer R., Radler J.O. Experiment and mathematical modeling of gene expression dynamics in a cell-free system. Integr. Biol. 2012;4:494–501. doi: 10.1039/c2ib00102k. [DOI] [PubMed] [Google Scholar]
- 52.Niederholtmeyer H., Xu L., Maerkl S.J. Real-time mRNA measurement during an in vitro transcription and translation reaction using binary probes. ACS Synth. Biol. 2013;2:411–417. doi: 10.1021/sb300104f. [DOI] [PubMed] [Google Scholar]
- 53.Sei-Iida Y., Koshimoto H., Kondo S., Tsuji A. Real-time monitoring of in vitro transcriptional RNA synthesis using fluorescence resonance energy transfer. Nucleic Acids Res. 2000;28:e59. doi: 10.1093/nar/28.12.e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siegal-Gaskins D., Tuza Z.A., Kim J., Noireaux V., Murray R.M. Gene circuit performance characterization and resource usage in a cell-free ‘breadboard’. ACS Synth. Biol. 2014;3:416–425. doi: 10.1021/sb400203p. [DOI] [PubMed] [Google Scholar]
- 55.Paige J.S., Wu K.Y., Jaffrey S.R. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim D.M., Swartz J.R. Efficient production of a bioactive, multiple disulfide-bonded protein using modified extracts of Escherichia coli. Biotechnol. Bioeng. 2004;85:122–129. doi: 10.1002/bit.10865. [DOI] [PubMed] [Google Scholar]
- 57.Yin G., Swartz J.R. Enhancing multiple disulfide bonded protein folding in a cell-free system. Biotechnol. Bioeng. 2004;86:188–195. doi: 10.1002/bit.10827. [DOI] [PubMed] [Google Scholar]
- 58.Siibak T., Remme J. Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA. 2010;16:2023–2032. doi: 10.1261/rna.2160010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


