Abstract
Objective
Late onset sepsis (LOS) contributes to mortality and morbidity in preterm infants. We tested the hypotheses that microbes causing LOS originate from the gut, and that distortions in the gut microbial community increases subsequent risk of LOS.
Study Design
We examined the gut microbial community in prospectively collected stool samples from preterm infants with LOS and an equal number of age-matched controls at two sites (Cincinnati, OH and Birmingham, AL), by sequencing the bacterial 16S rDNA. We confirmed our findings in a subset of infants by whole genome shotgun sequencing, and analyzed the data using R and LEfSe.
Results
Infants with LOS in Cincinnati, as compared to controls, had less abundant Actinobacteria in the first samples after birth (median 18 days before sepsis onset), and less abundant Pseudomonadales in the last samples collected prior to LOS (median 8 days before sepsis onset). Infants with LOS in Birmingham, as compared to controls, had no differences identified in the first sample microbial communities, but Lactobacillales was less abundant in the last samples prior to LOS (median 4 days before sepsis onset). Sequencing identified detectable levels of the sepsis-causative organism in stool samples prior to disease onset for 82% of LOS cases.
Conclusions
Translocation of gut microbes may account for the majority of LOS cases. Distortions in the fecal microbiota occur prior to LOS, but the form of distortion depends on timing and site. The microbial composition of fecal samples does not predict LOS onset in a generalizable fashion.
Introduction
Late-onset neonatal sepsis (LOS) is a major cause of morbidity and mortality in preterm infants. LOS occurs in more than 20% of very low birth weight infants [1] and is associated with increased risk of mortality [2]. Survivors of LOS exhibit worse long-term outcomes, with increased risk of cerebral palsy, vision, and hearing impairment [3]. Given the seriousness of LOS, diagnostic and predictive biomarkers are needed [4].
Two-thirds of LOS cases in preterm neonates have been attributed to indwelling vascular catheters; this statistic is based on co-occurrence of events, i.e., catheter use within 48 hours of sepsis onset [5]. However, LOS is often caused by organisms that are typically found colonizing the intestinal tract, suggesting that LOS may often originate from translocation of gut organisms across the intestinal lining into the blood stream. Indeed, several small studies have demonstrated that the majority of LOS cases, in particular, those caused by group B Streptococcus, Serratia marcescens, or Escherichia coli strains, are due to organisms that were detectable in infant stool samples collected prior to the occurrence of sepsis [6, 7]. Several investigators have applied 16S microbial community analysis to examine distortions in the intestinal microbiota in advance of neonatal sepsis. A study of six infants found that those who later developed sepsis had an intestinal microbiota distinct from that of healthy infants [8]; samples from infants who later developed LOS tended to have more Staphylococcus and Proteobacteria than healthy infants who exhibited greater microbial diversity and more anaerobic bacteria [8]. Another study, of 10 LOS cases and 18 matched control infants, found a decrease in diversity and lower Bifidobacteria counts preceding LOS compared to control infants [9]. A third study, of 27 infants, found an increase in Staphylococcaceae associated with sepsis but did not report a difference in diversity between cases and controls [10].
The microbiota of the gut is a complex community, and dysbiosis within that community can result in disease. A clear example of this process is Clostridium difficile infection following antibiotic treatment: Antibiotic treatment may remove microbial community members that normally keep C. difficile in check, thereby allowing C. difficile to overgrow and cause disease. Restoration of the microbial community through fecal transplant is highly successful in treating refractory C. difficile disease [11]. Another example of microbial community dysbiosis predisposing to disease is seen in the different colonization patterns of preterm infants who develop necrotizing enterocolitis (NEC) compared to infants who remain healthy [12, 13]. The ability of a healthy microbial community to prevent disease may have implications for LOS. Indeed, mouse studies have demonstrated that microbiota are important to the barrier function of gastrointestinal epithelium and that alterations to microbiota influence intestinal permeability [14].
We therefore hypothesized that the composition of the intestinal microbiome influences the risk of LOS in premature infants. This study addresses two distinct but related questions: 1) What proportion of neonatal LOS are potentially attributable to translocation of gut microbes; and 2) is there an identifiable pattern of microbial community dysregulation that occurs in advance of neonatal sepsis?
Methods
Subjects
Study infants were selected from all infants <29 weeks gestational age who were enrolled in an ongoing cohort study of novel biomarkers for NEC. Infants were enrolled from three level III Neonatal Intensive Care Units (NICUs), including one in Birmingham, AL and two in Cincinnati, OH. LOS was defined as development of blood culture-proven sepsis more than72 hours after birth. When blood culture indicated the cause of sepsis as coagulase-negative Staphylococcus or diphtheroid Bacilli, infants had to have been either blood culture positive for an additional organism or treated with antibiotics for at least 5 days beginning within 72 hours of the time of the blood draw for the culture. To be eligible for inclusion as a case, infants had to have at least one stool sample collected and successfully processed prior to sepsis onset. Eligibility for selection as a control required that an infant was born during the time period between the birth of the first case and the birth of the last case at each site, and survive free of proven NEC and sepsis. Additionally, to be eligible as a control, an infant had to have at least one successfully sequenced stool sample. Controls were frequency matched to cases on study site and gestational age. The Cincinnati Children’s Hospital Medical Center IRB, the University of Alabama IRB, the TriHealth IRB, and the University of Cincinnati IRB approved this study. Written consent was obtained from parents or guardians of study subjects.
Sample Collection
Serial stool samples were collected from infants on scheduled collection postnatal days 5, 8, 11, 14, and 21 plus or minus two days. For this study, the first successfully sequenced stool sample and the last successfully sequenced sample prior to LOS onset from each infant were included. For the last sample analysis, control samples were frequency matched to case samples on collection postnatal days. Frequency matching on the postnatal day of sample collection was done to account for the known normal progression of intestinal microbiota that occurs with increasing time since birth [15]. Samples were collected from soiled diapers, immediately refrigerated at 4°C in the NICU and transported to the laboratory where they remained in the refrigerator until processing and storage at -80°C. All Birmingham samples were stored without buffer, as facilities for processing samples were limited. In Cincinnati, most samples were processed and stored with thioglycollate buffer. However, the protocol for use of buffer changed over time as efforts were made to optimize sample storage: Thus, for the first sample analysis, 4 of 22 control samples and 7 of 22 case samples were stored without thioglycollate (Fisher’s Exact Test, p = 0.49). In Cincinnati for the last sample analysis, 1 of 22 control samples and 5 of 22 case samples were stored without thioglycollate (Fisher’s Exact Test, p = 0.19). From previous methodologic studies, we found that samples stored without thioglycollate to be enriched in phylum Bacteroidetes, Proprionibacterineae (a family belonging to class Actinomycetales), and Leuconostocaeae (a family belonging to class Bacilli)[16].
Stool Extraction and 16S rDNA Sequencing
Stool extractions were completed using the methods described in Morrow et al. 2013 [12]. The DNA was then sequenced and processed as described in Taft et al. 2014 [16]. The OTU table was rarified to 2000 reads per sample for analysis.
Whole Genome Sequencing (WGS)
For half of the sepsis cases, sequence data was available to determine whether the sepsis-causing organism identified by the clinical laboratory was also present in the infant’s intestinal tract, as determined by WGS. The WGS data consisted of 101-base, paired-end reads on the Illumina HiSeq2000 platform producing an average of 3.2E+7 reads per sample. A minimal read length filter of 80nt was applied to all data used in analysis. Relative taxonomic abundances were determined from WGS data using MetaPhlAn v2.0 (https://bitbucket.org/biobakery/metaphlan2) [17].
Statistical Analysis
Analysis focused on the 16S sequencing data as the number of cases and controls with WGS data was limited. Ordination was conducted using non-metric multi-dimensional scaling (NMDS),which did not indicate distinct microbial communities between the two sites, but the linear discriminant analysis effect size tool, LEfSe [18], found specific differences by site, as reported previously for this cohort [16](S1 and S2 Figs) The two Cincinnati NICUs had similar colonization patterns, and so were combined into one analysis. The initial analyses used the first available stool sample from each infant prior to LOS. We then analyzed the last sample prior to sepsis in cases compared to a sample at a matched postnatal timing from controls. To select the appropriate sample from controls for this analysis, the sample collection day of life was frequency matched to the sample collection day of life for the sepsis cases by sampling without replacement. We first created a table including the day of life of collection of all samples from control infants and a table of collection day of life of the last sample prior to sepsis for all case samples. The first control sample was selected by identifying the collection day of life of a case sample on which the fewest control samples were available. Those two samples and all additional samples from that control and case infants were then removed from the pool eligible for selection. The selection of the next control sample was made by again identifying the collection day of life of a case sample with the fewest control samples to choose from. This procedure was repeated until only a single case sample and a single control infant remained. The control sample collected closest to the collection day of life of the final case sample was selected for inclusion.
Alpha diversity was calculated using the Chao1 index and the Simpson index with the Vegan package in R; the Kruskal-Wallis test was used to test for differences in alpha diversity between cases and controls. Beta diversity was examined using non-metric multidimensional scaling, based on weighted and unweighted Uni-Frac distance as described in Morrow et al. [12] The linear discriminant analysis effect size tool (LEfSe) was used to screen for differences at all taxonomic levels between the LOS case and control samples [18]. Classification trees were generated to provide additional information on differences in microbial composition between the two sites. To generate trees for the Cincinnati analysis, the rpart package in R [19] was used with default settings. For Birmingham, the minimum number to split changed from 20 to 10 due to the small number of subjects. Logistic regression using a forward inclusion approach conducted in R was used in combination with the taxa difference cut-points identified by the classification tree, to test for significance of the models created by the classification trees. Covariates considered for inclusion in the model were delivery mode, sex, gestational age, birthweight, birth length, history of chorioamnionitis, infant exposure to antibiotics in the first week of life, mother’s marital status, preeclampsia, and gravida. In Cincinnati, extraction protocol and thioglycollate storage protocol were also considered. Race was not included in the model because there were no differences in race between cases and controls in Birmingham, and in Cincinnati controls had only one more infant identified as black than did controls. Since the number of subjects included at each site was small, the models of Cincinnati data were limited to 4 covariates at a time and the models of Birmingham data were limited to two covariates at a time. All models are reported for sites and time points with multiple models possible.
Finally, we analyzed the most common cause of LOS, coagulase-negative Staphylococcus (CONS), compared to all controls to determine whether LOS caused by a single organism has a distinct form of dysbiosis. No other cause of LOS was numerous enough to analyze as a subgroup.
Results
Study subjects
Included for study were 13 infants in Birmingham, AL and 20 infants in Cincinnati, OH who developed LOS and had at least one successfully sequenced stool sample that was collected prior to disease onset. Also included were an equal number of control infants who did not develop LOS. Controls were frequency matched to cases by site and gestational age. LOS cases and controls were well matched (Table 1) and we noted no significant differences between cases and controls at either site in any clinical or demographic variables. In Birmingham, the mean birth weight was 736 grams for infant with LOS and 804 grams for control infants. In Cincinnati, the mean birth weight was 795 grams for infant with LOS and 851 grams for controls. The median gestational age at birth was 24 weeks for Birmingham cases, 25 weeks for Birmingham controls and Cincinnati cases, and 25.5 weeks for Cincinnati controls. However, potentially important differences were noted between sites: Intrapartum antibiotics were given to 92% of Birmingham mothers, whereas in Cincinnati, intrapartum antibiotics were given to 75% of mothers. Infant antibiotic use was significantly higher in Birmingham than Cincinnati (p = 0.051, Kruskal-Wallis test). Furthermore, the rate of infant formula use was lower in Cincinnati (where none received formula prior to day of life 15) compared to Birmingham (p = 0.005, Fisher’s exact test).
Table 1. Comparison of cases and controls in Birmingham and Cincinnati.
| Characteristic | Birmingham | Cincinnati | ||
|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |
| n = 13 | n = 13 | n = 20 | n = 20 | |
| Multiple Birth | 3 (23%) | 2 (15%) | 8 (40%) | 5 (25%) |
| C-section | 5 (38%) | 6 (46%) | 11 (55%) | 11 (55%) |
| Sex, female | 7 (54%) | 7 (54%) | 7 (35%) | 11 (55%) |
| Hispanic | 1 (8%) | 0 (0%) | 2 (10%) | 0 (0%) |
| Race, Black, other | 7 (54%), 0 (0%) | 7 (54%), 0 (0%) | 7(35%), 0(0%) | 9(45%), 1(5%) |
| Maternal antibiotics given | 12 (92%) | 12 (92%) | 17 (85%) | 13 (65%) |
| Infant birthweight (mean), g | 736 | 804 | 795 | 851 |
| Infant gestation, weeks (median) | 24 | 25 | 25 | 25.5 |
| Days on antibiotics in first 14 (median)* | 6 | 8 | 6 | 7 |
| Any formula prior to last included stool sample** | 5 (38%) | 4 (31%) | 2 (10%) | 0 (0%) |
Comparing all subjects between sites:
*p = 0.05;
**p = 0.005
Twelve (92%) of 13 infants with LOS in Birmingham had Gram-positive sepsis (median day of life 13 for sepsis onset) and 1 had Gram-negative sepsis on day of life 12. Twelve (60%) of 20 infants with LOS in Cincinnati had Gram-positive sepsis (median day of life 16) and 6 (30%) had Gram-negative sepsis (median day of life 33), while 2 (10%) infants were culture positive for both Gram-positive and Gram-negative organisms at the time sepsis was diagnosed (Table 2). To examine the concordance between the clinical LOS isolate and the presence of a matching organism in infant stool, we utilized whole genome sequencing data that was available for samples prior to sepsis in 17 of the 33 cases. If a species-level match was identified from the stool metagenome, we considered it a match to the clinical isolate. For example, if the causative organism was identified as coagulase negative Staphylococcus, we considered a match to occur if the stool metagenome indicated the presence of at least one species of coagulase negative Staphylococcus. If no information below the genus level was available from the clinical laboratory, we considered a match to have occurred if DNA from at least one species belonging to that genus was detected in stool. Of these 17 cases, 14 (82%) had at least 1 stool sample with identifiable abundance of the causative organism as identified by the clinical laboratory; results matched to the lowest taxonomic level for which clinical and stool data were available (genus or species, Table 2). The three infants whose clinical isolate was not matched by identifying the same organism in stool sample had LOS caused by Klebsiella pneumoniae, Group B Streptococcus, or Staphylococcus aureus.
Table 2. Sepsis cause, day of life of blood collection, and the number of stool samples collected prior to sepsis.
| Case No. | City | Causative Organisms identified by clinical laboratories | Day of life of blood culture | No. samples prior to sepsis | Closest related organism identified in WGS data | Scored as match to cultured isolate^ |
|---|---|---|---|---|---|---|
| 1 | C | Coagulase negative Staphylococcus | 13 | 1 | Staphylococcus epidermidis | Yes |
| 8 | C | Coagulase negative Staphylococcus | 10 | 3 | Staphylococcus epidermidis | Yes |
| 9 | C | Coagulase negative Staphylococcus | 20 | 3 | Staphylococcus epidermidis | Yes |
| 11 | C | Coagulase negative Staphylococcus | 13 | 2 | Staphylococcus epidermidis Staphylococcus hominis | Yes |
| 12 | C | Coagulase negative Staphylococcus | 29 | 2 | Staphylococcus epidermidis Staphylococcus warneri | Yes |
| 16 | C | Coagulase negative Staphylococcus | 13 | 2 | Staphylococcus epidermidis | Yes |
| 22 | B | Coagulase negative Staphylococcus | 9 | 1 | Staphylococcus epidermidis | Yes |
| 33 | B | Enterococcus faecalis | 21 | 2 | Enterococcus faecalis | Yes |
| 21 | B | Streptococcus viridans | 19 | 2 | Streptococcus thermophilus | Yes * |
| 5 | C | Diphtheroid bacilli | 19 | 1 | Corynebacterium striatum Corynebacterium pyruviciproducens | Yes |
| 2 | C | Enterobacter cloacae | 27 | 3 | Enterobacter cloacae | Yes |
| 13 | C | Escherichia coli | 20 | 5 | Escherichia coli | Yes |
| 15 | C | Escherichia coli | 35 | 3 | Escherichia coli | Yes |
| 4 | C | Klebsiella | 41 | 4 | Klebsiella unclassified | Yes |
| 14 | C | Staphylococcus aureus | 6 | 1 | Staphylococcus epidermidis | No |
| 7 | C | Group B Streptococcus | 27 | 1 | Streptococcus salivarius | No |
| 3 | C | Klebsiella pneumoniae | 31 | 4 | Klebsiella oxytoca Klebsiella unclassified | No |
| No WGS data available prior to sepsis | ||||||
| 6 | C | Serratia marcescens Enterococcus faecium Enterococcus faecalis | 64 | 1 | ||
| 10 | C | Group B Streptococcus | 28 | 2 | ||
| 17 | C | Group B Streptococcus | 72 | 1 | ||
| 18 | C | Escherichia coli | 38 | 1 | ||
| 19 | C | Pseudomonas Coagulase negative StaphylococcusGroup B Streptococcus | 27 | 2 | ||
| 20 | C | Coagulase negative Staphylococcus | 13 | 1 | ||
| 23 | B | Enterococcus faecalis | 18 | 2 | ||
| 24 | B | Coagulase negative Staphylococcus | 9 | 1 | ||
| 25 | B | Escherichia coli | 12 | 2 | ||
| 26 | B | Coagulase negative Staphylococcus | 10 | 1 | ||
| 27 | B | Enterococcus faecalis | 14 | 2 | ||
| 28 | B | Staphylococcus aureus (methicillin resistant) | 9 | 1 | ||
| 29 | B | Coagulase negative Staphylococcus | 40 | 2 | ||
| 30 | B | Staphylococcus aureus (methicillin resistant) | 105 | 2 | ||
| 31 | B | Coagulase negative Staphylococcus | 11 | 1 | ||
| 32 | B | Coagulase negative Staphylococcus | 12 | 2 | ||
*Streptococcus viridans is a group of organisms that includes Streptococcus thermophilus.
^WGS results were scored as a match to the clinical isolate if the same taxonomy was identified at the species level, or if the species identified by WGS was an member of the genus noted clinically when clinical results were not conclusive at the species level.
Microbial Diversity
To examine the microbial community pattern prior to LOS, we examined alpha and beta diversity in the first sample after birth and the last sample available prior to LOS onset.
First Sample Analysis
The timing of first sample did not differ significantly between cases and controls within or between Birmingham and Cincinnati sites. For Birmingham infants, the median sample collection day of life was day 7 (range, 4–10) for LOS cases and day 8 (range, 6–12) for controls; the median number of days from first sample collection day until sepsis onset was 8 days (range, 1–96). For Cincinnati infants, the median sample collection day of life was day 8 (range, 3–20) for LOS cases and day 8 (range, 4–21) for controls; the median number of days from first sample collection day until sepsis onset was 18 days (range was 2–56).
In Birmingham, cases had a significantly (p<0.02) lower alpha diversity than controls by both the Simpson and Chao1 metrics. In Cincinnati, however, there were no significant differences by either alpha diversity index between cases and controls. For both Birmingham and Cincinnati analyses, there was no clustering of cases or controls in the non-metric multidimensional scaling ordinations, whether using weighted or unweighted UniFrac measures (data not shown.)
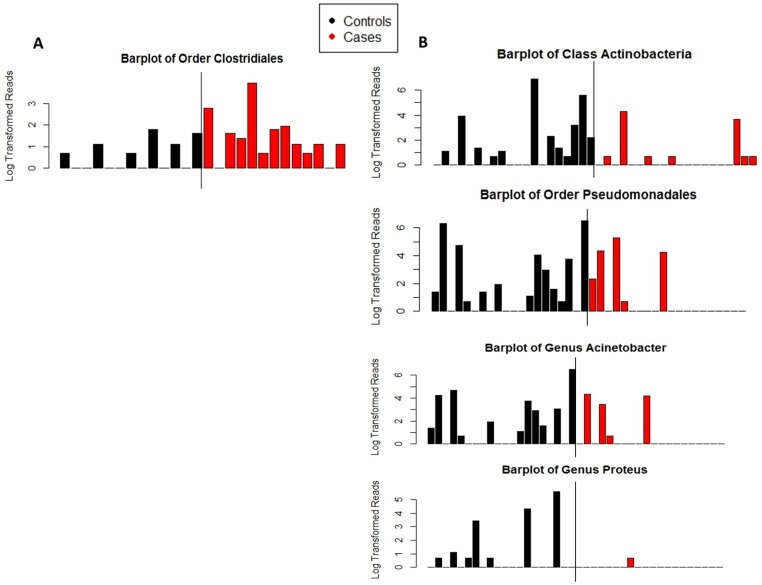
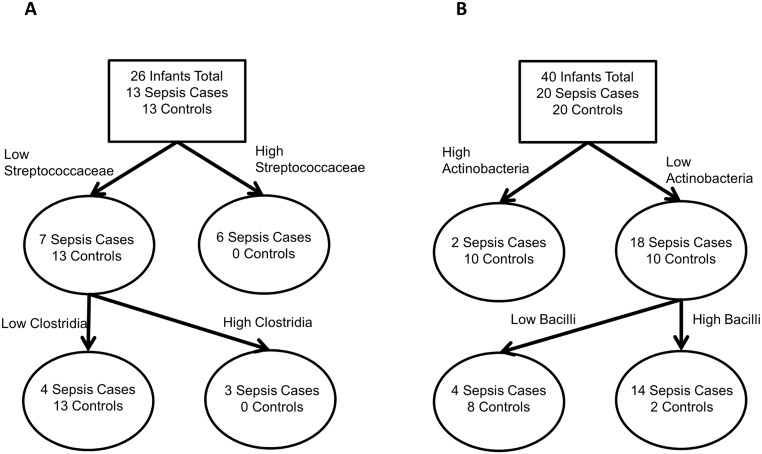
For Birmingham samples, LEfSe analysis indicated a significant association of Clostridia and Clostridiales abundance in infants who later developed LOS compared to control infants (Fig 1A). Clostridia and Clostridiales more frequently colonized cases and were present in higher abundance. The classification tree model confirmed that Clostridia differed between cases and controls (cases were more likely to have greater than 0.3% relative abundance of Clostridia), but also found a signal from Streptococcaceae (cases were more likely to have greater than 0.1% relative abundance of Streptococcaceae (Fig 2A). The misclassification rate of the tree was 15%. However, logistic regression models did not find a significant association between the cut points identified in the classification tree and sepsis. The final model did not include any other covariates, all dropped out of the model as non-significant (model not shown).
Fig 1. Barplots of taxa identified as differential abundant in cases and controls.
Y-axis was transformed by taking the base ten logarithm of the number of reads plus one. Samples were rarefied to 2000 reads per sample prior to creating the plots. A) Taxa differentially abundant by LEfSe in the first sample analysis in Birmingham. Median number of reads of Clostridiales in controls was 0, range was 0 to 5. Median number of reads of Clostridiales in cases was 2, the range was 0 to 50. Class Clostridia was also significantly different between cases and controls, it is not shown here because class Clostridia and order Clostridiales had a correlation of 1. B) Taxa differentially abundant by LEfSe in the first sample analysis in Cincinnati. Median number of reads of class Actinobacteria in controls was 1.5, range was 0 to 985. Median number of reads of class Actinobacteria in cases was 0, range was 0 to 74. Median number of reads of Pseudomonadales in controls was 2.5, range was 0 to 661. Median number of reads of Pseudomonadales in cases was 0, range was 0 to 193. Median number of reads of Acinetobacter in controls was 1.5, range was 0 to 660. Median number of reads of Acinetobacter in cases was 0, range was 0 to 75. Median number of reads of Proteus in controls was 0, range was 0 to 267. Median number of reads of Proteus in cases was 0, range was 0 to 1, Phyla Actinobacteria was also significantly different between cases and controls, it is not shown here because it has a correlation 0.9996 with class Actinobacteria and was visually indistinguishable. Family Moraxellaceae was also significantly different between cases and controls; it is not shown here because it had a correlation of 1 with genus Acinetobacter.
Fig 2. First sample classification tree results.
(A) Birmingham results, showing infants colonized with Streptococcaceae and infants with higher levels of Clostridia are at greater risk of sepsis. (B) Cincinnati results, showing that infants colonized with greater relative abundance of Actinobacteria and infants with lower relative abundance of Bacilli are protected from sepsis.
For Cincinnati samples, LEfSe analysis indicated a statistically significant association between higher levels of phylum Actinobacteria and genus Proteus in control infants compared to LOS infants. In addition, control infants were more likely to be colonized with order Pseudomonadales, family Moraxellaceae, and genus Acinetobacter (Fig 1B). As with the Birmingham samples, examination of the differential feature plot (not shown) found the differences observed were not driven by outliers. The classification tree confirmed that phylum Actinobacteria differed between cases and controls, and detected a further signal. In the subset of infants lacking Actinobacteria, the classification tree found that infants whose stool samples contained at least 3% Bacilli by relative abundance (65 of 2000 rarefied reads) were more likely to be cases than were infants whose stool samples had fewer reads (Fig 2B). The misclassification rate of the tree in Cincinnati was 20%. Logistic regression was again used to determine the significance of the cut points identified in the classification tree at p<0.05; only the Actinobacteria cut point was significant. All covariates dropped out of the model as non-significant (Table 3).
Table 3. Logistic modeling results of LOS by site using cut points defined by classification trees.
| Model | Other covariates+ | Independent Variables | beta | p-value |
|---|---|---|---|---|
| Cincinnati—first sample | none | Actinobacteria > = 0.1% | -2.67 | 0.018 |
| Bacilli < 3.25% | -1.39 | 0.39 | ||
| Cincinnati—last sample | Gravida (p = 0.098) | Pseudomonadales > = 0.1% | -2.65 | 0.029 |
| Clostridiaceae > = 0.35% | 2.43 | 0.030 | ||
| Proteobacteria < 72.3% | -2.59 | 0.011 | ||
| Birmingham—last sample Model 1 | Infant GA (p = 0.087) | Lactobacillales present | -3.6 | 0.017 |
| Birmingham—last sample Model 2 | Chorioamnionitis (p = 0.070) | Lactobacillales present | -3.2 | 0.010 |
Modeling confirmed the statistical significance of cut points identified from classification tree analysis except for the Birmingham first sample tree (not shown) and the unknown Bacillales in the Birmingham last sample tree.
+Covariates that remained in each model are noted along with their p-value.
Last Sample Analysis
There were no significant differences in day of life of sample collection in either Birmingham or Cincinnati. In both Birmingham and Cincinnati samples, one-third of samples included in the last sample analysis were also included in the first sample analysis. In Birmingham, the median sample collection day of life was 10 days (range, 4–14) for both LOS cases and controls; the median number of days from last sample collection day of life until sepsis onset was 4 days (range, 1–91). In Cincinnati, the median sample collection day of life was 13 days (range, 4–21) for both cases and controls; the median number of days from last sample collection day of life until sepsis onset was 8 days (range, 1–56). In Birmingham, cases had a significantly (p<0.001) lower alpha diversity than controls by the Simpson index but no significant difference by the Chao1 index. As in the first sample analysis, there were no significant differences between cases and controls in alpha diversity by either metric in Cincinnati. There was no visible separation between cases and controls on the non-metric multidimensional scaling ordinations using either UniFrac metric at either site (data not shown.)
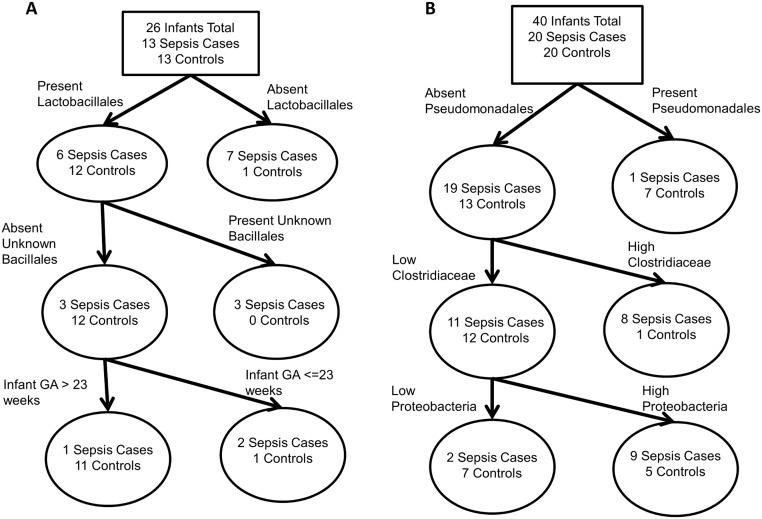
For Birmingham, LEfSe did not detect significant differences in microbial colonization above the OTU level between cases and controls. The OTUs enriched in both cases and controls were almost entirely classified as Enterobacteriaceae (data not shown). A classification tree found that the presence or absence of Lactobacillales, the presence or absence of an OTU of Bacillales of unknown family, and infant gestational age less than or equal to 23 weeks were the best predictors of sepsis status (Fig 3A). The misclassification rate was 12%. Logistic regression confirmed the significance of the presence of Lactobacillales as protective against sepsis, but the unknown Bacillales had a p-value >0.99 in all possible models and so was excluded. Infant gestational age (model 1) and chorioamnionitis (model 2) were borderline significant (Table 3).
Fig 3. Last sample classification tree results for (A) Birmingham showing infants without Lactobacillales, with the unknown OTU of Bacillales, or born at younger gestational ages were more at risk of sepsis and (B) Cincinnati showing that infants with high levels of Enterobacteriales or high levels of Firmicutes were at increased risk of sepsis.
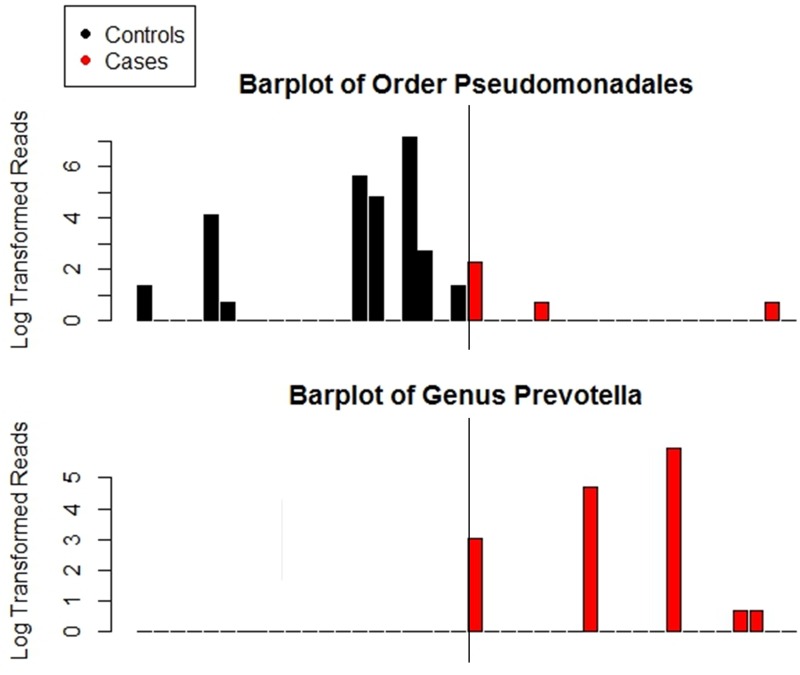
For Cincinnati, we observed a significant association of Prevotellaceae, and Prevotella with cases and Pseudomonadales with controls (Fig 4). Examination of the differential feature plots revealed that Prevotella and Prevotellaceae were detected in 5 (25%) of the cases and never in controls. Pseudomonadales was present in in only 3 (15%) cases and 8 (40%) controls. A classification tree agreed with the Pseudomonadales finding, splitting first on Pseudomonadales but then split on Clostridiaceae and Proteobacteria instead of either Prevotellaceae or Prevotella (Fig 3B). The misclassification rate was 23%. Logistic regression confirmed that the cut points created by the classification tree were significant predictors of sepsis after controlling for the borderline significant covariate, gravida (Table 3).
Fig 4. Barplots of taxa identified as differential abundant in cases and controls at genus level and higher in Cincinnati.
Y-axis was transformed by taking the base ten logarithm of the number of reads plus one. Samples were rarefied to 2000 reads per sample prior to creating the plots. Median number of reads of Pseudomonadales in controls was 0, range was 0 to 1251. Median number of reads of Pseudomonadales in cases was 0, range was 0 to 9. No control samples had reads of Prevotella. Median number of reads of Prevotella in cases was 0, range was 0 to 384. Family Prevotellaceae was also significantly different between cases and controls, it is not shown here because it had a correlation of 1 with genus Prevotella. All other differences identified by LEfSe were at the OTU level. All differences identified by LEfSe in the Birmingham last sample analysis were at the OTU level.
Cause-specific analysis of LOS dysbiosis
To examine the possibility that dysbiosis prior to sepsis is influenced by the causative organism, we focused on coagulase negative Staphylococcus (CONS), which constituted 42% of all LOS cases in our study, and was the most abundant causative organism. In the first sample after birth, the Birmingham CONS cases exhibited higher levels of Actinobacteria than in controls; but no other differences were identified between the groups. Whereas, analysis of first samples of Cincinnati CONS cases found lower Actinobacteria in cases than controls (S3 Fig) Analysis of last samples of Cincinnati CONS cases versus controls demonstrated enrichment in Prevotella and Prevotellaceae. Overall, the stool microbiome patterns identified with CONS cases are similar to those found for LOS cases overall. Staphylococcaceae were not found more abundantly in infant stool samples prior to CONS-associated sepsis.
Analysis using whole genome sequencing
Though we had whole genome sequencing available only for a subset of Cincinnati LOS cases and controls, we examined the WGS data to determine consistency with 16S results. For the first sample analysis, we obtained WGS data for 12 LOS and 15 controls. Controls were enriched in Actinomycetales (an order of phylum Actinobacteria), Streptococcus, and Streptococcaceae. Actinomycetales enrichment is consistent with the 16S results showing enrichment of phylum Actinobacteria. WGS evidence of Streptococcus enrichment was not identified in the 16S data; it is, however, consistent with the 16S classification tree results insofar as Streptococcus belongs to class Bacilli. For the last sample analysis, WGS data were available for 10 LOS and 11 control samples. Similar to the 16S analysis, we found the Prevotella and Prevotellaceae enriched in cases compared to controls. Altogether, while 16S and WGS results were not identical, they were generally concordant.
Discussion
Late onset sepsis in preterm infants is associated with very high mortality [20] and disproportionately high morbidities among survivors [21–23], including poor neurodevelopmental outcomes [3]. While the source of microbes causing LOS is not well established, reports suggest that the gut is often the source of microbes causing LOS in preterm neonates [7]. We used high-throughput sequencing to address the relationship between gut colonization and neonatal LOS. Consistent with previous reports, we identified a concordance of 82% between the causative LOS organism and its identification in stool samples prior to LOS; previous studies have reported concordance ranging from 64% [7] to 95% [6]. In this and an earlier publication [16], we have reported differences in colonization between our two study sites [16], which was the basis for analyzing the gut microbiota at each site separately. Site-specific analyses identified differences in microbial-community patterns in advance of LOS compared to controls. In Cincinnati, we found lower levels of Actinobacteria in early samples from infants who later developed LOS compared to controls, similar to a report from Mai et al, in which lower levels of Bifidobacteria, a genus of the phylum Actinobacteria, were observed in infants who later developed LOS [9]. In Birmingham, we found lower microbial-diversity in infants with LOS compared to controls, similar to reports by Madan et al. [8] and Mai et al. [9]. In Birmingham, the presence of Lactobacillales in the last sample was associated with protection against LOS, whereas in Cincinnati, the presence of Lactobacillales in the first sample, but not the last sample, was associated with protection against LOS. Thus, while we found significant site-specific differences in gut-microbial patterns in advance of LOS, we failed to detect a specific and generalizable gut-microbial colonization pattern in advance of LOS. Furthermore, the observed differences were dependent not only on site, but also on the timing of sample collection. Previous studies at single sites have reported associations with gut microbiota prior to the onset of sepsis, but also like the current study, the reported associations are not consistent between sites: One study found an association between increased abundance of Staphylococcaceae [10] and sepsis, while another found an association between decreased abundance of Clostridia and sepsis [8]; and some, but not all studies reported lower alpha diversity in sepsis cases [8, 9].
What could explain the site-specific differences in the gut-microbial pattern in advance of LOS? We sought to minimize the potential for chance associations by reporting only significant differences found at the bacterial family level in most cases and genus level in cases where OTUs were successfully classified at the genus level. Acquisition of intestinal microbiota, which begins immediately after birth [24], is impacted by gestational age [25], delivery mode [26], the use of antibiotics [27], feeding mode (breast milk versus formula) [28], the neonate’s genetic background [29], and the circulating microbiota of the environment [16]. In this study, we used site-, age-, birth weight- and sex-matched-controls thus minimizing the impact of confounding variables. However, the sites did significantly differ in their use of infant formula and the use of antibiotics (both were higher in Birmingham). The failure to find a consistent form of intestinal dysbiosis in prior to LOS between the sites could thus be due to site differences in initiating and advancing feeds, use of breast milk and use of antibiotics. Differences in the environmental microbiota could also potentially explain site-specific forms of dysbiosis.
Preterm infants undergo frequent and abrupt shifts in microbial community composition [15]. We analyzed two time points—the first collected sample after delivery and last sample prior to LOS onset—which identified different associations with LOS, similar to findings reported by Mai et al. [9]. We therefore speculate that to observe consistent within and between-site differences between cases and controls, more frequent (e.g., daily) sampling may be needed.
The sepsis-causing organism is frequently detectable in stool prior to disease onset but such pathogens are not typically found in infants who remain healthy [7]. We speculate that these pathogens are able to colonize the intestinal tract due to dysbiosis of the microbiome, and that these distortions vary by pathogen. Therefore, we compared stool from control infants to that of infants with LOS due to coagulase negative staphylococcus, the most common cause of neonatal LOS in our study. These sepsis-associated microbes did not occur more abundantly in infant stool samples in advance of LOS compared to controls. We posit that microbial-composition of the stool samples may not accurately represent the adherent gut microbial composition in advance of sepsis. Indeed, several groups have demonstrated that stool microbiota correlates poorly with the adherent microbiota identified by intestinal biopsy of specific intestinal sites [30, 31]. As the microbes that translocate to cause disease are likely to be adherent [32], analysis of stool samples may not adequately reflect the relevant adherent microbes and their activity.
LOS has a diverse etiology, with coagulase negative Staphylococcus and Gram-negative microbes such as E. coli accounting for majority of the cases. This complexity in etiology presents a substantial challenge to our understanding of LOS and underscores the inherent heterogeneity in how the LOS is defined in preterm infants. It is suggested that LOS due to coagulase negative Staphylococcus originates from the skin/indwelling catheters [33], while Gram negative LOS is thought to reflect the microbial translocation across the gut epithelium. However, our data suggest that LOS due to coagulase negative Staphylococcus may in some cases originate in the gut. Nevertheless, LOS due to Gram negative bacteria reflects a distinct group, with different etiology and pathobiology [34], as compared to LOS due to coagulase negative Staphylococcus.
In conclusion, our findings support the hypothesis that neonatal LOS is frequently caused by organisms that translocate from the intestinal tract, as 82% of LOS causative organisms were identified in infant stool samples prior to LOS onset. Within each center, we found differences in infant microbial communities preceding LOS compared to control infants. However, the differences in microbiota between cases and controls were not consistent between the sites. Our data suggest that factors besides intestinal microbial composition may underlie the risk of LOS. As yet unexplored in the context of LOS is dysfunction or dysregulation of the microbiota, or most likely, of host-microbial interactions. Such functional studies are key to advancing the understanding of the microbiota prior to sepsis.
Supporting Information
Red dots indicate samples from Cincinnati infants and black dots indicate samples from Birmingham infants. Panel A) All f (both cases and controls) from the first sample analysis. Panel B) First samples from controls only. Panel C) All subjects (cases and controls) from the last sample analysis. Panel D) Last samples from controls only. There is no clear separation of samples from the two sites in either the first or the last sample analysis.
(TIF)
Panel A) All first samples (cases and controls) were included in the analysis. Cincinnati infants had higher levels of Lactobacillales and Enterococcaceae among others. Panel B) All last samples (cases and controls) were included in the analysis. Birmingham infants had higher levels of Bacteroidetes among others.
(TIF)
A) Taxa identified as different between Birmingham coagulase negative Staphylococcus cases and controls. Phyla Actinobacteria was also significantly different between cases and controls, it is not shown here because it has a correlation 0.9996 with class Actinobacteria and was visually indistinguishable. Family Corynebacteriaceae was also significantly different, it is not shown here because it has a correlation of 1 with genus Corynebacterium. Family Moraxellaceae was also significantly different between cases and controls, it is not shown here because it had a correlation of 1 with genus Acinetobacter. B) Taxa identified as different between Cincinnati coagulase negative Staphylococcus cases and controls. Phyla Actinobacteria was also significantly different between cases and controls, it is not shown here because it has a correlation 0.9996 with class Actinobacteria and was visually indistinguishable. Family Corynebacteriaceae was also significantly different, it is not shown here because it has a correlation of 1 with genus Corynebacterium. Family Moraxellaceae was also significantly different between cases and controls, it is not shown here because it had a correlation of 1 with genus Acinetobacter.
(TIF)
(CSV)
File contains variable name and description of variable for all variables included in the metadata file.
(CSV)
(XLSX)
Acknowledgments
We gratefully acknowledge Estelle Fischer and the Perinatal Institute clinical research core for clinical data collection and data entry, Barbara Davidson for sample collection planning and management oversight, Myra Johnson, Amy LeFevers, Maria Hughes for sample collection and management, and Donna Wuest for assistance with manuscript preparation. This project has been funded in whole or in part with Federal funds from the National Institute of Child Health and Human Development, National Institutes of Health (grant numbers R01HD059140, P01HD13021, and HD27853); National Center for Research Resources, National Institutes of Health (grant numbers 5UL1RR026314-03, U54HG004969); National Human Genome Research Institute, National Institutes of Health (HG005969); Danone Research (PLF-5972-GD); National Institute of Environmental Health Sciences, National Institutes of Health (T32 ES 10957–11); and the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200900018C.
Data Availability
Data is available on NCBI SRA, Accession PRJNA63661. Additional metadata and the metadata dictionary are available as a supplementary file to this article. This file contains all the metadata used in this article. The OTU table and taxa assignments after rarefaction are also available as supplementary file to this article. Whole genome sequencing data is deposited under the NCBI bioproject ID 63661.
Funding Statement
This project has been funded in whole or in part with Federal funds from the National Institute of Child Health and Human Development, National Institutes of Health (grant numbers R01HD059140, P01HD13021, and HD27853); National Center for Research Resources, National Institutes of Health (grant numbers 5UL1RR026314-03, U54HG004969); National Human Genome Research Institute, National Institutes of Health (HG005969); Danone Research (PLF-5972-GD); National Institute of Environmental Health Sciences, National Institutes of Health (T32 ES 10957-11); and the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200900018C. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Boghossian NS, Page GP, Bell EF, Stoll BJ, Murray JC, Cotten CM, et al. Late-onset sepsis in very low birth weight infants from singleton and multiple-gestation births. The Journal of pediatrics. 2013;162(6):1120–4, 4 e1 Epub 2013/01/18. 10.1016/j.jpeds.2012.11.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hornik CP, Fort P, Clark RH, Watt K, Benjamin DK Jr., Smith PB, et al. Early and late onset sepsis in very-low-birth-weight infants from a large group of neonatal intensive care units. Early human development. 2012;88 Suppl 2:S69–74. Epub 2012/05/29. 10.1016/s0378-3782(12)70019-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stoll BJ, Hansen NI, Adams-Chapman I, Fanaroff AA, Hintz SR, Vohr B, et al. Neurodevelopmental and growth impairment among extremely low-birth-weight infants with neonatal infection. JAMA: the journal of the American Medical Association. 2004;292(19):2357–65. Epub 2004/11/18. 10.1001/jama.292.19.2357 . [DOI] [PubMed] [Google Scholar]
- 4. Ng PC, Lam HS. Biomarkers for late-onset neonatal sepsis: cytokines and beyond. Clinics in perinatology. 2010;37(3):599–610. Epub 2010/09/04. 10.1016/j.clp.2010.05.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaplan HC, Lannon C, Walsh MC, Donovan EF. Ohio statewide quality-improvement collaborative to reduce late-onset sepsis in preterm infants. Pediatrics. 2011;127(3):427–35. Epub 2011/02/23. 10.1542/peds.2010-2141 . [DOI] [PubMed] [Google Scholar]
- 6. Graham PL 3rd, Della-Latta P, Wu F, Zhou J, Saiman L. The gastrointestinal tract serves as the reservoir for Gram-negative pathogens in very low birth weight infants. The Pediatric infectious disease journal. 2007;26(12):1153–6. Epub 2007/11/29. 10.1097/INF.0b013e31814619d4 . [DOI] [PubMed] [Google Scholar]
- 7. Carl MA, Ndao IM, Springman AC, Manning SD, Johnson JR, Johnston BD, et al. Sepsis from the gut: the enteric habitat of bacteria that cause late-onset neonatal bloodstream infections. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2014;58(9):1211–8. Epub 2014/03/22. 10.1093/cid/ciu084 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Madan JC, Salari RC, Saxena D, Davidson L, O'Toole GA, Moore JH, et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Archives of disease in childhood Fetal and neonatal edition. 2012;97(6):F456–62. Epub 2012/05/09. 10.1136/fetalneonatal-2011-301373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mai V, Torrazza RM, Ukhanova M, Wang X, Sun Y, Li N, et al. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PloS one. 2013;8(1):e52876 Epub 2013/01/24. 10.1371/journal.pone.0052876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stewart CJ, Marrs EC, Magorrian S, Nelson A, Lanyon C, Perry JD, et al. The preterm gut microbiota: changes associated with necrotizing enterocolitis and infection. Acta paediatrica (Oslo, Norway: 1992). 2012;101(11):1121–7. Epub 2012/08/01. 10.1111/j.1651-2227.2012.02801.x . [DOI] [PubMed] [Google Scholar]
- 11. Ley RE. Harnessing microbiota to kill a pathogen: the sweet tooth of Clostridium difficile. Nature medicine. 2014;20(3):248–9. Epub 2014/03/08. 10.1038/nm.3494 . [DOI] [PubMed] [Google Scholar]
- 12. Morrow AL, Lagomarcino AJ, Schibler KR, Taft DH, Yu Z, Wang B, et al. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome. 2013;1(1):13 Epub 2014/01/24. 10.1186/2049-2618-1-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Torrazza RM, Ukhanova M, Wang X, Sharma R, Hudak ML, Neu J, et al. Intestinal microbial ecology and environmental factors affecting necrotizing enterocolitis. PloS one. 2013;8(12):e83304 Epub 2014/01/05. 10.1371/journal.pone.0083304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Candela M, Maccaferri S, Turroni S, Carnevali P, Brigidi P. Functional intestinal microbiome, new frontiers in prebiotic design. International journal of food microbiology. 2010;140(2–3):93–101. Epub 2010/05/18. 10.1016/j.ijfoodmicro.2010.04.017 . [DOI] [PubMed] [Google Scholar]
- 15. La Rosa PS, Warner BB, Zhou Y, Weinstock GM, Sodergren E, Hall-Moore CM, et al. Patterned progression of bacterial populations in the premature infant gut. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(34):12522–7. Epub 2014/08/13. 10.1073/pnas.1409497111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taft D, Ambalavanan N, Schibler K, Yu Z, Newburg D, Ward D, et al. Intestinal microbiota of preterm infants differ over time and between hospitals. Microbiome. 2014;2(1):36 10.1186/2049-2618-2-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nature methods. 2012;9(8):811–4. Epub 2012/06/13. 10.1038/nmeth.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12(6):R60 Epub 2011/06/28. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Therneau T, Atkinson B, Ripley B. rpart: Recursive Partitioning2011.
- 20. Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, et al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics. 2002;110(2 Pt 1):285–91. Epub 2002/08/08. . [DOI] [PubMed] [Google Scholar]
- 21. Silveira RC, Procianoy RS, Dill JC, da Costa CS. Periventricular leukomalacia in very low birth weight preterm neonates with high risk for neonatal sepsis. Jornal de pediatria. 2008;84(3):211–6. Epub 2008/04/22. 10.2223/JPED.1777 . [DOI] [PubMed] [Google Scholar]
- 22. Polin RA. Systemic infection and brain injury in the preterm infant. Jornal de pediatria. 2008;84(3):188–91. Epub 2008/06/07. 10.2223/JPED.1784 . [DOI] [PubMed] [Google Scholar]
- 23. Shah DK, Doyle LW, Anderson PJ, Bear M, Daley AJ, Hunt RW, et al. Adverse neurodevelopment in preterm infants with postnatal sepsis or necrotizing enterocolitis is mediated by white matter abnormalities on magnetic resonance imaging at term. The Journal of pediatrics. 2008;153(2):170–5, 5 e1 Epub 2008/06/07. 10.1016/j.jpeds.2008.02.033 . [DOI] [PubMed] [Google Scholar]
- 24. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS biology. 2007;5(7):e177 Epub 2007/06/28. 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schwiertz A, Gruhl B, Lobnitz M, Michel P, Radke M, Blaut M. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatric research. 2003;54(3):393–9. Epub 2003/06/06. 10.1203/01.pdr.0000078274.74607.7a . [DOI] [PubMed] [Google Scholar]
- 26. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(26):11971–5. Epub 2010/06/23. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Greenwood C, Morrow AL, Lagomarcino AJ, Altaye M, Taft DH, Yu Z, et al. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of Enterobacter. The Journal of pediatrics. 2014;165(1):23–9. Epub 2014/02/18. 10.1016/j.jpeds.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. Journal of pediatric gastroenterology and nutrition. 2000;30(1):61–7. Epub 2000/01/12. . [DOI] [PubMed] [Google Scholar]
- 29. Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nature reviews Microbiology. 2011;9(4):279–90. Epub 2011/03/17. 10.1038/nrmicro2540 . [DOI] [PubMed] [Google Scholar]
- 30. Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell host & microbe. 2014;15(3):382–92. Epub 2014/03/19. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. American journal of physiology Gastrointestinal and liver physiology. 2012;303(6):G675–85. Epub 2012/07/24. 10.1152/ajpgi.00152.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakayama M, Yajima M, Hatano S, Yajima T, Kuwata T. Intestinal adherent bacteria and bacterial translocation in breast-fed and formula-fed rats in relation to susceptibility to infection. Pediatric research. 2003;54(3):364–71. Epub 2003/06/06. 10.1203/01.pdr.0000077482.28990.2d . [DOI] [PubMed] [Google Scholar]
- 33. Clarke DE, Raffin TA. Infectious complications of indwelling long-term central venous catheters. Chest. 1990;97(4):966–72. Epub 1990/04/01. . [DOI] [PubMed] [Google Scholar]
- 34. Sriskandan S, Cohen J. Gram-positive sepsis. Mechanisms and differences from gram-negative sepsis. Infectious disease clinics of North America. 1999;13(2):397–412. Epub 1999/05/26. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Red dots indicate samples from Cincinnati infants and black dots indicate samples from Birmingham infants. Panel A) All f (both cases and controls) from the first sample analysis. Panel B) First samples from controls only. Panel C) All subjects (cases and controls) from the last sample analysis. Panel D) Last samples from controls only. There is no clear separation of samples from the two sites in either the first or the last sample analysis.
(TIF)
Panel A) All first samples (cases and controls) were included in the analysis. Cincinnati infants had higher levels of Lactobacillales and Enterococcaceae among others. Panel B) All last samples (cases and controls) were included in the analysis. Birmingham infants had higher levels of Bacteroidetes among others.
(TIF)
A) Taxa identified as different between Birmingham coagulase negative Staphylococcus cases and controls. Phyla Actinobacteria was also significantly different between cases and controls, it is not shown here because it has a correlation 0.9996 with class Actinobacteria and was visually indistinguishable. Family Corynebacteriaceae was also significantly different, it is not shown here because it has a correlation of 1 with genus Corynebacterium. Family Moraxellaceae was also significantly different between cases and controls, it is not shown here because it had a correlation of 1 with genus Acinetobacter. B) Taxa identified as different between Cincinnati coagulase negative Staphylococcus cases and controls. Phyla Actinobacteria was also significantly different between cases and controls, it is not shown here because it has a correlation 0.9996 with class Actinobacteria and was visually indistinguishable. Family Corynebacteriaceae was also significantly different, it is not shown here because it has a correlation of 1 with genus Corynebacterium. Family Moraxellaceae was also significantly different between cases and controls, it is not shown here because it had a correlation of 1 with genus Acinetobacter.
(TIF)
(CSV)
File contains variable name and description of variable for all variables included in the metadata file.
(CSV)
(XLSX)
Data Availability Statement
Data is available on NCBI SRA, Accession PRJNA63661. Additional metadata and the metadata dictionary are available as a supplementary file to this article. This file contains all the metadata used in this article. The OTU table and taxa assignments after rarefaction are also available as supplementary file to this article. Whole genome sequencing data is deposited under the NCBI bioproject ID 63661.