Abstract
Gametophytic self-incompatibility in Japanese pear (Pyrus pyrifolia Nakai) is controlled by the single, multi-allelic S-locus. Information about the S-genotypes is important for breeding and the selection of pollen donors for fruit production. Rapid and reliable S-genotype identification system is necessary for efficient breeding of new cultivars in Japanese pear. We designed S allele-specific PCR primer pairs for ten previously reported S-RNase alleles (S1–S9 and Sk) as simple and reliable method. Specific nucleotide sequences were chosen to design the primers to amplify fragments of only the corresponding S alleles. The developed primer pairs were evaluated by using homozygous S-genotypes (S1/S1–S9/S9 and S4sm/S4sm) and 14 major Japanese pear cultivars, and found that S allele-specific primer pairs can identify S-genotypes effectively. The S allele-specific primer pairs developed in this study will be useful for efficient S-genotyping and for marker-assisted selection in Japanese pear breeding programs.
Keywords: Pyrus pyrifolia, self-incompatibility, S-genotyping, S-RNase
Introduction
Self-incompatibility is a widespread genetic mechanism that prevents inbreeding in plants (de Nettancourt 1997). Self-incompatibility systems can be classified into two major classes: gametophytic self-incompatibility (GSI), in which the S phenotype of the pollen (male gametophyte) is determined by its own haploid S-genotype, and sporophytic self-incompatibility (SSI), in which the S phenotype of the pollen is determined by the diploid S-genotype of its parent plant (sporophyte) (Brennan et al. 2011, Hiscock and McInnis 2003). SSI is observed in the families Brassicaceae and Asteraceae (Brennan et al. 2011, Takasaki et al. 2000) and results in fertilization arrest when one or both S haplotypes of pollen coincide with the S haplotype of the pistil. On the other hand, GSI is observed in the Rosaceae, Solanaceae, and Scrophulariaceae (Wang et al. 2003). In these families, GSI is controlled by a single multi-allelic locus (S locus). If the pollen S allele matches one of the two pistil S alleles, pollen tube growth in the style is arrested and fertilization is prevented. Simple GSI systems have been found in species of the subfamily Amygdaloideae of the Rosaceae, such as sweet cherry (Prunus avium (L.) L.), sour cherry (Prunus cerasus L.), Japanese plum (Prunus mume (Sieb.) Sieb. et Zucc.), and almond (Prunus dulcis (Mill.) D.A. Webb). In these species, the pistil-S determinant is a single ribonuclease (S-RNase) and the pollen-S determinant is a protein encoded by a single S haplotype-specific F-box gene (SFB) (Entani et al. 2003, Ushijima et al. 2003, Yamane et al. 2003). It has been hypothesized that a general inactivation mechanism detoxifies non-self S-RNases, whereas SFB protects self S-RNase (Sonneveld et al. 2005). In the Solanaceae, the pistil-S determinant is also a single S-RNase, but the pollen-S determinants—multiple S-locus F-box (SLF) proteins—collaboratively recognize and inactivate non-self S-RNases (Kubo et al. 2010). Similarly, in Japanese pear and in apple (Malus × domestica Borkh.), which belong to the subfamily Maloideae, the pollen-S determinants are multiple S haplotype-specific F-box brothers (SFBBs), which collaboratively recognize and inactivate non-self S-RNases (Kakui et al. 2011).
Since GSI-limited self-fertilization decreases fruit setting and promotes the production of low-quality parthenocarpic fruits, pollination with compatible pollen is essential for the commercial fruits production. In commercial fruit tree orchards, cross-compatible cultivars harboring different S-genotypes (combinations of two S alleles) and having similar flowering time are planted together to ensure successful cross-pollination and marketable fruits. Thus, identifying the S-genotype of Japanese pear cultivars and forecasting mutual pollination compatibility can make a proper selection of cross-compatible cultivars and parental genotypes in breeding programs.
In Japanese pear, one self-compatible S allele (S4sm) and ten self-incompatible S alleles (S1 to S9 and Sk) have been identified. Up to now, ten cDNAs encoding S-RNases (S1 to S9 and Sk) have been isolated from self-incompatible S-genotype cultivars and sequenced (Castillo et al. 2002, Ishimizu et al. 1999, Kim et al. 2007, Sawamura et al. 2002). S-RNases of the Rosaceae, including Japanese pear, share five conserved regions (C1, C2, C3, RC4, and C5); two catalytic domains have been also found; this domain structure is similar to those of fungal T2-type ribonucleases (Yamane and Tao 2009). A single hypervariable region may mediate the S-RNase interaction with its functional partners (Matsuura et al. 2001). The S-RNase alleles in Japanese pear have an intron within the hypervariable region. Up to now, S-genotypes (S alleles) in Japanese pear have been identified by using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis based on sequence polymorphisms of hypervariable regions and introns of S-RNase (Ishimizu et al. 1999, Kim et al. 2004, Takasaki et al. 2004). PCR-RFLP combines PCR amplification using common primer pair and digestion by each S allele-specific restriction endonucleases. Although this method is reliable, digestion by restriction endonucleases is complex and costly (Long et al. 2010). In addition, this approach has several disadvantages. For example, no specific restriction endonucleases are known for the S3 and S7 alleles, and two restriction endonucleases are needed to distinguish the S3 and S5 alleles (Ishimizu et al. 1999, Takasaki et al. 2004). In the latter case, one endonuclease digests the S5 allele, whereas the other one digests both S3 and S5 alleles, and the S3 allele is distinguished by the absence of digestion (negative reaction). A similar negative reaction is used to distinguish S7 from S6. Incomplete digestion may compromise the accuracy of these assays.
Recently, the S allele of the self-compatible cultivar ‘Osa-Nijisseiki’ (a mutant of the self-incompatible cultivar ‘Nijisseiki’) has been identified. Self-compatibility of ‘Osa-Nijisseiki’ is controlled by the S4sm allele; this allele lacks a 236-kbp genomic region that includes the S4 RNase-coding region (Okada et al. 2008). It is difficult to identify S4sm allele using PCR-RFLP system because S4sm allele is a null allele of S4 RNase, and there is no amplification using PCR-RFLP system. Thus, the S4sm allele has been identified by S4sm allele-specific PCR using primer set designed to recognize the regions flanking the deletion (Okada et al. 2008). In the same way, each S allele-specific PCR system is promising as a simple, reliable and low-cost alternative method.
An S allele-specific PCR system may detect length polymorphisms of the S-RNase or SFB intron(s) with common PCR primers or may amplify each S allele with a specific primer pair. In sweet cherry, length polymorphisms of S-RNase and SFB introns (Guerra et al. 2012) or two introns of S-RNase (Sonneveld et al. 2003) were used. In Japanese pear, this approach cannot be used because SFBB has no introns and the length of the single S-RNase intron is similar in different alleles: 167 bp in S1 vs. 168 bp in S4 and 179 bp in both S3 and S5. On the other hand, S allele-specific PCR amplification system has been successfully developed for apple and sweet cherry on the basis of differences in nucleotide sequences among S-RNase alleles (Broothaerts 2003, Broothaerts et al. 1996, 2001, Janssens et al. 1995, Long et al. 2010, Sonneveld et al. 2003, van Nerum et al. 2001, Verdoodt et al. 1998).
In the present study, we established an S allele-specific PCR amplification system for rapid, reliable and inexpensive S-genotyping of Japanese pear, which uses ten primer pairs based on the S-RNase nucleotide sequences.
Materials and Methods
Plant materials
A total of 26 Japanese pear accessions were used. Ten S allele homozygotes (Im-3, 312-9, 312-6, 421-6, 421-24, Im-18, 420-50, 548-1, S9 homozygote and Chukanbohon Nashi nou 1 gou) and a cultivar and an accession with the Sk allele (‘Kinchaku’ and 314-32) were used for the development of an S-genotype identification technique, and 14 major leading cultivars (‘Akizuki’, ‘Chojuro’, ‘Gold-Nijisseiki’, ‘Hosui’, ‘Imamuraaki’, ‘Kosui’, ‘Meigetsu’, ‘Nansui’, ‘Niitaka’, ‘Nijisseiki’, ‘Okusankichi’, ‘Osa-Gold’, ‘Shinko’ and Tsukuba 56 gou) were used for its evaluation (Table 1). Tsukuba 56 gou is line name of new cultivar applying for breed registry in Japan on 12/5/2014. Nine S allele homozygotes were derived from self-pollinated progeny: Im-3 (S1/S1) was derived from ‘Imamuraaki’, 312-9 (S2/S2) and 312-6 (S3/S3) from ‘Chojuro’, 421-6 (S4/S4) and 421-24 (S5/S5) from ‘Shinsui’, Im-18 (S6/S6) from ‘Imamuraaki’, 420-50 (S7/S7) from ‘Okusankichi’, 548-1 (S8/S8) from ‘Heiwa’, and the S9 homozygote (S9/S9) from ‘Shinko’. The S4sm/S4sm homozygote ‘Chukanbohon Nashi nou 1 gou’ was established from self-pollinated progeny of ‘Osa-Nijisseiki’. The S4sm allele of ‘Osa-Nijisseiki’, which was derived from ‘Nijisseiki’ (S2/S4), lacks a 236-kbp genomic region including the S4-RNase coding region (Okada et al. 2008). ‘Kinchaku’, 314-32 and Tsukuba 56 gou were used for analysis of the Sk allele, because an Sk homozygote has not been established yet. Thirteen major cultivars represent several S allele combinations. All plant materials were maintained and cultivated at the NARO Institute of Fruit Tree Science (Tsukuba, Japan).
Table 1.
Japanese pear accessions used in this study
| Accession name | Origin | S-genotypes | Germplasm accession no. |
|---|---|---|---|
| Im-3a | Imamuraaki × Imamuraaki | S1/S1 | |
| 312-9b | Chojuro × Chojuro | S2/S2 | |
| 312-6b | Chojuro × Chojuro | S3/S3 | |
| 421-6c | Shinsui × Shinsui | S4/S4 | |
| 421-24c | Shinsui × Shinsui | S5/S5 | |
| Im-18a | Imamuraaki × Imamuraaki | S6/S6 | |
| 420-50c | Okusankichi × Okusankichi | S7/S7 | |
| 548-1a | Heiwa × Heiwa | S8/S8 | |
| S9 homozygoted | Shinko × Shinko | S9/S9 | |
| 314-32 | Kinchaku × Hosui | Sk/S3 | |
| Chukanbohon Nashi nou 1 gou | Osa-Nijisseiki × Osa-Nijisseiki | S4sm/S4sm | JP238479 |
| Akizuki | (Niitaka × Hosui) × Kosui | S3/S4 | JP118538 |
| Chojuro | Indigenous, unknown parentage | S2/S3 | JP113575 |
| Gold-Nijisseiki | Mutant of Nijisseiki | S2/S4 | JP110823 |
| Hosui | Kosui × Hiratsuka 1 gou | S3/S5 | JP113598 |
| Imamuraaki | Indigenous, unknown parentage | S1/S6 | JP113600 |
| Kinchaku | Indigenous, unknown parentage | Sk/S4 | JP113613 |
| Kosui | Kikusui × Wasekozo | S4/S5 | JP113619 |
| Meigetsu | Indigenous, unknown parentage | S1/S8 | JP113626 |
| Nansui | Shinsui × Echigo | S4/S9 | JP115742 |
| Niitaka | Amanogawa × Chojuro | S3/S9 | JP113630 |
| Nijisseiki | Indigenous, unknown parentage | S2/S4 | JP113631 |
| Okusankichi | Indigenous, unknown parentage | S5/S7 | JP113634 |
| Osa-Gold | Mutant of Osa-Nijisseiki | S2/S4sm | JP110825 |
| Shinko | Nijisseiki × Amanogawa | S4/S9 | JP113657 |
| Tsukuba 56 gou | 314-32 × Akiakari | Sk/S5 |
Designing the S allele-specific primer pairs
S allele-specific PCR primer pairs were designed on the basis of the nucleotide sequences of S-RNase alleles: S1 (DDBJ accession nos. AB002139 and DQ515793), S2 (AB545982), S3 (AB025421), S4 (AB308360), S5 (AB045711), S6 (Kim et al. 2002), S7 (Kim et al. 2002), S8 (AB104908), S9 (Sawamura et al. 2002), and Sk (AB284262 and AB284263). Nucleotide sequences of ten S-RNase alleles were aligned in CLC Main Workbench v. 6.9.1 software (Qiagen GmbH, Hilden, Germany). The alignment was visually inspected to find the divergent regions, and these regions were used to design specific primer sets. Primer GC content was designed to be 35% to 65% (Supplemental Fig. 1). To ensure the specificity of each primer set, we chose 3 bases at the 3′ end of at least one primer to be specific for target S-RNase. Primer pairs were designed to amplify fragments longer than 150 bp and of different lengths for each S allele. The specificity of the primer sets was confirmed by BLAST searches against S-RNase alleles. The S4sm-specific primer set SM was described previously (Okada et al. 2008); S4sm showed no significant sequence similarity with the ten S-RNase gene sequences.
PCR analysis to validate the S allele-specific primers
Young leaves were collected, frozen in liquid nitrogen, and then homogenized in a Shake Master shaker (Bio Medical Science, Tokyo, Japan). Genomic DNA was isolated using the Genomic-tip 20/G and Genomic DNA Buffer Set (Qiagen GmbH) as described by Yamamoto et al. (2006). The PCR mixture (10 μL) contained 5 μL of GoTaq Colorless Master Mix (Promega, Madison, WI, USA), 10 pmol each of forward and reverse primers, and 5 ng of genomic DNA. PCR was performed on a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) using the following program: an initial denaturing step at 94°C for 2 min; 35 cycles of denaturation at 94°C for 1 min, annealing at either 52°C, 55°C, 58°C, or 61°C for 1 min, and extension at 72°C for 1 min; and a final extension step at 72°C for 3 min. PCR products were separated in 2% (w/v) agarose gel in TAE buffer and stained with 0.5 μg/mL ethidium bromide or SYBR green I (TaKaRa bio, Shiga, Japan). Amplified fragment sizes were estimated against a 100-bp DNA Ladder (Toyobo, Osaka, Japan).
Results
Design of S allele-specific primer pairs
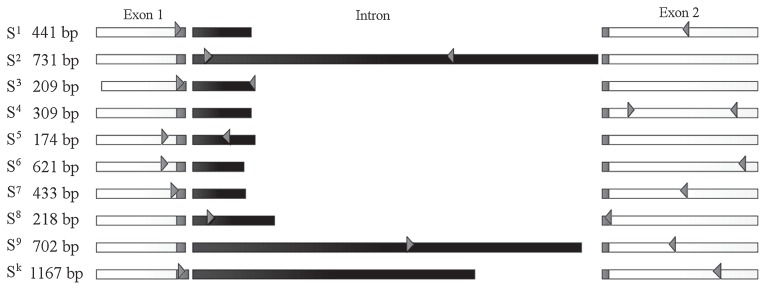
We aligned ten nucleotide sequences of previously identified S-RNase alleles, viz., S1 to S9 and Sk, and selected specific primer pairs for each allele (Fig. 1, Table 2, Supplemental Fig. 2). Our primer design flow chart is shown in Supplemental Fig. 1. Hypervariable regions and introns were preferentially selected because they were abundant in each S-RNase-specific sequence (Supplemental Fig. 2). Primer pairs were re-designed if (1) amplification of the target allele was absent or weak, or (2) a non-target allele or a non-specific fragment was amplified. In the case of redesign failure, a conserved exon sequence was used for one or both primers. In the primer pair PpS4 and PpS6, forward and reverse primers were in conserved region of exon. In the primer pair PpS1, PpS5, PpS7, PpS9 and PpSk, one primer was in conserved region of exon, and the other in hypervariable region or intron. In the primer pairs PpS2, PpS3 and PpS8, both primers were in hypervariable region or intron. The identities between the coding sequences of S-RNases ranged from 74% to 95% (Supplemental Table 1). Most coding sequences were around 80% identical and had several allele-specific regions. However, sequence identity within the pairs S1 vs. S4 and S3 vs. S5 was 94% and 95%, respectively. To achieve primer specificity within these groups, at least one primer was designed to have specific sequence on 3 bp from 3′ end (Supplemental Fig. 1). The SM primer pair recognizes the S4sm allele regions flanking the missing 236-kbp region (Okada et al. 2008).
Fig. 1.
Designed primers specific for 10 S-RNase (S) alleles. The names of the S allele and the expected sizes of the PCR fragments are shown on the left. White boxes show the conserverd region of the S-RNase gene; gray boxes show the hypervariable region; the black box shows the intron. Primer binding sites are indicated by gray arrowheads.
Table 2.
Characteristics of S allele-specific primer pairs
| S allele | Primer pair | Sequence (5′-3′) | Size of PCR product (bp) |
|---|---|---|---|
| S1 | PpS1 | F: AATGTAAGACTACAGCCCTG R: TCCACCAGTGGCCTGTTTG |
441 |
| S2 | PpS2 | F: TCCTTCCATCAAATTCTCCCCAGCA R: GGGGTACACCGTGCGTCCAT |
731 |
| S3 | PpS3 | F: TGCCCGATAAAGAATATTCG R: CTCTGGTATGCACAAGAGAG |
209 |
| S4 | PpS4 | F: TCTGGGAAAGAGAGTGGCTC R: GGCAATTTATGAACTTAGTC |
309 |
| S5 | PpS5 | F: TTGTGGCCCTCAAGCATGGC R: CGTGCATGAAAATCTATGTTTGAGGAC |
174 |
| S6 | PpS6 | F: GTTTGTGGCCTTCAAACGACG R: GTGATCCTTTAAAAGAACTGC |
621 |
| S7 | PpS7 | F: TCACCCAGAAAATTGCACTAATGC R: CCAGTGGCCTTTGTATTCCCAA |
433 |
| S8 | PpS8 | F: GTCATTGACGGGGTTTGAACCC R: CCAACTGGGCTTTGAGTGAT |
218 |
| S9 | PpS9 | F: CAAAAATGTACCCATGTTTGGT R: CGCCTTTGAGAGGATTTCAG |
702 |
| Sk | PpSk | F: GAAAACCAAGGTGCCTCAGGC R: CTCAACCAATTCAATAGTCCC |
1167 |
| S4sm | SM* | F: TCGTCTTAGGGATTTCCAATGC R: GCCTTAAGGGTTCATTGGGC |
666 |
Development of S allele-specific primer pairs
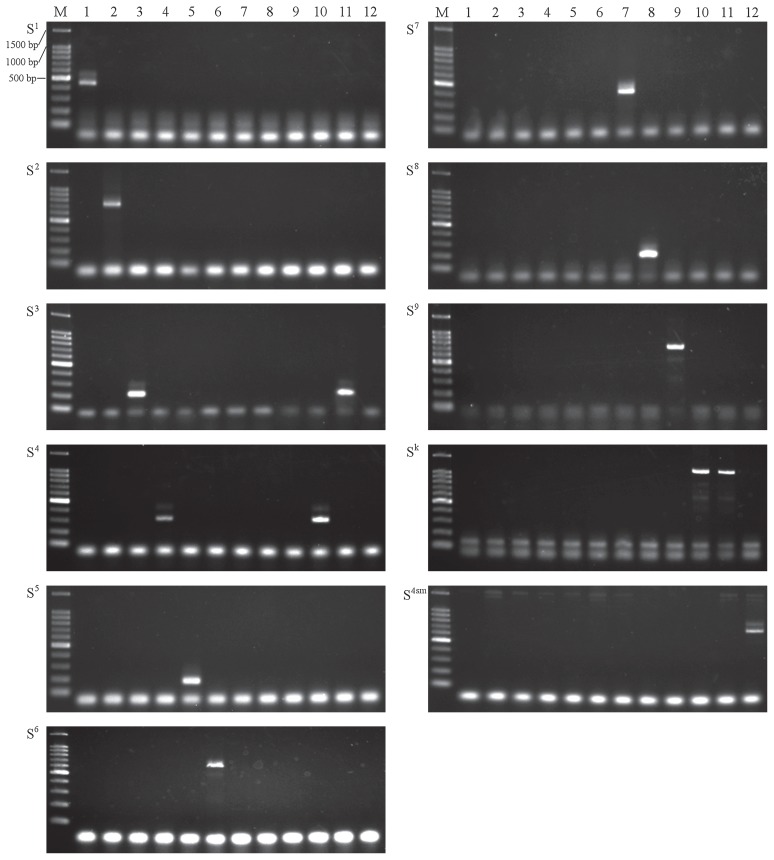
Ten newly designed S allele-specific primer pairs and the SM primer pair were evaluated using ten homozygous accessions (S1/S1–S9/S9, and S4sm/S4sm). Because an Sk homozygous line has not yet been established, we used two accessions carrying the Sk allele, ‘Kinchaku’ (Sk/S4) and 314-32 (Sk/S3). All primer pairs clearly amplified fragments of the expected length (Fig. 2). The primer pair PpSk showed clear fragment amplification in ‘Kinchaku’ (Sk/S4) and 314-32 (Sk/S3), but not in 312-6 (S3/S3) or 421-6 (S4/S4). Annealing at 55, 58, and 61°C resulted in one clear band for each primer pair. Some minor bands for PpS3, PpS6, PpS7, PpS9, and PpSk were observed at 52°C, whereas annealing at 61°C sometimes resulted in weak amplification (data not shown). Therefore, the optimum annealing temperatures appear to be between 55 and 61°C. We chose 58°C for further experiments. Unexpected fragments were observed in the primer pair PpSk between 100–200 bp in all templates and under 100 bp in all primer pairs in all templates (Fig. 2). Since these fragments were similarly stained using ethidium bromide or SYBR Green I (data not shown), they were suggested as primer dimers or non-specifically amplified DNA. However, they did not seem to affect S-genotyping because their fragment sizes were quite different from expected fragment sizes. Thus, 11 S allele-specific primer pairs were successfully developed. The use of homozygous accessions facilitated the evaluation of primer specificity.
Fig. 2.
Validation of S allele-specific primers. PCR fragments were amplified from 10 S allele homozygotes and two accessions with the Sk allele by using the designed primer pairs. M: 100 bp DNA ladder (Toyobo); 1: Im-3 (S1/S1); 2: 312-9 (S2/S2); 3: 312-6 (S3/S3); 4: 421-6 (S4/S4); 5: 421-24 (S5/S5); 6: Im-18 (S6/S6); 7: 420-50 (S7/S7); 8: 548-1 (S8/S8); 9: S9-homozygote (S9/S9); 10: ‘Kinchaku’ (Sk/S4); 11: 314-32 (Sk/S3); 12: ‘Chukanbohon Nashi nou 1 gou’ (S4sm/S4sm).
Application of S allele-specific primer pairs to Japanese pear cultivars
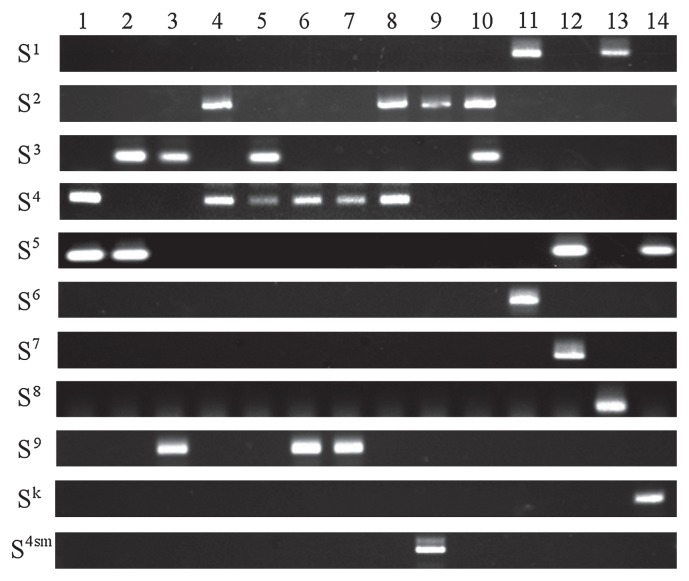
Next, we compared the performance of the primer pairs using accessions heterozygous for S alleles. To evaluate the practicability of the developed technique, we chose 13 major leading Japanese pear cultivars and one accession evaluated them using the 11 specific primer pairs (Fig. 3). The expected S allele combinations were identified for ‘Kosui’ (S4/S5), ‘Hosui’ (S3/S5), ‘Niitaka’ (S3/S9), ‘Nijisseiki’ (S2/S4), ‘Akizuki’ (S3/S4), ‘Shinko’ (S4/S9), ‘Nansui’ (S4/S9), ‘Gold-Nijisseiki’ (S2/S4), ‘Osa-Gold’ (S2/S4sm), ‘Chojuro’ (S2/S3), ‘Imamuraaki’ (S1/S6), ‘Okusankichi’ (S5/S7), ‘Meigetsu’ (S1/S8), and Tsukuba 56 gou (S5/Sk).
Fig. 3.
Identification of S-genotypes of 13 major Japanese pear cultivars and 1 accession by PCR with S allele-specific primer pairs. S allele names and specific primers are shown to the left of each electrophoretogram. 1: ‘Kosui’ (S4/S5); 2: ‘Hosui’ (S3/S5); 3: ‘Niitaka’ (S3/S9); 4: ‘Nijisseiki’ (S2/S4); 5: ‘Akizuki’ (S3/S4); 6: ‘Shinko’ (S4/S9); 7: ‘Nansui’ (S4/S9); 8: ‘Gold-Nijisseiki’ (S2/S4); 9: ‘Osa-Gold’ (S2/S4sm); 10: ‘Chojuro’ (S2/S3); 11: ‘Imamuraaki’ (S1/S6); 12: ‘Okusankichi’ (S5/S7); 13: ‘Meigetsu’ (S1/S8); 14: Tsukuba 56 gou (Sk/S5).
Discussion
We developed a simple, low-cost PCR-based method which can identify 11 S alleles, including S4sm, on the basis of the presence or absence of specific bands. For the development of an S allele-specific PCR system, SFBB sequences and the intergenic regions in the S-locus could be considered as primer design sites. However, sequence similarities of SFBB-gamma in Japanese pear are high (98% to 99.5%; Supplemental Table 2), and SFBB paralogs also have similar sequences (Kakui et al. 2011). Intergenic region sequences other than S2 and S4 allele were not available in the DNA Data Bank of Japan (DDBJ) as of 16 January 2015. Thus, we focused on the development of a PCR amplification system for S-RNase alleles in Japanese pear.
Among S-RNase alleles, introns show lower sequence similarities (mean 43%, range 27%–95%) than exons (74%–95%; Supplemental Tables 1, 3). However, the contents of A (35%) and T (40%) are much higher than the contents of G (12%) and C (13%) in introns than in exons (A, 33%; T, 25%; G, 22%; C, 20%) (data not shown). In exons, hypervariable regions are highly polymorphic, but several hypervariable regions show high sequence similarity and these regions are short (48–56 bp). As we could not design all primer pairs using sequences of introns or hypervariable regions, we designed several primers using sufficiently polymorphic exon sequences. Several S allele-specific primer pairs were redesigned several times owing to poor or non-specific amplification. For example, we designed three forward and three reverse primers for S6, and only an exon-specific primer pair was satisfactory (data not shown). Our approach to primer design (Supplemental Fig. 1) may be also applicable to other species, in particular, self-incompatible Maloideae species such as Chinese pear (Pyrus bretschneideri Rehd.), European pear (Pyrus communis L.), and loquat (Eriobotrya japonica (Thunb.) Lindl.).
S allele-specific primers designed in this study were not identical to known nucleotide sequences from other Pyrus species, such as Chinese pear and European pear. Since S allele-specific PCR relies on the presence of specific bands to detect the S alleles, it is important to avoid misidentification due to false-positive or false-negative results. A possibility of false-positive results is unlikely, because no bands other than the expected ones were detected in any samples at all tested annealing temperatures. On the other hand, we should pay attention to false-negative results. We confirmed that S allele-specific primer pairs could reliably amplify the target fragments at annealing temperatures between 55 and 61°C. Otherwise, it is effective to use internal PCR amplification controls such as chloroplast DNA markers atpB–rbcL (intergenic spacer between ATPase B subunit and ribulose 1,5-bisphosphate carboxylase/oxygenase large subunit genes), trnL–trnF (intergenic spacer between tRNA-Leu and tRNA-Phe genes), or both (Yamamoto et al. 2006) with one-tenth of the primer concentration used for S allele-specific markers.
To facilitate the evaluation of primer specificity, we initially used S allele-homozygous accessions, which makes it unnecessary to consider non-specific amplification of other S alleles. If S allele-heterozygous accessions are to be used for evaluation of primer specificities, in addition to the accession carrying the target S allele, accessions carrying all non-target S alleles but not the target one should be examined for all primer pairs.
The temperature control of PCR thermal cyclers may differ among manufacturers and models. Some thermal cyclers show a difference of around 1.6°C between the programmed and actual annealing temperature (Kim et al. 2008). In this study, stable amplification at an annealing temperature of 58 ± 3°C was observed, and we expect that 58°C would be suitable for different thermal cycler models.
Unexpected fragments were observed under 100 bp in all primer pairs. Because these fragments were stained similarly using SYBR green I, which stains double-stranded nucleic acid rather than single-stranded nucleic acid, these fragments were assumed as primer dimers, not unreacted primers (data not shown). Although formation of primer dimers reduces target fragment amplification, we confirmed that target fragments were stably amplified between 55 to 61°C. It is possible that low input of template DNA induces false-negative results. However, there are few possibilities that formation of primer dimers induce false-negative results if sufficient template DNA is input.
Our rapid and reliable method for identification of S-genotypes would be useful in marker-assisted selection (MAS). MAS can accelerate selection and reduce the progeny size and the cost of raising individuals to maturity in the field (Luby and Shaw 2001). Molecular markers associated with self-incompatibility (Ishimizu et al. 1999), ethylene production (Itai et al. 2003), pear scab resistance (Terakami et al. 2006), and black spot resistance (Terakami et al. 2007) have been developed, and some of them have been used for selection in Japanese pear breeding programs. Because identification of S-genotypes is also important for breeding of Japanese pear, we expect that our developed S allele-specific markers to be applied for MAS.
Supplementary Material
Acknowledgments
We thank Ms. F. Hosaka, H. Oshino, N. Shigeta, and N. Yagihashi for their technical help. This work was partially supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Development of mitigation and adaptation techniques to global warming in the sectors of agriculture, forestry, and fisheries, C-3-2010).
Literature Cited
- Brennan, A.C., Tabah, D.A., Harris, S.A. and Hiscock, S.J. (2011) Sporophytic self-incompatibility in Senecio squalidus (Asteraceae): S allele dominance interactions and modifiers of cross-compatibility and selfing rates. Heredity 106: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broothaerts, W., Verdoodt, L., Keulemans, J., Janssens, G.A. and Broekaert, W.F. (1996) The self-incompatibility gene in apple and determination of the S-genotype of apple cultivars by PCR. Acta Hort. 423: 103–109. [Google Scholar]
- Broothaerts, W., Wiersma, P.A. and Lane, W.D. (2001) Lane multiplex PCR combining transgene and S-allele control primers to simultaneously confirm cultivar identity and transformation in apple. Plant Cell Rep. 20: 349–353. [Google Scholar]
- Broothaerts, W.. (2003) New findings in apple S-genotype analysis resolve previous confusion and request the re-numbering of some S-alleles. Theor. Appl. Genet. 106: 703–714. [DOI] [PubMed] [Google Scholar]
- Castillo, C., Takasaki, T., Saito, T., Norioka, S. and Nakanishi, T. (2002) Cloning of the S8-RNase (S8-allele) of Japanese pear (Pyrus pyrifolia Nakai). Plant Biotech. 19: 1–6. [Google Scholar]
- de Nettancourt, D.. (1997) Incompatibility in angiosperms. Sex. Plant Reprod. 10: 185–199. [Google Scholar]
- Entani, T., Iwano, M., Shiba, H., Che, F.S., Isogai, A. and Takayama, S. (2003) Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8: 203–213. [DOI] [PubMed] [Google Scholar]
- Guerra, E.M., López-Corrales, M. and Wünsch, A. (2012) Improved S-genotyping and new incompatibility groups in Japanese plum. Euphytica 186: 445–452. [Google Scholar]
- Hiscock, S.J. and McInnis, S.M. (2003) The diversity of self-incompatibility systems in flowering plants. Plant Biol. 5: 23–32. [Google Scholar]
- Ishimizu, T., Inoue, K., Shimonaka, M., Saito, T., Terai, O. and Norioka, S. (1999) PCR-based method for identifying the S-genotypes of Japanese pear cultivars. Theor. Appl. Genet. 98: 961–967. [Google Scholar]
- Itai, A., Kotaki, T., Tanabe, K., Tamura, F., Kawaguchi, D. and Fukuda, M. (2003) Rapid identification of 1-aminocyclopropane-1-carboxylate (ACC) synthase genotypes in cultivars of Japanese pear (Pyrus pyrifolia Nakai) using CAPS markers. Theor. Appl. Genet. 106: 1266–1272. [DOI] [PubMed] [Google Scholar]
- Janssens, G.A., Goderis, I.J., Broekaert, W.F. and Broothaerts, W. (1995) A molecular method for S-allele identification in apple based on allele-specific PCR. Theor. Appl. Genet. 91: 691–698. [DOI] [PubMed] [Google Scholar]
- Kakui, H., Kato, M., Ushijima, K., Kitaguchi, M., Kato, S. and Sassa, H. (2011) Sequence divergence and loss-of-function phenotypes of S locus F-box brothers genes are consistent with non-self recognition by multiple pollen determinants in self-incompatibility of Japanese pear (Pyrus pyrifolia). Plant J. 68: 1028–1038. [DOI] [PubMed] [Google Scholar]
- Kim, H.T., Hirata, Y. and Nou, I.S. (2002) Identification of self-incompatibility alleles by S-RNases sequencing and PCR-RFLP analysis in Korean-bred pear (Pyrus pyrifolia) strains. Acta Hort. 587: 467–476. [Google Scholar]
- Kim, H., Hirata, Y., Shin, Y.U., Hwang, H.S., Hwang, J.H., Shin, I.S., Kim, D.I., Kang, S.J., Kim, H.J., Shin, D.Y.et al. (2004) A molecular technique for selection of self-compatible varieties of Japanese pear (Pyrus pyrifolia Nakai). Euphytica 138: 73–80. [Google Scholar]
- Kim, H.T., Kakui, H., Koba, T., Hirata, Y. and Sassa, H. (2007) Cloning of a new S-RNase and development of a PCR-RFLP system for the determination of the S-genotypes of Japanese pear. Breed. Sci. 57: 159–164. [Google Scholar]
- Kim, Y.H., Yang, I., Bae, Y.S. and Park, S.R. (2008) Performance evaluation of thermal cyclers for PCR in a rapid cycling condition. Biotechniques 44: 495–505. [DOI] [PubMed] [Google Scholar]
- Kubo, K., Entani, T., Takara, A., Wang, N., Fields, A.M., Hua, Z., Toyoda, M., Kawashima, S., Ando, T., Isogai, A.et al. (2010) Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330: 796–799. [DOI] [PubMed] [Google Scholar]
- Long, S., Li, M., Han, Z., Wang, K. and Li, T. (2010) Characterization of three new S-alleles and development of an S-allele-specific PCR system for rapidly identifying the S-genotype in apple cultivars. Tree Genet. Genomes 6: 161–168. [Google Scholar]
- Luby, J.J. and Shaw, D.V. (2001) Does marker-assisted selection make dollars and sense in a fruit breeding program? HortScience 36: 872–879. [Google Scholar]
- Matsuura, T., Sakai, H., Unno, M., Ida, K., Sato, M., Sakiyama, F. and Norioka, S. (2001) Crystal structure at 1.5-Å resolution of Pyrus pyrifolia pistil ribonuclease responsible for gametophytic self-incompatibility. J. Biol. Chem. 276: 45261–45269. [DOI] [PubMed] [Google Scholar]
- Okada, K., Tonaka, N., Moriya, Y., Norioka, N., Sawamura, Y., Matsumoto, T., Nakanishi, T. and Takasaki-Yasuda, T. (2008) Deletion of a 236 kb region around S4-RNase in a stylar-part mutant S4sm-haplotype of Japanese pear. Plant Mol. Biol. 66: 389–400. [DOI] [PubMed] [Google Scholar]
- Saito, T., Sawamura, Y., Takada, N., Shoda, M., Terai, O., Abe, K. and Kotobuki, K. (2005) Breeding of homozygotes of self-incompatible haplotype in Japanese pear (Pyrus pyrifolia Nakai). Acta Hort. 671: 233–238. [Google Scholar]
- Sassa, H., Kakui, H., Miyamoto, M., Suzuki, Y., Hanada, T., Ushijima, K., Kusaba, M., Hirano, H. and Koba, T. (2007) S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics 175: 1869–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura, Y., Saito, T., Shoda, M., Yamamoto, T., Sato, Y., Hayashi, T. and Kotobuki, K. (2002) A new self-incompatible allele in Japanese pear ‘Shinsei’ and ‘Shinkou’. J. Japan. Soc. Hort. Sci. 71: 342–347. [Google Scholar]
- Schopfer, C.R., Nasrallah, M.E. and Nasrallah, J.B. (1999) The male determinant of self-incompatibility in Brassica. Science 286: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Sonneveld, T., Tobutt, K.R. and Robbins, T.P. (2003) Allele-specific PCR detection of sweet cherry self-incompatibility (S) alleles S1 to S16 using consensus and allele-specific primers. Theor. Appl. Genet. 107: 1059–1070. [DOI] [PubMed] [Google Scholar]
- Sonneveld, T., Tobutt, K.R., Vaughan, S.P. and Robbins, T.P. (2005) Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17: 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki, T., Hatakeyama, K., Suzuki, G., Watanabe, M., Isogai, A. and Hinata, K. (2000) The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403: 913–916. [DOI] [PubMed] [Google Scholar]
- Takasaki, T., Okada, K., Castillo, C., Moriya, Y., Saito, T., Sawamura, Y., Norioka, N., Norioka, S. and Nakanishi, T. (2004) Sequence of the S9-RNase cDNA and PCR-RFLP system for discriminating S1- to S9-allele in Japanese pear. Euphytica 135: 157–167. [Google Scholar]
- Terai, O., Sato, Y., Saito, T., Abe, K. and Kotobuki, K. (1999) Identification of homozygotes of self-incompatibility gene (S-gene), as useful tool to determine the S-genotype in Japanese pear, Pyrus pyrifolia Nakai. Bull. Natl. Inst. Fruit Tree Sci. 32: 31–38. [Google Scholar]
- Terakami, S., Shoda, M., Adachi, Y., Gonai, T., Kasumi, M., Sawamura, Y., Iketani, H., Kotobuki, K., Patocchi, A., Gessler, C.et al. (2006) Genetic mapping of the pear scab resistance gene Vnk of Japanese pear cultivar Kinchaku. Theor. Appl. Genet. 113: 743–752. [DOI] [PubMed] [Google Scholar]
- Terakami, S., Adachi, Y., Iketani, H., Sato, Y., Sawamura, Y., Takada, N., Nishitani, C. and Yamamoto, T. (2007) Genetic mapping of genes for susceptibility to black spot disease in Japanese pears. Genome 50: 735–741. [DOI] [PubMed] [Google Scholar]
- Ushijima, K., Sassa, H., Dandekar, A.M., Gradziel, T.M., Tao, R. and Hirano, H. (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond (Prunus dulcis): identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nerum, I., Geerts, M., van Haute, A., Keulemans, J. and Broothaerts, W. (2001) Re-examination of the self-incompatibility genotype of apple cultivars containing putative ‘new’ S alleles. Theor. Appl. Genet. 103: 584–591. [Google Scholar]
- Verdoodt, L., van Haute, A., Goderis, I.J., De Witte, K., Keulemans, J. and Broothaerts, W. (1998) Use of the multi-allelic self-incompatibility gene in apple to assess homozygosity in shoots obtained through haploid induction. Theor. Appl. Genet. 96: 294–300. [Google Scholar]
- Wang, Y., Wang, X., Skirpan, A.L. and Kao, T.H. (2003) S-RNase-mediated self-incompatibility. J. Exp. Bot. 54: 115–122. [DOI] [PubMed] [Google Scholar]
- Yamamoto, T., Kimura, T., Hayashi, T. and Ban, Y. (2006) DNA profiling of fresh and processed fruits in pear. Breed. Sci. 56: 165–171. [Google Scholar]
- Yamane, H., Ikeda, K., Ushijima, K., Sassa, H. and Tao, R. (2003) A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol. 44: 764–769. [DOI] [PubMed] [Google Scholar]
- Yamane, H. and Tao, R. (2009) Molecular basis of self-(in)compatibility and current status of S-genotyping in Rosaceous fruit trees. J. Japan. Soc. Hort. Sci. 78: 137–157. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.