Abstract
Until now, design of the annual influenza vaccine has relied on phylogenetic or whole-sequence comparisons of the viral coat proteins hemagglutinin and neuraminidase, with vaccine effectiveness assumed to correlate monotonically to the vaccine-influenza sequence difference. We use a theory from statistical mechanics to quantify the non-monotonic immune response that results from antigenic drift in the epitopes of the hemagglutinin and neuraminidase proteins. The results explain the ineffectiveness of the 2003–2004 influenza vaccine in the United States and provide an accurate measure by which to optimize the effectiveness of future annual influenza vaccines.
Keywords: Influenza vaccine, Original antigenic sin, Antigenic drift
1 Introduction
Antigenic variation constitutes one mechanism employed by influenza viruses to evade the adaptive response of the host immune system. This antigenic drift of the recognized, epitope regions of the viral surface proteins hemagglutinin (HA) and neuraminidase (NA) constitutes a major challenge to effective vaccine design, where historical experience and phylogenetic analysis of HA and NA protein sequences from circulating human strains are used to decide the components of the annual influenza vaccine. Here we introduce a theory to guide this important public health decision. Application of the theory could help prevent critical situations such as occurred with 2003–2004 influenza epidemic [1], whence the administered A/Panama/2007/99 H3N2 vaccine gave unexpectedly [2] low protection against the mutant strain A/Fujian/411/2002. A model from statistical mechanics is used to evaluate the non-linear decrease of the immune response due to mutations in the viral epitope region sequences. We propose that this epitope analysis be regularly used as a measure of the immunological distance between mutant strains in the annual design of the influenza vaccine.
Influenza A virus infections and posterior complications, such as pneumonia, are a major cause of human morbidity and mortality. The 2003–2004 influenza epidemic was mainly due to the proliferation of the new H3N2 subtype strain A/Fujian/411/2002, an antigenic drift mutant of A/Panama/2007/99. According to the February 2003 WHO report [2], after comparing the whole hemagglutinin (HA) sequences of both strains, the CDC council members concluded that both proteins were similar enough to expect a significant degree of cross protection, and decided to include the Panama strain in the H3N2 component of the vaccine. No information concerning the neuraminidase sequence for the Fujian strain was used. Recent clinical results, as stated in the 16 January 2004 CDC Morbidity and Mortality Report [1], show the vaccine provided essentially no protection against infection during the 2003–2004 season.
2 Methods
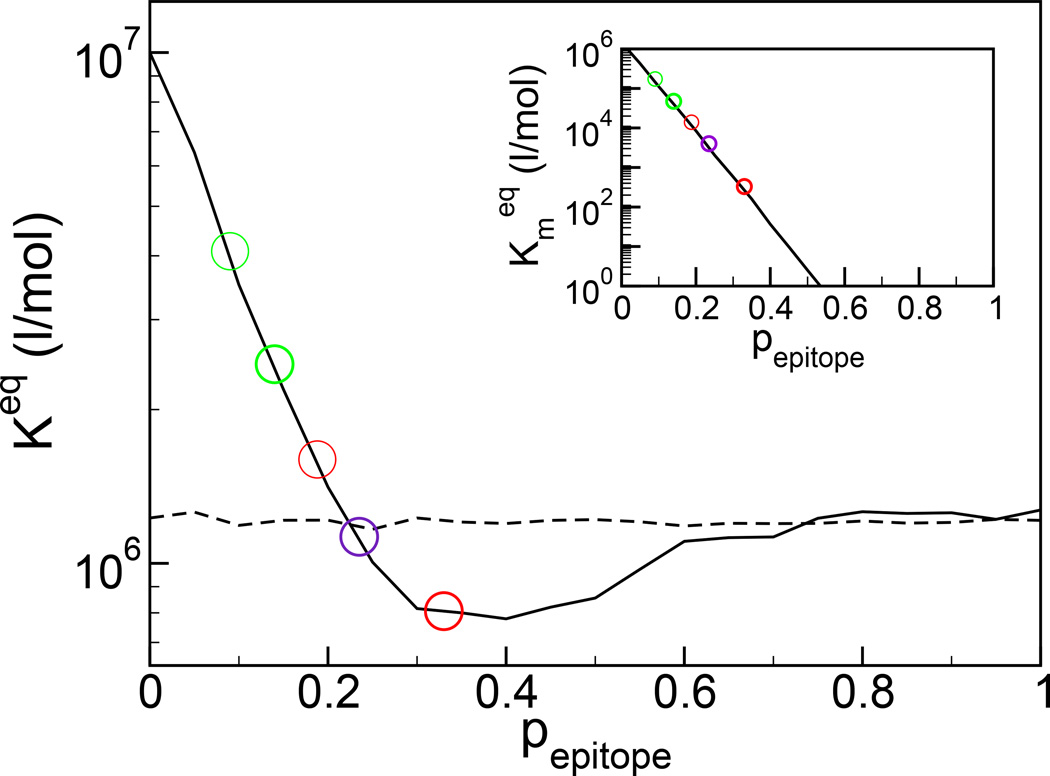
We have developed a theory of the immune response to an antigenic drift strain after vaccination based on statistical mechanics, Figure 1 [3]. The model predicts the affinity constant values
| (1) |
for a second antigen, after exposure to an original antigen whose epitope region differs by probability pepitope- The key measure of antigenic drift in the theory is pepitope, the fractional change between the dominant epitope regions of the vaccine and the circulating strain, defined by the equation
| (2) |
Fig. 1.
The evolved affinity constant to a second antigen after exposure to an original antigen whose epitope region differs by probability pepitope (solid line). The dashed line represents the affinity constant without previous exposure. In green are shown the responses at the values of the differences between the A/Panama/2007/99 (vaccine) and A/Fujian/411/2002 (circulating) strains for the B and E hemagglutinin epitopes. In red are shown the responses at the values of the difference between the A/Panama/2007/99 (vaccine) and A/Fujian/411/2002 (circulating) strains for the A and B neuraminidase epitopes. Dominant epitopes are shown in bold. The clinical outcome is an average of the response to the HA and NA proteins, and in purple is shown the immune response at the average difference for the dominant HA and NA epitopes (see text). The effectiveness of the 2003–2004 flu vaccine was marginal at best. In inset is shown the cross affinity of the memory sequences for the mutated antigen, often measured biochemically and distinct from the evolved immune response. As if often found, the cross activity decreases exponentially with antigenic drift [32].
This characterization of antigenic drift in our theoretical model emphasizes the experimental fact that only the epitope regions are significantly involved in immune recognition, as shown by immunoassays and crystallographic images [4].
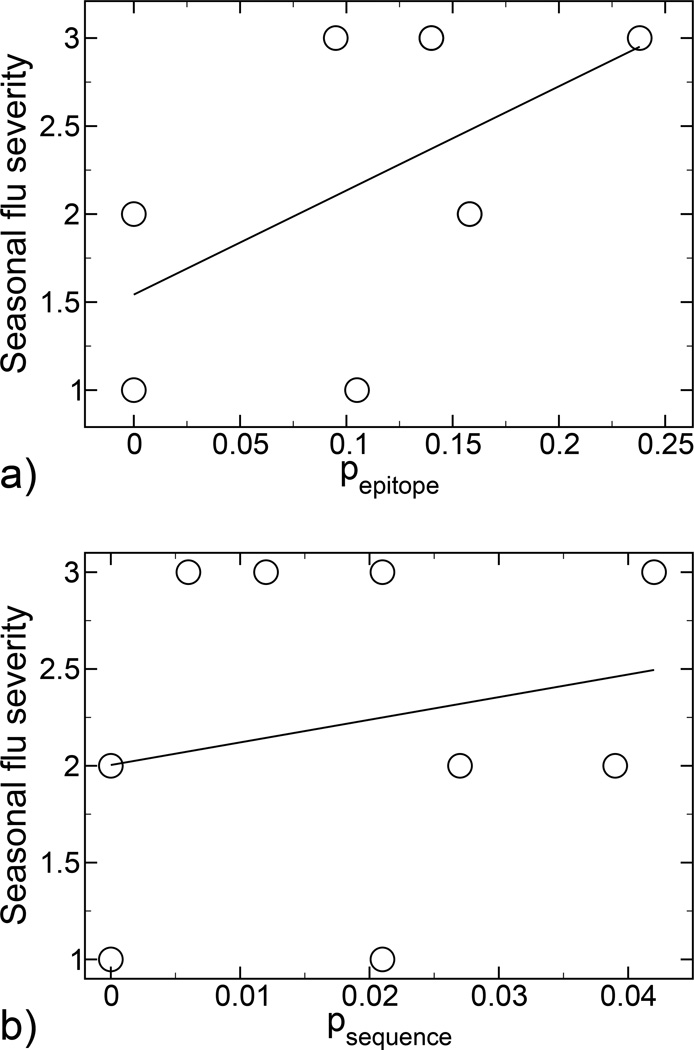
In the theory, it is the percent of the epitope that changes that characterizes antigenic drift. To provide additional empirical support for the theory, we performed an historical analysis of the influenza seasons between 1991–2000 when the H3N2 virus suptype was dominant [5–13]. For every season, hemagglutinin sequences [14] were compared between the vaccine strain and the predominant circulating strain. A quantitative scale was defined to measure the seasonal flu severity as follows: low (1), mild (2) and high (3). The values of seasonal flu severity were correlated with the calculated pepitope values. In addition, to make clear that the epitope region is primarily responsible for immune recognition, we also correlated the seasonal flu severity with antigenic drift of the entire hemagglutinin sequence, normalized by the total number of amino acids in the protein psequence. The results presented in Figure 2 show that seasonal flu severity is correlated with pepitope rather than psequence, thus favoring the epitope analysis approach. Therefore, it is both logical and consistent with the observed data to characterize antigenic drift by the number of mutations within the epitope regions, as we do in the present work.
Fig. 2.
a) Correlation between influenza seasonal severity (see text) and hemagglutinin antigenic drift, calculated by epitope analysis. Least-squares regression analysis yields the linear fit y = 1.5425 + 5.9162 pepitope, with a correlation coefficient R = 0.54432. b) Correlation between influenza seasonal severity (see text) and hemagglutinin antigenic drift, calculated by whole sequence analysis. Least-squares regression analysis yields the linear fit y = 2.0041 + 11.684 psequence, with a correlation coefficient R = 0.2183.
According to historic clinical experience, and to our model, when the antigenic drift between the vaccine and circulating strain, characterized by the pepitope value, is small, exposure to the vaccine antigen leads to a higher affinity constant than without exposure. This result is why immune system memory and vaccination are generally effective. When the antigenic drift between the vaccine and circulating strain is large, the vaccine antigen is uncorrelated with the circulating strain antigen, and so immune system memory does not play a role. When the antigenic drift between the vaccine and circulating strain epitopes is modest (0.23 < pepitope < 0.6), our theory predicts that memory response may be worse than the naive response (the solid curve lies below the dashed curve in Figure 1), which means that the immunological memory from the vaccine exposure actually gives worse protection, i.e., a lower affinity constant, than would no vaccination whatsoever. This result is the original antigenic sin phenomena for influenza: vaccination creates memory sequences that for some mutation rates of influenza may increase susceptibility to future exposure [15,16]. Parenthetically, not every infectious disease exhibits original antigenic sin, with measles one such example. The measles virus does not undergo antigenic drift, and despite approximately eight different subtypes that have been identified to date [17], the HA and NA genetic variation among them does not exceed 7% on a nucleotide basis [17], or roughly 2% on an amino acid basis. Accordingly, within the context of our model, there is no possibility of original antigenic sin for measles (since pepitope < 0.02, see Figure 1).
Original antigenic sin stems from localization of the immune system response in antibody sequence space. This localization is a result of the roughness in sequence space of the evolved antibody affinity constant for antigen. Interestingly, there appears to have been a modest degree of original antigenic sin, termed negative vaccine effectiveness in the CDC Morbidity and Mortality report [1], associated with the 2003–2004 influenza vaccine.
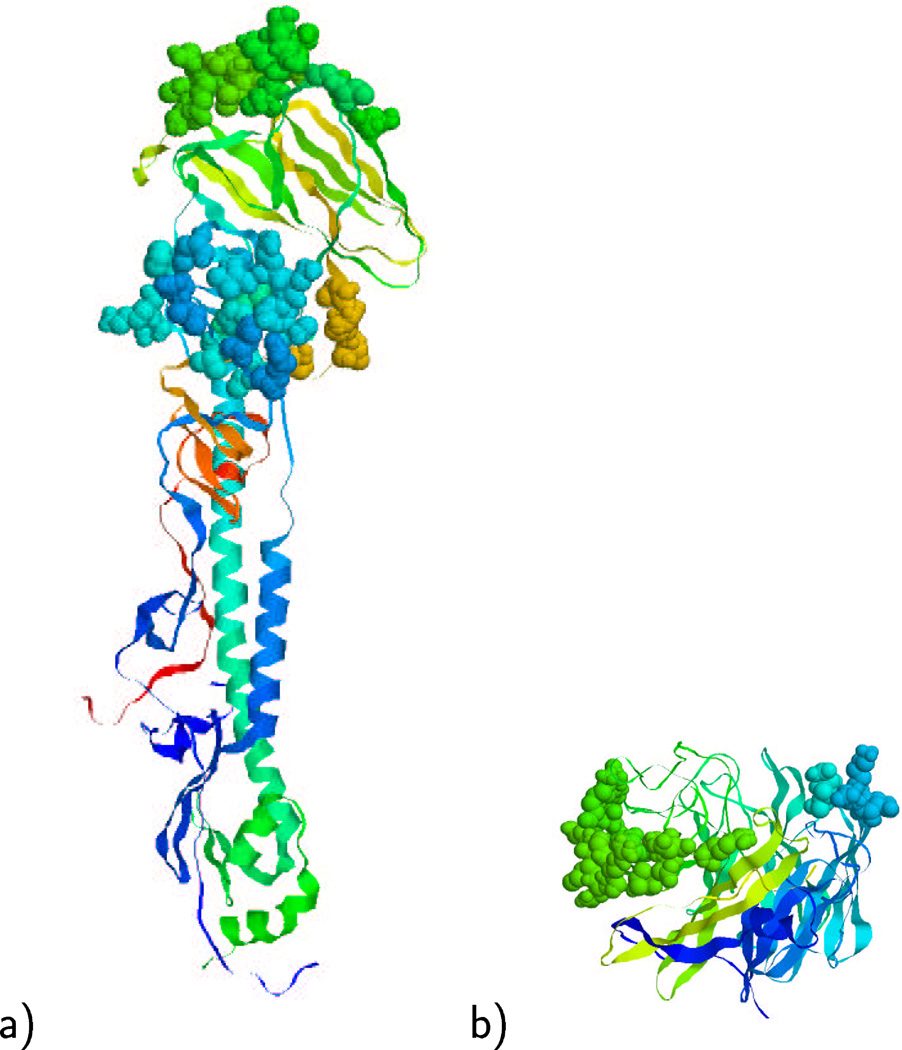
Human influenza A viruses are classified in different subtypes according to the neuraminidase and hemagglutinin proteins. The current influenza A vaccine includes both the H1N1 and H3N2 subtypes, and the consensus sequence for the HA and NA proteins corresponding to each subtype requires annual update due to continuous antigenic drift. Variations due to point mutations in the residues in the epitope regions can considerably reduce the immune response, despite biochemical cross activity between strains related by antigenic drift. The epitope regions of the HA and NA proteins are shown in Figure 3. We propose that antigenic drift mutants be compared not by the whole sequences of the HA and NA proteins, but more precisely by the sequences of the dominant epitopes for the proposed vaccine strain, by calculating the pepitope parameter of our theory. According to the definition (2), a different value of pepitope is obtained for each epitope region in both hemagglutinin and neuraminidase viral proteins. We propose to include in the analysis only the pepitope values corresponding to the dominant epitopes in both proteins. Since both hemagglutinin and neuraminidase participate in the immune recognition process, it is some combination of the immune recognition of these two proteins that contributes to reducing the seasonal flu severity. We, thus, define an approximate total response as the binding constant at the average of the pepitope values for the dominant HA and NA epitopes:
Fig. 3.
a) Shown are the dominant B (top) and subdominant E (middle) epitope of the hemagglutinin protein in space-filling format [19]. b) Shown are the dominant B (right) and subdominant A (left) epitope of the neuraminidase protein in space-filling format [30]. The rest of the proteins are shown in ribbon format.
3 Results
We compared the epitope sequences of the HA protein to look for mutations in the A/Fujian/411/2002 strain [14] with respect to the A/Panama/2007/99 strain [18]. The hemagglutinin H3 protein has five epitope regions (A, B, C, D, E) that have been identified and sequenced [19], among which A and B are usually dominant [20,21]. There exists experimental and clinical evidence that epitope regions mutate much faster than other regions in the viral proteins, presumably due to antibody selective pressure [22–24], with the dominant epitopes mutating most rapidly [25]. Therefore, in the absence of more detailed information, we take an observed high mutation rate (i.e., a high pepitope value) in a given epitope to correlate with dominance. Epitope A (residues 122, 124, 126, 130–133, 135, 137, 138, 140, 142–146, 150, 152, 168) presents one point mutation at residue 131. The calculated value pepitope = 1/19 = 0.053. Epitope B (residues 128, 129, 155–160, 163, 165, 186–190, 192–194, 196–198) presents three point mutations at residues 155, 156, and 186. The calculated value pepitope = 3/21 = 0.14. Epitope C (residues 44–48, 50, 51, 53, 54, 273, 275, 276, 278–280, 294, 297, 299, 300, 304, 305, 307–312) presents one point mutation at residue 50. The calculated value pepitope = 1/27 = 0.037. Epitope D (residues 96,102,103,117,121,167,170–177,179,182, 201, 203, 207–209, 212–219, 226–230, 238, 240, 242, 244, 246–248) presents no mutations. Epitope E (residues 57, 59, 62, 63, 67, 75, 78, 80–83, 86–88, 91, 92, 94, 109, 260–262, 265) presents two point mutations at residues 75 and 83. The calculated value pepitope = 2/22 = 0.09. In absence of further information, which is the typical case for the annual task of influenza vaccine design, we conclude that likely B epitope is dominant and E epitope is subdominant for the A/Panama/2007/99 HA protein, whereas the other epitopes are cryptic. The dominance of epitope B is in accordance with observed data [26]. Note that by looking at the antigenic drift within the dominant epitope, rather than the drift of the whole protein sequence, we obtain a larger and much more accurate estimate of the degree to which the immune response to A/Fujian/411/2002 and A/Panama/2007/99 will differ.
We also calculated the values for antigenic drift in the NA epitopes between the A/Fujian/411/2002 [27] and A/Panama/2007/99 [28] strains. The neuraminidase N2 protein has been completely sequenced and crystallized [29,30]. Mutational studies with monoclonal antibodies identified three regions (A,B,C) in NA N2 that are important for recognition [4,31], of which only the surface residues can be within the epitopes. Regions A and B are usually dominant [31]. Epitope A (residues 383–387, 389–394, 396, 399, 400, 401, 403) presents three point mutations in at residues 385, 399, and 403. The calculated value pepitope = 3/16 = 0.188. Epitope B (residues 197–200, 221, 222) presents two point mutations at residues 197 and 217 (although this residue is not present in the epitope, its mutation will affect the epitope due to physical proximity). The calculated value pepitope = 2/6 = 0.33. Epitope C (residues 328–332, 334, 336, 338, 339, 341–344, 346, 347, 357–359, 366–370) presents one point mutation at residue 370. The calculated value pepitope = 1/23 = 0.043. We conclude that likely epitope B is dominant, and epitope A is subdominant for the A/Panama/2007/99 NA protein, whereas epitope C is cryptic.
4 Discussion
In Figure 1 are shown the predicted immune responses to the A/Fujian/411/2002 strain for the hemagglutinin (green) and neuraminidase (red) dominant epitopes after vaccination to A/Panama/2007/99. The predicted values for hemagglutinin lie in the region of moderate immune response, and so consistent with the WHO data [2] one would expect some degree of cross-strain protection. However, the predicted immune response to the dominant neuraminidase epitope is in the region of original antigenic sin. In the design of the 2003–2004 influenza vaccine, neither the cross activity nor the immune response were measured for the A/Fujian/411/2002 NA protein in response to A/Panama/2007/99 NA vaccination [2]. Upon analysis of actual effectiveness of the 2003–2004 vaccine, there does appear to have been a modest degree of original antigenic sin, or negative vaccine effectiveness [1].
Summarizing our findings, it would appear that for the hemagglutinin protein of A/Panama/2007/99, epitope B is dominant, and vaccination gives modest protection to the A/Fujian/411/2002 strain. For the neuraminidase protein of A/Panama/2007/99, it would appear that epitope B is also dominant, and vaccination may increase the susceptibility to the A/Fujian/411/2002 strain. Taken in aggregate, these results, Figure 1, suggest that the 2003–2004 flu vaccine would have essentially no effect against the circulating A/Fujian/411/2002 strain, in agreement with clinical findings [1] and in disagreement with early expectations [2].
In conclusion, we suggest that strains related by antigenic drift be compared by measuring differences in the epitope regions of the hemagglutinin and neuraminidase proteins and not by differences in the whole sequence or phylogeny as is presently done. This particular point is supported by the correlations of seasonal flu severity with epitope antigenic drift shown in Figure 2. Thus, there is a need for a detailed characterization of the epitope regions in the different influenza strains. In particular, a precise determination of which epitopes are dominant in the proposed vaccine strain and an experimental measure of pepitope and of cross activity between the proposed vaccine strain and the circulating strains would be highly productive. In absence of this determination, we suggest to estimate the dominant epitope as that which shows the most antigenic drift [22–25], as we do in the present work. From either epitope sequence drift or cross activity, Figure 1 can be used to estimate the degree of the immune response, which is a non-linear and non-monotonic function of the measured data. We believe that this quantitative epitope analysis should be incorporated as part of the regular protocol for construction of the annual influenza vaccine.
Acknowledgments
We thank Nancy Cox, Director of the WHO Collaborating Center for Surveillance, Epidemiology and Control of Influenza, Centers for Disease Control and Prevention (Atlanta), for helpful discussions. We also thank Xijan Xu from the Strain Surveillance Section, Influenza Branch G16, Centers for Disease Control and Prevention (Atlanta), for providing the A/Fujian/411/2002 neuraminidase sequence. This research was supported by the U. S. National Institutes of Health.
ETM carried out the sequence and structural analysis. MWD conceived of the study and participated in its design and coordination. All authors read and approved the final manuscript.
References
- 1.Dolan S, Nyquist AC, Ondrejka D, Todd J, Gershman K, Alexander J, Bridges C, Copeland J, David F, Euler G, Gargiullo P, Kenyan K, Moore Z, Seward J, Jain N. Preliminary assessment of the effectiveness of the 2003–4 inactivated influenza vaccine - Colorado, Denver. 2003 Centers for Disease Control and Prevention Morbidity and Mortality Weekly Report. 2004;53(1):8–11. [PubMed] [Google Scholar]
- 2.Cox N, Balish A, Brammer L, Fukuda K, Hall H, Klimov A, Lindstrom S, Mabry J, Perez-Oronoz G, Postema A, Shaw M, Smith C, Subbarao K, Wallis T, Xijan X. Information for the vaccines and related biological products advisory committee, CBER, FDA, 2003. WHO Collaborating Center for Surveillance Epidemiology and Control of Influenza [Google Scholar]
- 3.Deem MW, Lee H-Y. Sequence space localization in the immune system response to vaccination and disease. Phys. Rev. Lett. 2003;91:068101. doi: 10.1103/PhysRevLett.91.068101. [DOI] [PubMed] [Google Scholar]
- 4.Air GM, Els MC, Brown LE, Laver WG, Webster RG. Location of antigenic sites on the three-dimensional structure of the influenza N2 virus neuraminidase. Virology. 1985;145:237–248. doi: 10.1016/0042-6822(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control. Update: Influenza activity — United States and worldwide, 1991–92 season, and composition of the 1992–93 influenza vaccine. Centers for Disease Control and Prevention Morbidity and Mortality Weekly Report. 1992;41(18):315–317. 323. [PubMed] [Google Scholar]
- 6.Centers for Disease Control. Current trends update: Influenza activity — United States and worldwide, 1992–93 season, and composition of the 1993–94 influenza vaccine. Centers for Disease Control and Prevention Morbidity and Mortality Weekly Report. 1993;42(10):177–180. [Google Scholar]
- 7.Centers for Disease Control. Current trends update: Influenza activity — United States and worldwide, 1993–94 season, and composition of the 1993–94 influenza vaccine. Centers for Disease Control and Prevention Morbidity and Mortality Weekly Report. 1994;43(10):179–183. [PubMed] [Google Scholar]
- 8.Centers for Disease Control. Update: Influenza activity — United States and worldwide, 1994–95 season, and composition of the 1995–96 influenza vaccine. Centers for Disease Control and Prevention Morbidity and Mortality Weekly Report. 1995;44(15):292–295. [PubMed] [Google Scholar]
- 9.Centers for Disease Control. Update: Influenza activity — United Sates and worldwide, 1995–96 season, and composition of the 1996–97 influenza vaccine. Centers for Disease Control and Prevention Morbidity and Mortality Weekly Report. 1996;45(16):326–329. [PubMed] [Google Scholar]
- 10.Centers for Disease Control. Update: Influenza activity — United States and worldwide, 1996–97 season, and composition of the 1997–98 influenza vaccine. Centers for Disease Control and Prevention Morbidity and Mortality Weekly Report. 1997;46(15):325–330. [PubMed] [Google Scholar]
- 11.Centers for Disease Control. Update: Influenza activity — United States and worldwide, 1997–98 season, and composition of the 1998–99 influenza vaccine. Centers for Disease Control and Prevention Morbidity and Mortality Weekly Report. 1998;47(14):280–284. [PubMed] [Google Scholar]
- 12.Centers for Disease Control. Update: Influenza activity — United States and worldwide, 1998–99 season, and composition of the 1998–99 influenza vaccine. Centers for Disease Control and Prevention Morbidity and Mortality Weekly Report. 1999;48(18):374–378. [PubMed] [Google Scholar]
- 13.Centers for Disease Control. Update: Influenza activity — United States and worldwide, 1999–2000 season, and composition of the 2000–01 influenza vaccine. Centers for Disease Control and Prevention Morbidity and Mortality Weekly Report. 2000;49(17):375–381. [PubMed] [Google Scholar]
- 14.Macken C, Lu H, Goodman J, Boykin L. The value of a database in surveillance and vaccine selection. In: Osterhaus ADME, Cox N, Hampson AW, editors. Options for the Control of Influenza IV. Elsevier Science; 2001. accession number ISDN38157. http://www.flu.lanl.gov/. [Google Scholar]
- 15.Fazekas de St. Groth S, Webster RG. Disquisition on original antigenic sin: Evidence in man. J. Exp. Med. 1966;124:331–345. doi: 10.1084/jem.124.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davenport FM, Hennessy AV, Francis T. Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J. Exp. Med. 1953;98:641–656. doi: 10.1084/jem.98.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellini WJ, Rota AP. Genetic diversity of wild-type measles viruses: Implications for global measles elimination programs. Emerging Infectious Diseases. 1998;4:29–35. doi: 10.3201/eid0401.980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Accession number ISDNCDA001 from the Influenza Sequence Database[14].
- 19.Hemagglutinin H3 epitope structural mapping from the Influenza Sequence Database[14].
- 20.Bush RM, Fitch WM, Bender CA, Cox NJ. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 1999;16:1457–1465. doi: 10.1093/oxfordjournals.molbev.a026057. [DOI] [PubMed] [Google Scholar]
- 21.Cox NJ, Bender CA. The molecular epidemiology of influenza viruses. Semin. Virol. 1995;6:359–370. [Google Scholar]
- 22.Fitch WM, Leiter JM, Li X, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc. Natl. Acad. Sci. USA. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. Predicting the evolution of human influenza A. Science. 1999;286:1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- 24.Plotkin JB, Dushoff J. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc. Natl. Acad. Sci. USA. 2003;100:7152–7157. doi: 10.1073/pnas.1132114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitch WM, Butch RM, Bender CA, Subbarao K, Cox NJ. Predicting the evolution of human influenza A. The Journal of Heredity. 2000;91:183–185. doi: 10.1093/jhered/91.3.183. [DOI] [PubMed] [Google Scholar]
- 26.Kostolanský F, Varečkova E, Betáková T, Mucha V, Russ G, Wharton SA. The strong positive correlation between effective affinity and infectivity neutralization of highly cross-reactive monoclonal antibody IIB4, which recognizes antigenic site B on influenza A virus haemagglutinin. Journal of General Virology. 2000;81:1727–1735. doi: 10.1099/0022-1317-81-7-1727. [DOI] [PubMed] [Google Scholar]
- 27.Personnal communication, Xu Xijan.
- 28.Accession number AJ457937 from the Influenza Sequence Database[14].
- 29.Tulip WR, Varghese JN, Baker AT, van Donkelaar A, Laver WG, Webster RG, Colman PM. Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants. J. Mol. Bio. 1992;221:487–497. doi: 10.1016/0022-2836(91)80069-7. PDB accession number 3NN9. [DOI] [PubMed] [Google Scholar]
- 30.Jedrzejas JJ, Singh S, Brouillette WJ, Laver WG, Air GM, Luo M. Structures of aromatic inhibitors of influenza A virus neuraminidase. Biochemistry. 1995;34:3144–3151. doi: 10.1021/bi00010a003. PDB accession number 1IVD. [DOI] [PubMed] [Google Scholar]
- 31.Gulati U, Hwang C-C, Venkatramani L, Gulati S, Stray S, Lee JT, Laver WG, Bochkarev A, Zlotnick A, Air G. Antibody epitopes on the neuraminidase of a recent H3N2 influenza virus (A/Memphis/31/98) Journal of Virology. 2002;76:12274–12280. doi: 10.1128/JVI.76.23.12274-12280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith DJ, Forrest S, Ackley DH, Perelson AS. Variable efficacy of repeated annual influenza vaccination. Proc. Natl. Acad. Sci. USA. 1999;96:14001–14006. doi: 10.1073/pnas.96.24.14001. [DOI] [PMC free article] [PubMed] [Google Scholar]