Abstract
Objective
Nucleated red blood cells (NRBCs) have been identified in maternal circulation and potentially provide a resource for the monitoring and diagnosis of maternal, fetal, and neonatal health and disease. Past strategies used to isolate and enrich for NRBCs are limited to complex approaches that result in low recovery and less than optimal cell purity. Here we report the development of a high-throughput and highly efficient microfluidic device for isolating rare NRBCs from maternal blood.
Material and Methods
NRBCs were isolated from the peripheral blood of 58 pregnant women using a microfluidic process that consists of a microfluidic chip for size-based cell separation and a magnetic device for hemoglobin-based cell isolation.
Results
The microfluidic–magnetic combination removes nontarget red blood cells and white blood cells at a very high efficiency (∼99.99%). The device successfully identified NRBCs from the peripheral blood of 58/58 pretermination samples with a mean of 37.44 NRBC/mL (range 0.37–274.36 NRBC/mL). These results were compared with those from previous studies.
Conclusion
The microfluidic device results in an approximate 10- to 20-fold enrichment of NRBCs over methods described previously. The reliability of isolation and the purity of the NRBC product have the potential to enable the subsequent application of molecular diagnostic assays.
Keywords: microfluidics, nucleated red blood cells, noninvasive prenatal diagnosis
Introduction
It is well established that fetal cells are present in the peripheral blood of pregnant women, and for more than three decades, researchers have focused on their use for the development of noninvasive prenatal diagnosis (Purwosunu et al., 2006a). Since the early 1990s, significant attention has been focused on fetal nucleated red blood cells (fNRBCs) as a source for diagnostic purposes since they reflect the current pregnancy, do not persist for long periods of time in maternal circulation, have limited replicating capacity, have biochemical characteristics amenable to cell enrichment, and are detectable early in pregnancy (Bianchi et al., 1990). However, the selection of fNRBCs as a candidate fetal cell type has been deterred, in part, because of estimates that ∼50% of nucleated red blood cells (NRBCs) are maternal in origin (Troeger et al., 1999a). Therefore, optimization of NRBC enrichment techniques is critical for fetal cell isolation and analysis. It has also been suggested that the total number of NRBCs present in maternal circulation, whether of fetal or maternal origin, alone or in combination with biochemical and ultrasonographic markers, may be an indicator of fetal anomalies and/or pregnancy complications (Prieto et al., 2002; Falcidia et al., 2004; Mavrou et al., 2007). In both applications, the challenge is to remove the overwhelmingly large background of maternal non-NRBCs and white blood cells (WBCs) without loss or damage of the target cells of interest (NRBCs).
The most frequently used methods to enrich or sort NRBCs from the whole blood of pregnant women have included density gradient separation (Kuo and Guo, 1999; Sekizawa et al., 1999; Samura et al., 2000; Smits et al., 2000), fluorescence-activated cell sorting (FACS)-based positive selection (Iverson et al., 1981; Bianchi et al., 1990; Price et al., 1991; Lewis et al., 1996; Wang et al., 2000), magnetic-activated cell sorting (MACS)-based depletion or enrichment (Ganshirt-Ahlert et al., 1992, 1993; Busch et al., 1994; Campagnoli et al., 1997; Valerio et al., 1997; Troeger et al., 1999b; Mavrou et al., 2007), micromanipulation (Takabayashi et al., 1995; Sekizawa et al., 2007), selective lysis (de Graaf et al., 1999), and galactose-lectin-based methods (Babochkina et al., 2005; Purwosunu et al., 2006b; Sekizawa et al., 2007). Both FACS and MACS rely on sequential steps of preliminary separation of background cells, generally by density gradient centrifugation or cell lysis, followed by antigen–antibody recognition using NRBCspecific monoclonal antibodies. However, those techniques are limited by the specificity of the available cell surface markers as they are also expressed on significant (>0.5%) subpopulations of maternal blood cells. In addition, while they remove nontarget background cells, they result in low cell recovery efficiency (FACS) or less than optimal cell purity (MACS) (Bianchi et al., 2002). More recent studies have used galactose-specific lectin for the isolation of NRBCs. The method is based primarily on the observation that erythroid precursors express large numbers of galactose molecules on their cell surface. While that method has resulted in a greater NRBC recovery when compared to other technologies, it is time-consuming, labor-intensive, and results in the coisolation of a large number of non-nucleated erythrocytes, thus decreasing purity.
Herein, we describe the development and application of a highly sensitive microfluidic device that efficiently and reproducibly results in the high recovery and purity of NRBCs from maternal blood. The technology capitalizes on the intrinsic characteristics of NRBCs using a two-step enrichment process (Figure 1a). The cell separation microchip (CSM) was designed to separate blood cells based on cell size and their ability to deform. The hemoglobin enrichment (HE) module was developed to separate cell fractions based on the premise that NRBCs are the only cells in whole blood that contain both a nucleus and hemoglobin. Accordingly, the CSM separates blood into a nucleated fraction (nucleus+) and a non-nucleated fraction (nucleus−), and serves as a debulking process to remove the vast number of red blood cells (RBCs) from whole blood samples (Figure 1a and b). After the collection of nucleus+ cells, the CSM product is separated by the second stage HE module into a hemoglobin-containing fraction (nucleus+/Hb+) and a non–hemoglobin-containing fraction (nucleus+/Hb−) (Figure 1a and b). The HE step takes advantage of paramagnetic properties of NRBCs whereby hemoglobin is converted to methemoglobin following treatment with 50 mm sodium nitrite (NaNO2), thus rendering all Hb+ cells paramagnetic. WBCs and NRBCs are then passed through a magnetic column (Miltenyi BioTech, Germany) under a magnetic field of 1.4 T, where Hb+ cells (NRBCs) are retained. The separation does not rely on any antibody, and is not subject to the purity and yield limitations of affinity enrichment. However, it is necessary to eliminate a vast majority of non-NRBCs prior to HE separation to prevent the magnetic matrices used to generate the required field gradient to be saturated and lose their ability to retain NRBCs. The power of the two-step system is that both dimensions of separation can be optimized independently to result in an ideal separation mechanism.
Figure 1.
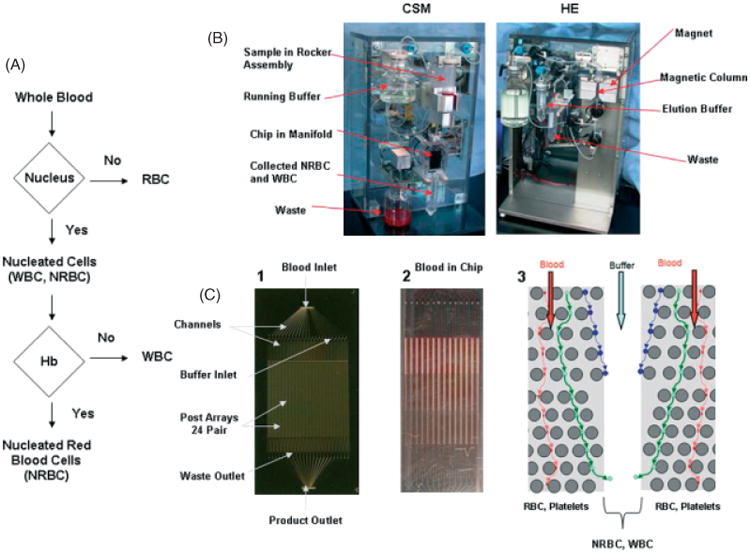
Isolation of NRBCs from whole blood using a microfluidic device. (a) Schematic of flow logic of the microfluidic system. (b) The CSM (left) separates cells based on size through the microfluidic chip into nucleus+ and nucleus− fractions. The blood sample is continually mixed on a rocker, and pumped through the chip using a pneumatic-pressure regulated pump. The HE module (right) collects hemoglobin-containing cells (NRBCs) on a magnetic column that are eluted off the column when the run is complete. (c) (1) A photograph of the microfluidic device etched in silicon, (2) an example of whole blood running through the device, and (3) a schematic of one of 24 sets of buffer inlets and paired array sets on the microfluidic device showing the flow stream of NRBCs and WBCs, deflected by microposts (circles), into the buffer stream. RBCs are small and run into the waste outlets at the bottom of each chip. Colored lines with arrows represent the flow direction of RBCs (red lines), NRBCs (blue lines), and WBCs (green lines)
To effectively remove RBCs, the microfluidic chip was designed on the basis of ‘deterministic lateral displacement’ (DLD), first published by Huang (Huang et al., 2004), and exploits the hydrodynamic size differences between nucleated and non-nucleated cells. Maternal RBCs and platelets are ∼2 μm (thickness), whereas NRBCs and WBCs are larger than 5 to 10 μm. Whole blood is passed through an array of microposts under laminar flow at a small angle relative to the array (Figure 1c). Cells that are small with respect to the flow stream (RBCs and platelets) drift along the flow and stay in the stream. In contrast, cells that are large compared to the flow stream (NRBCs and WBCs) are displaced out of the stream when they encounter a micropost. Hence, the array acts like a size filter, in which the pore size of the array is not the spacing between posts, but is dictated by the width of the flow stream, which is much smaller than the micropost spacing. The array enables large gaps to separate small cells at a reasonable flow rate, overcomes shear stress on cells, and avoids chip clogging. Optimal capacity and minimal shear stress were achieved by designing high-throughput arrays and multiplexing. Microposts were engineered to manipulate the flow and to minimize shear stress in the device. The wide gap size and optimized micropost design allowed the device to be fabricated deeper (∼150 μm), further increasing throughput. Each microfluidic chip device comprises a single blood inlet and 24 buffer inlets that feed blood and buffer into 24 identical pairs of arrays, where nucleated and non-nucleated cells are separated. The cells are collected in phosphate buffer solution, and harvested via a single outlet (Figure 1c). Collectively, 48 arrays are multiplexed on a single 64 × 32 × 1 mm chip, allowing blood to sort at ∼107 cell/s, two orders of magnitude faster than the current state-of-the-art flow cytometers.
Methods
Blood samples
After informed consent was obtained under protocols approved by the Western Columbia University, Tufts-New England Medical Center, or Women & Infants Hospital IRB, ∼20 to 30 mL of venous peripheral blood was collected in K2EDTA vacutainers from 58 pregnant women. Median maternal age was 35.5 years (range 18–44 years), and gestational age ranged from 9 to 22 weeks (median 18 weeks). Blood processing began within 6 h of draw, and all samples were drawn prior to an invasive procedure. In 21/58 cases, the karyotype was determined, and in cases where the karyotype was not known, fetal gender identification was performed on plasma and/or fetal tissue by polymerase chain reaction (PCR) analysis using a probe for the Y chromosome only. In addition, 30 mL peripheral blood was obtained from each of five nonpregnant donors for the optimization of ideal chip throughput.
Blood preparation
Once received in the laboratory, blood samples were inverted three times to mix, and filtered into a 50-mL tube using a 30 μm Pre-Sep filter (Miltenyi) to remove any visible clots. An equal volume of incomplete calcium- and magnesium-free Dulbecco's phosphate buffer solution (iDPBS) was added to the blood, and mixed. A complete blood count was performed on each sample.
Microfluidic chip
Microfluidic chips were fabricated in silicon using standard microfabrication processes. Fluidic structures were defined on silicon wafers using photolithography, and etched to ∼150-μm deep using deep reactive ion etch (DRIE). Five nanometers of thermal oxide was grown on the etched wafer to make the channels hydrophilic, and the wafer was subsequently anodic-bonded to a glass wafer to form enclosed fluidic channels. Holes for fluid access were fabricated before bonding using KOH wet etch. The wafer was then diced into separate devices. Microfluidic chips were designed and fabricated with 18-, 20-, and 24-μm gaps.
Cell separation microchip (CSM)
A microfluidic chip assembly was mounted on the CSM machine with all tubing attached. A solution of running buffer, consisting of 1% bovine serum albumin (BSA) and 2-mm EDTA in iDPBS, was degassed using a vacuum degassing system (Fisher) stirring at 800 rpm for 2 min, and then mounted on the CSM. A 250-mL Nalgene bottle was placed on the CSM to collect CSM waste, and a 50-mL tube was placed under the chip assembly to collect the cell product. The CSM was then switched to the ‘on’ position and the pressure was set to 1.0 psi for 1 min, and then at 3.0 psi for 4 min to purge air bubbles from the system. The pressure was returned to 1.0 psi to completely process the sample. The 50-mL tube containing the blood sample and iDPBS was poured into a syringe attached to the CSM. A system rocker was used to ensure continuous mixing of the blood/buffer solution, and to prevent sample settling. The run was considered complete when all of the blood had passed through the chip.
Hemoglobin enrichment (HE) module
After samples were passed through the CSM, they were centrifuged at 300 g for 10 min, aspirated to 5 mL, and incubated with 50 mm NaNO2 for 10 min at room temperature to render hemoglobin paramagnetic. A Miltenyi LS magnetic column was attached to the tubing and then to the HE module magnet. Hanks' balanced salt solution (HBSS) was placed in a 50-mL tube that was mounted on the module along with the running buffer. Once all the tubing was attached, the HE system was primed with running buffer at 0.35 psi. The pressure was then brought to 1.4 psi and the sample was passed through the column magnetized at 1.4 T, and rinsed in 3 mL of iDPBS and 1% BSA. After rinsing, the sample was eluted at a higher flow rate with HBSS. To reduce nonspecific binding of WBC to the column, the rinse and elution process was repeated three times.
Slide preparation
Collected cells from the HE module were diluted in HBSS. Slides were prepared using ThermoShandon Megafunnel slides. Cells were placed in Shandon EZ Megafunnel Disposable Sample Chambers and spun onto slides using a Shandon Cytospin 4 Cyto Centrifuge at 1900 rpm for 5 min at medium acceleration. The cell concentration was 1 × 106 total cells/slide. After centrifugation, slides were incubated in 1% BSA for 10 min at room temperature. Slides were then air dried for 1 h in a vertical position, followed by a final methanol fixation.
Wright/Giemsa stain
Slides were prefixed in methanol for 10 min at room temperature, allowed to air dry, and then exposed to Wright/Giemsa stain using standard methods in an automated Sakura Hematology Slide Stainer.
Bioview duet bright field scanning
Stained slides were placed on a BioView Duet automated scanner and were completely scanned using an algorithm established to identify cells that contain a nucleus.
NRBC enumeration
Once slide scans were complete, a gallery of target cells was manually prescreened on a BioView Solo Review Station. Target NRBCs that showed a low nucleus to cytoplasm ratio, a small dense nucleus, and orthochromatic nongranular cytoplasm were confirmed manually on the BioView Duet microscope. Cells were classified as typical NRBCs, atypical NRBCs with nonstandard morphology, and not NRBCs. At least two scorers were assigned to each case, and NRBC counts were compared for potential variance. The final number of NRBC/mL of sample was calculated by dividing the total number of NRBCs identified across all slides by the amount of whole blood processed.
Benzidine stain
Wright/Giemsa-stained slides were destained in methanol prior to exposure to 0.25% benzidine/methanol for 3 min. Slides were then developed in 1.25 mL 30% hydrogen peroxide in 50 mL 50% ethanol for 1.5 min, followed by two rinses in dH2O. Subsequently, slides were placed in 50 mL Wright/1.5 mL Giemsa stain for 10 min, followed by three rinses in dH2O. All steps were carried out at room temperature. Stained slides were reexamined on a BioView Duet automated microscope. Each NRBC target identified from the initial Wright/Giemsa scan was relocated and examined as positive or negative for benzidine stain. All cells were photographed. Benzidine was purchased from Sigma-Aldrich (#3503-1G).
Results
To optimize separation between nucleus+ and nucleus− fractions, three microchip designs were initially fabricated with 18-, 20-, or 24-μm gaps. Peripheral blood from five nonpregnant donors was split equally into three aliquots of ∼10 mL each, and run through each of the three chip designs in parallel at ∼1.0 psi. The nucleus+ and nucleus− fractions were characterized using a hematology analyzer (Coulter AcT diff, Beckman Coulter). All three designs resulted in excellent separation, eliminating RBCs to within 0.056, 0.010, and 0.004%, using 18-, 20-, and 24-μm chips, respectively. The median, and 20th and 80th percentiles for each device are shown in Figure 2a. WBC cell retention was 99.9, 99.9, and 99.7%, using 18-, 20-, and 24-μm devices, respectively (Figure 2a). The final microfluidic chip was designed with wide gaps of ∼20 μm to maximize NRBC recovery while maintaining a reasonable flow speed, and to reduce shear stress of the cells. Retention of Hb+ cells was optimized by performing a flow titration using donor blood samples. The results showed that at a flow rate of 27 mL/h (flow rate−1 135 s/mL), greater than 95% of the Hb+ cells were retained. As the flow rate was reduced to 13 mL/h (270 s/mL), essentially no Hb+ cells were detected in the Hb− fraction (Figure 2b). The purity of the Hb+ fraction was optimized by applying one, two, and three cycles of magnetic cell separation. The median, 20th, and 80th percentiles are shown in Figure 2c. Overall, the results show ∼99.99% depletion of the maternal Hb− cells after three cycles of separation. Under optimal conditions, enrichment performance of the microfluidic device and CSM/HE modules resulted in the recovery of ∼1 to 275 NRBCs postprocess. Thus, ∼99.99% of RBCs are removed with ∼5 000 000 remaining in the final product; WBCs are removed with an efficiency of ∼99.90 to 99.99%, and ∼8000–8 000 000 WBCs remained in the final cell product from 1 mL whole blood (Table 1). Postprocess examination of the enriched cell fraction by Wright/Giemsa-staining showed isolation of several cell types including WBC, RBC, and NRBC (Figure 3). Only cells that showed a low nucleus to cytoplasm ratio, a small dense nucleus, and orthochromatic nongranular cytoplasm were considered NRBCs. To confirm the identification of NRBCs postenumeration, slides from a portion of patients were sequentially stained with benzidine. Benzidine is a highly selective and efficient method used to demonstrate the presence of hemoglobin in erythroid cells, and differentially stains the cytoplasm of hemoglobin-containing cells golden brown while non–hemoglobin-containing cells appear blue. In all samples tested, cells identified as NRBCs using Wright/Giemsa stain were positive by benzidine staining (Figure 4).
Figure 2.
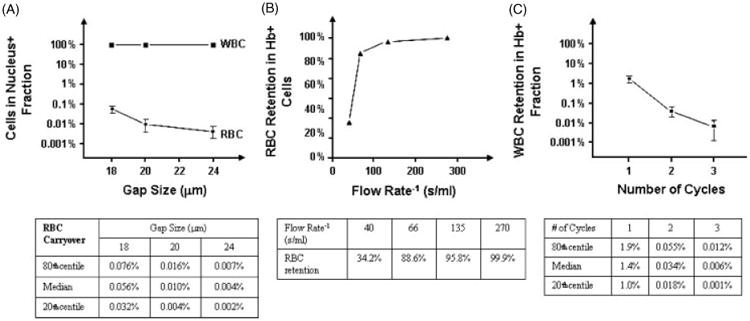
Engineering optimization of system performance. (a) Cell separation performance of the microfluidic chip shows WBC and RBC retention for three devices of differing gap size. (b) Flow rate dependence of RBC retention during magnetic hemoglobin separation. (c) Retention of WBCs in the NRBC product after 1, 2, and 3 cycles of magnetic hemoglobin separation
Table 1. Enrichment performance of microfluidic device.
| NRBC | RBC | WBC | Platelet | |
|---|---|---|---|---|
| Whole blood | 100% | 100% | 100% | 100% |
| After enrichment | Unknown | 0.01% | 0.01–0.10% | 0% |
| Number of cells/mL remaining | 1–275 | 5 000 000 | 8 000–80 000 | 0 |
NRBC, nucleated red blood cell; RBC, red blood cell; WBC, white blood cell.
Figure 3. Cell types identified after microfluidic cell separation using Wright/Giemsa stain.
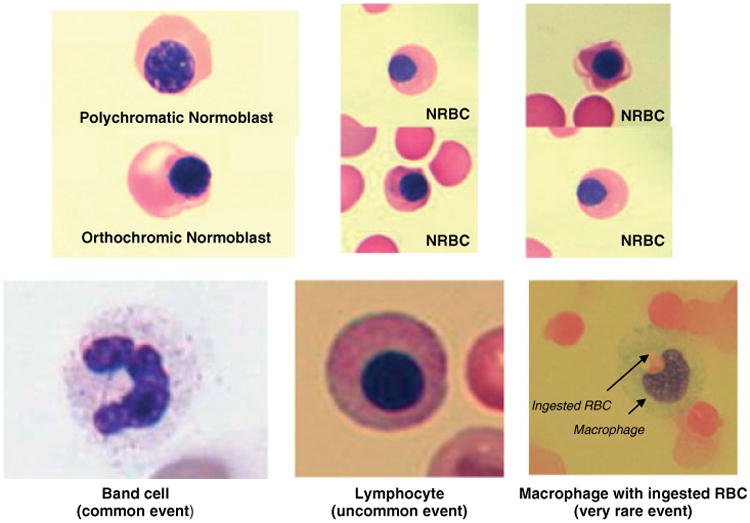
Figure 4.
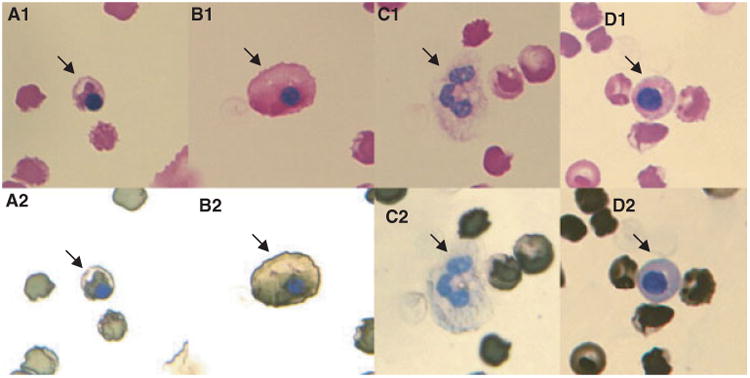
Confirmation of NRBCs by differential staining with Wright/Giemsa followed by benzidine stain. All cells labeled ‘1’ are Wright/Giemsa, and photos labeled ‘2’ are Wright/Giemsa/Benzidine. Cells with a golden brown cytoplasm are hemoglobin-containing cells (NRBC and RBC), and nonhemoglobin-containing cells (WBC and platelets) have a blue cytoplasm. Cells ‘A’ and ‘B’ are representative NRBCs, cell ‘C’ is a polymorphonuclear neutrophil, and cell ‘D’ is an apoptotic WBC
Having optimized the microfluidic device with control samples, we tested its capacity to isolate NRBCs from peripheral blood from pregnant women. Examination of the enriched cell product after Wright/Giemsa staining showed that NRBCs were isolated from 58/58 blood samples tested (Table 2). Of the 58 samples, 37 were from singleton pregnancies with limited outcome information. No invasive procedure was performed, and it was not known whether the fetus was chromosomally abnormal, or if there were other pregnancy complications that may have affected the NRBC count including preeclampsia, intrauterine growth restriction (IUGR), gestational diabetes, or anemia. The remaining 21 pre-procedural samples were from women who were subsequently identified as carrying a chromosomally abnormal fetus. Among the 37 pregnancies with unknown follow-up, a mean of 37.68 NRBC/mL, median of 21.08, and a range of 0.37 to 168.2 was identified. These results were compared with those of previously reported studies using alternative enrichment processes (Table 2) (Slunga-Tallberg et al., 1995; Takabayashi et al., 1995; Babochkina et al., 2005; Purwosunu et al., 2006b; Kwon et al., 2007; Mavrou et al., 2007; Sekizawa et al., 2007). Mean NRBC/mL in those studies ranged from 0.40 to 5.40 vs 37.68 in the current study. Of the 21 cases known to carry an aneuploid fetus, the mean number of NRBC/mL was 37.20, median was 18.13, and the range was 0.99 to 274.36. Among 13 cases of trisomy 21, the mean number was 52.81 NRBC/mL (range 5.65–274.36) and a median of 31.82. One case with trisomy 13 showed 4.94 NRBC/mL, whereas five cases of trisomy 18 showed a mean number of 14.97 NRBC/mL, a range of 0.98 to 31.16, and a median of 12.29. Two cases of 69,XXY/XXX showed a mean number of 7.41 NRBC/mL, median of 7.41, and a range of 6.67 to 8.14 (Table 3). Of the abnormal cases, fetuses with trisomy 21 showed the highest mean number of NRBCs. This finding is consistent with previous studies (Bianchi et al., 1997; Krabchi et al., 2006; Mavrou et al., 2007) that suggest more NRBCs are detected in maternal blood when the fetus is aneuploid, particularly trisomy 21. The maternal blood of pregnancies with trisomy 18 fetuses also had a higher number of NRBC/mL compared to trisomy 13 and triploid fetuses. However, the sample numbers were low and require further investigation.
Table 2. Technology comparison of NRBC enrichment efficiencies.
| Study | Pregnancy status | Number of cases | Gestational age (weeks) | Mean cells/mL | Median cells/mL | Range cells/mL | Method |
|---|---|---|---|---|---|---|---|
| Artemis Health | Singleton | 37 | 9–22 | 37.68 | 21.08 | 0.37–168.27 | CSM/HE |
| Abnormal | 21 | 11–21 | 37.20 | 18.13 | 0.99–274.36 | CSM/HE | |
| Slunga-Tallberg et al., 1995 | Singleton | 37/40 | 6–17 | 0.63 | 0.20 | 0.05–11.50 | miniMACS |
| Takabayashi et al., 1995 | Not Given | 33/60 | 4–40 | 2.05 | — | 0.50–11.00 | Density gradient |
| Babochkina et al., 2005 | Not Given | 11 | 11–40 | — | 16.00 | 2.00–59.00 | SBA-Lectin |
| 11 | 11-40 | — | 2.00 | 0.00–6.00 | MACS | ||
| Purwosunu et al., 2006b | Singleton | 22 | 9–34 | — | 2.08 | 0.16–13.66 | Galactose–Lectin |
| Post-termination | 23 | 6–19 | — | — | — | Galactose–Lectin | |
| Mavrou et al., 2007 | Normal (preamnio) | 282 | 14–28 | 0.40 | — | 0.05–0.60 | MACS |
| Abnormal (postamnio) | 23 | 14–28 | 1.75 | — | 0.35–5.65 | MACS | |
| Carriers B-thal | 26 | 14–28 | 2.10 | — | 1.10–7.90 | MACS | |
| Abnormal Doppler | 20 | 14–28 | 0.75 | — | 0.10–3.75 | MACS | |
| Sekizawa et al., 2007 | Singleton | 20 | 10–18 | 0.91 | 0.97 | 0.17–1.69 | Galactose–Lectin |
| Kwon et al., 2007 | Normal | 24 | 11–24 | 1.95 | 1.55 | 0.20–4.80 | Double density gradient/MACS |
| Abnormal | 1 | 13 | 5.40 | — | — |
CSM, cell separation microchip; HE, hemoglobin enrichment; MAC, magnetic-activated cell sorting.
Table 3. Number of NRBCs in 21 fetuses with aneuploidy.
| Abnormality | Number of cases | Gestational age (weeks) | Mean NRBC/mL | Median NRBC/mL | Range NRBC/mL |
|---|---|---|---|---|---|
| +13 | 1 | 16 | 4.94 | N/A | N/A |
| +18 | 5 | 11–21 | 14.97 | 12.29 | 0.98–31.16 |
| +21 | 13 | 11–21 | 52.81 | 31.82 | 5.65–274.36 |
| XXY/XXX | 2 | 18–19 | 7.41 | 7.41 | 6.67–8.14 |
NRBC, nucleated red blood cell.
Discussion
We have shown the development and application of a robust microfluidic device that can effectively, efficiently, and reproducibly isolate NRBCs from the whole blood of pregnant women. From a technical perspective, our approach of combining a microfluidic device and hemoglobin enrichment is unique in that it takes advantage of the intrinsic characteristics of NRBCs, including cell size, presence of a nucleus, and hemoglobin content. The process is easy to perform compared to the most frequently used approaches to enrich or sort for NRBCs. The CSM/HE system has the ability to process 5 to 20 mL of maternal whole blood in 2 to 6 h, automated scanning is ∼25 min/slide, and manual enumeration is ∼30 min/slide. Only one other report of ‘biochip’ technology has been described for the identification of fetal cells from maternal blood (Mohamed et al., 2007). In that study, the device was created to separate fetal cells from maternal cells in cord blood using density gradient separation prior to microchip sorting based only on differences in cell size and deformation characteristics. The results showed a very low processing throughput (0.35 mL/h), and chip clogging. The current design of our microfluidic device is unique in that it sorts NRBCs directly from whole blood without other pre-enrichment processes or prelabeling, and is not subject to the purity and yield limitations of affinity enrichment methods. The microfluidic device effectively eliminates ∼99.99% of RBCs in the first CSM step. The stationary capture of NRBCs on a magnetic column during the second HE step allows depletion of nonspecifically bound WBCs at ∼99.90 to 99.99% efficiency.
Applying this approach to the isolation of NRBCs from 58 pregnant women, we detected a mean number of 37.68 NRBC/mL and 37.20 NRBC/mL in singleton and abnormal pregnancies, respectively. The range was 0.37 to 274.36 NRBC/mL for all cases examined. This is in contrast with means ranging from 0.58–5.4 NRBC/mL identified by MACS density gradient, and in two of three studies using the galactose-lectin method. In one study, as many as 2.00 to 59.00 NRBCs were identified using soybean agglutinin galactose-lectin (Babochkina et al., 2005). However, the current method revealed an approximate 10- to 20-fold increase in NRBC counts over any other previously reported method.
In summary, the current microfluidic technology is the most successful approach for the isolation and enrichment of NRBCs reported to date. It has been suggested that an increase in the total number of circulating NRBCs may be an indicator of fetal aneuploidies or pregnancy complications (Prieto et al., 2002; Falcidia et al., 2004; Mavrou et al., 2007), and may be used to assess pregnancies at risk. A larger clinical study with appropriate clinical outcome information is in progress, and may provide further information regarding correlations for the monitoring and diagnosis of maternal, fetal, and neonatal health. In addition, the ability to effectively, reliably, and efficiently isolate a large number of NRBCs with high purity has the potential to enable molecular analyses for prenatal screening or for noninvasive prenatal diagnosis. Finally, a redesign of the current two-stage system into a single device with automated cell counting has significantly reduced sample processing time, and may enable point-of-care testing.
Acknowledgments
We wish to thank Drs. Bruce Carvalho, Darren Gray, Maury Cosman, Herman Wyandt, Kirby Johnson, and Lawrence Zuckerberg for their scientific input, and Alex French, Edmund Golasky, John Walsh, Laura Romonosky, Genevieve O'Connell, Paul Vernucci, Kristina Larsen, Zina Jarrah, Helene Stroh, and Dong Kai Zhen for their superior technical assistance.
Footnotes
Disclosures: Diana W. Bianchi, MD, is a member of the Artemis Health Inc. Clinical Advisory and the Scientific Advisory Boards. She had no role in either review or editorial processing of this manuscript. Mehmet Toner, PhD, Martin A. Schmidt, PhD, and Ronald G. Tompkins, MD, are members of the Artemis Health Inc. Scientific Advisory Board. Diana W. Bianchi, Thomas A. Barber, Martin A. Schmidt, and Ravi Kapur have an equity interest in Artemis Health Inc. Ronald G. Tompkins and Mehmet Toner divested their financial interests in Artemis Health Inc. per Harvard guidelines.
References
- Babochkina T, Mergenthaler S, Lapaire O, et al. Evaluation of a soybean lectin-based method for the enrichment of erythroblasts. J Histochem Cytochem. 2005;53:329–330. doi: 10.1369/jhc.4B6408.2005. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Flint AF, Pizzimenti MF, Knoll JH, Latt SA. Isolation of fetal DNA from nucleated erythrocytes in maternal blood. Proc Natl Acad Sci U S A. 1990;87:3279–3283. doi: 10.1073/pnas.87.9.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi DW, Simpson JL, Jackson LG, et al. Fetal gender and aneuploidy detection using fetal cells in maternal blood: analysis of NIFTY I data. Prenat Diagn. 2002;22:609–615. doi: 10.1002/pd.347. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Williams JM, Sullivan LM, Hanson FW, Klinger KW, Shuber AP. PCR quantitation of fetal cells in maternal blood in normal and aneuploid pregnancies. Am J Hum Genet. 1997;61:822–829. doi: 10.1086/514885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch J, Huber P, Pfluger E, Miltenyi S, Holtz J, Radbruch A. Enrichment of fetal cells from maternal blood by high gradient magnetic sorting (double MACS) for PCR-based genetic analysis. Prenat Diagn. 1994;14:1129–1140. doi: 10.1002/pd.1970141206. [DOI] [PubMed] [Google Scholar]
- Campagnoli C, Multhaupt HA, Ludomirski A, Haut MJ, Warhol MJ. Noninvasive prenatal diagnosis: use of density gradient centrifugation, magnetically activated cell sorting and in situ hybridization. J Reprod Med. 1997;42:193–199. [PubMed] [Google Scholar]
- de Graaf IM, Jakobs ME, Leschot NJ, Ravkin I, Goldbard S, Hoovers JM. Enrichment, identification and analysis of fetal cells from maternal blood: evaluation of a prenatal diagnosis system. Prenat Diagn. 1999;19:648–652. doi: 10.1002/(sici)1097-0223(199907)19:7<648::aid-pd600>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Falcidia E, Parano E, Grillo A, et al. Fetal cells in maternal blood: a six-fold increase in women who have undergone amniocentesis and carry a fetus with Down syndrome: a multicenter study. Neuropediatrics. 2004;35:321–324. doi: 10.1055/s-2004-830365. [DOI] [PubMed] [Google Scholar]
- Ganshirt-Ahlert D, Borjesson-Stoll R, Burschyk M, et al. Detection of fetal trisomies 21 and 18 from maternal blood using triple gradient and magnetic cell sorting. Am J Reprod Immunol. 1993;30:194–201. doi: 10.1111/j.1600-0897.1993.tb00620.x. [DOI] [PubMed] [Google Scholar]
- Ganshirt-Ahlert D, Burschyk M, Garritsen HS, et al. Magnetic cell sorting and the transferrin receptor as potential means of prenatal diagnosis from maternal blood. Am J Obstet Gynecol. 1992;166:1350–1355. doi: 10.1016/0002-9378(92)91603-8. [DOI] [PubMed] [Google Scholar]
- Huang LR, Cox EC, Austin RH, Sturm JC. Continuous particle separation through deterministic lateral displacement. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- Iverson GM, Bianchi DW, Cann HM, Herzenberg LA. Detection and isolation of fetal cells from maternal blood using the fluorescence-activated cell sorter (FACS) Prenat Diagn. 1981;1:61–73. doi: 10.1002/pd.1970010111. [DOI] [PubMed] [Google Scholar]
- Krabchi K, Gadji M, Samassekou O, Gregoire MC, Forest JC, Drouin R. Quantification of fetal nucleated cells in maternal blood of pregnant women with a male trisomy 21 fetus using molecular cytogenetic techniques. Prenat Diagn. 2006;26:28–34. doi: 10.1002/pd.1325. [DOI] [PubMed] [Google Scholar]
- Kuo PL, Guo HR. Nucleated red blood cells in maternal blood during pregnancy. J Obstet Gynaecol. 1999;94:464–468. doi: 10.1016/s0029-7844(99)00281-1. [DOI] [PubMed] [Google Scholar]
- Kwon KH, Jeon YJ, Hwang HS, et al. A high yield of fetal nucleated red blood cells isolated using optimal osmolarity and a double-density gradient system. Prenat Diagn. 2007;27:1245–1250. doi: 10.1002/pd.1888. [DOI] [PubMed] [Google Scholar]
- Lewis DE, Schober W, Murrell S, et al. Rare event selection of fetal nucleated erythrocytes in maternal blood by flow cytometry. Cytometry. 1996;23:218–227. doi: 10.1002/(SICI)1097-0320(19960301)23:3<218::AID-CYTO5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Mavrou A, Kouvidi E, Antsaklis A, Souke A, Kitsiou Tzeli S, Kolialexi A. Identification of nucleated red blood cells in maternal circulation: A second step in screening for fetal aneuploidies and pregnancy complications. Prenat Diagn. 2007;27:150–153. doi: 10.1002/pd.1640. [DOI] [PubMed] [Google Scholar]
- Mohamed H, Turner JN, Caggana M. Biochip for separating fetal cells from maternal circulation. J Chromatogr A. 2007;1162:187–192. doi: 10.1016/j.chroma.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Price JO, Elias S, Wachtel S, et al. Prenatal diagnosis with fetal cells isolated from maternal blood by multiparameter flow cytometry. Am J Obstet Gynecol. 1991;165:1731–1737. doi: 10.1016/0002-9378(91)90024-l. [DOI] [PubMed] [Google Scholar]
- Prieto B, Candenas M, Venta R, Ladenson JH, Alvarez FV. Isolation of fetal nucleated red blood cells from maternal blood in normal and aneuploid pregnancies. Clin Chem Lab Med. 2002;40:667–672. doi: 10.1515/CCLM.2002.114. [DOI] [PubMed] [Google Scholar]
- Purwosunu Y, Sekizawa A, Koide K, Okazaki S, Farina A, Okai T. Clinical potential for noninvasive prenatal diagnosis through detection of fetal cells in maternal blood. Taiwan J Obstet Gynecol. 2006a;45:10–20. doi: 10.1016/S1028-4559(09)60184-4. [DOI] [PubMed] [Google Scholar]
- Purwosunu Y, Sekizawa A, Farina A, et al. Enrichment of NRBC in maternal blood: a more feasible method for noninvasive prenatal diagnosis. Prenat Diagn. 2006b;26:545–547. doi: 10.1002/pd.1456. [DOI] [PubMed] [Google Scholar]
- Samura O, Sekizawa A, Zhen DK, Falco VM, Bianchi DW. Comparison of fetal cell recovery from maternal blood using a high density gradient for the initial separation step: 1.090 versus 1.119 g/ml. Prenat Diagn. 2000;20:281–286. doi: 10.1002/(sici)1097-0223(200004)20:4<281::aid-pd812>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Sekizawa A, Farina A, Zhen DK, et al. Improvement of fetal cell recovery from maternal blood: suitable density gradient for FACS separation. Fetal Diagn Ther. 1999;14:229–233. doi: 10.1159/000020927. [DOI] [PubMed] [Google Scholar]
- Sekizawa A, Purwosunu Y, Farina A, et al. Development of noninvasive fetal DNA diagnosis from nucleated erythrocytes circulating in maternal blood. Prenat Diagn. 2007;27:846–848. doi: 10.1002/pd.1792. [DOI] [PubMed] [Google Scholar]
- Slunga-Tallberg A, El-Rifai W, Keinanen M, et al. Maternal origin of nucleated erythrocytes in peripheral venous blood of pregnant women. Hum Genet. 1995;96:53–57. doi: 10.1007/BF00214186. [DOI] [PubMed] [Google Scholar]
- Smits G, Holzgreve W, Hahn S. An examination of different Percoll density gradients and magnetic activated cell sorting (MACS) for the enrichment of fetal erythroblasts from maternal blood. Arch Gynecol Obstet. 2000;263:160–163. doi: 10.1007/s004040050273. [DOI] [PubMed] [Google Scholar]
- Takabayashi H, Kuwabara S, Ukita T, Ikawa K, Yamafuji K, Igarashi T. Development of non-invasive fetal DNA diagnosis from maternal blood. Prenat Diagn. 1995;15:74–77. doi: 10.1002/pd.1970150116. [DOI] [PubMed] [Google Scholar]
- Troeger C, Holzgreve W, Hahn S. A comparison of different density gradients and antibodies for enrichment of fetal erythroblasts by MACS. Prenat Diagn. 1999b;19:521–526. [PubMed] [Google Scholar]
- Troeger C, Zhong XY, Burgemeister R, et al. Approximately half of erythroblasts in maternal blood are of fetal origin. Mol Hum Reprod. 1999a;5:1162–1165. doi: 10.1093/molehr/5.12.1162. [DOI] [PubMed] [Google Scholar]
- Valerio D, Aiello R, Altieri V. Isolation of fetal erythroid cells from maternal blood based on expression of erythropoietin receptors. Mol Hum Reprod. 1997;3:451–455. doi: 10.1093/molehr/3.5.451. [DOI] [PubMed] [Google Scholar]
- Wang JY, Zhen DK, Falco VM, et al. Fetal nucleated erythrocyte recovery: fluorescence activated cell sorting-based positive selection using anti-gamma globin versus magnetic activated cell sorting using anti-CD45 depletion and anti-gamma globin positive selection. Cytometry. 2000;39:224–230. [PubMed] [Google Scholar]


