Abstract
The modification of erucic acid content in seeds is one of the major goals for quality breeding in oil-yielding Brassica species. However, few low erucic acid (LEA) resources are available, and novel LEA genetic resources are being sought. Fatty acid elongase 1 (FAE1) is the key gene that controls erucic acid synthesis. However, the mechanism for erucic acid synthesis in B. rapa lacks systematic study. Here, we isolated zero erucic acid lines from 1981 Chinese landraces of B. rapa and found that the formation of LEA is not attributable to variations in FAE1 coding sequences, as reported for B. napus, but may be attributable to the decrease in FAE1 expression. Moreover, the FAE1 promoter sequences of LEA and high erucic acid materials shared 95% similarity. Twenty-eight bases deletions (containing a 24-base AT-rich region) were identified approximately 1300 bp upstream from the FAE1 start codon in the LEA accessions. The genotype with the deletions co-segregated with the LEA trait in the segregating population. This study isolated an LEA B. rapa resource that can be exploited in Brassica cultivation. The promoter variations might modify the expression level of FAE1, and the results shed light on novel regulation mechanisms for erucic acid synthesis.
Keywords: erucic acid, FAE1, Brassica rapa, expression, promoter, deletions
Introduction
Brassica crops are important in global agriculture and oil production. High concentrations of erucic acid (C22:1) in the seed of oil-yielding Brassica species have been reported to be nutritionally undesirable (Badawy et al. 1994, Beare-Rogers 1971, Gopalan et al. 1974). Therefore, a major objective for breeding is to identify and apply genetic resources to produce low erucic acid (LEA) (<2%) seeds. In the 1960s, the first LEA variant was found in a feed rape cultivar called Liho (Downey and Craig 1964), and the first LEA B. napus cultivar, ORO, was bred using Liho (Downey and Craig 1964, Harvey and Downey 1964). Then, the first LEA B. rapa, SPAN, was bred (Downey 1964), and the LEA B. juncea resource Zem was isolated (Kirk and Oram 1978). Most LEA cultivars of the above species were developed by the introduction of recessive alleles from the donor varieties ORO, SPAN and Zem, or their derivative lines. This single LEA genetic resource has caused great concern about inbreeding effects and genetic erosion (Harvey and Downey 1964). B. rapa is the ancestral parent species of B. napus and B. juncea and has become increasingly attractive to biologists and plant breeders, largely due to its higher diversification and economic importance. Therefore, an exploration of novel zero erucic acid lines of B. rapa will expand the genetic resource of LEA genes and promote independent innovation capacity for quality improvement of oilseed via interspecific crosses.
The fatty acid elongase 1 (FAE1) gene encodes the first enzyme (β-ketoacyl-CoA synthase, KCS) in erucic acid biosynthesis and serves as the rate-limiting enzyme for this process in higher plants (James et al. 1995). This 1521-bp FAE1 gene, with no intron, has been isolated from Arabidopsis thaliana, B. napus, B. oleracea and B. rapa (Das et al. 2002, Fourmann et al. 1998, James et al. 1995). Many recent reports have addressed the relationship of FAE1 to erucic acid content. The two FAE1 genes in B. napus (FAE1.1 and FAE1.2) were found to be tightly linked to the E1 and E2 loci controlling erucic acid content (Barret et al. 1998, Fourmann et al. 1998, Jourdren et al. 1996). Subsequently, Roscoe et al. (2001) found that an absence of erucic acid (22: 1Δ13) in LEA rapeseed was correlated with a lack of acyl-CoA elongation activity. The LEA trait in B. napus can be attributed to the substitution of a single amino acid residue from serine to phenylalanine at position 282 of the encoded protein (Han et al. 2001, Katavic et al. 2002, 2004). Cys223 was considered to be another putative active site of FAE1 (Ghanevati and Jaworski 2001, 2002), and the conserved Asn424 and His391 residues were confirmed to be important in FAE1-KCS activity (Davies et al. 2000, Ghanevati and Jaworski 2001, Huang et al. 1998, Jez et al. 2000). However, all of these amino acids are present in both LEA and high erucic acid (HEA) rapeseed (Han et al. 2001, Katavic et al. 2002, Roscoe et al. 2001). More recently, Wu et al. (2008) attributed the LEA trait of rapeseed to a four-nucleotide deletion in the BnFAE1 gene. Similarly, four substitution-type single-nucleotide polymorphisms (SNPs), one in FAE1.1 and three in FAE1.2, were identified to distinguish low erucic types from high erucic types in B. juncea (Gupta et al. 2004, Xu et al. 2010). These results indicate that the LEA phenotype was due to variations in the FAE1 coding sequence (CDS) in B. napus and B. juncea. Despite these recent advances in the biochemistry of seed elongases and cloning of FAE1, the nature of the mutations that characterize the agriculturally important LEA trait remains obscure, particularly in B. rapa. Sequence alignment of the FAE1 gene between the HEA and LEA B. rapa revealed three SNPs due to transition-type base substitutions at positions 591 (G/A), 735 (C/T), and 968 (C/T) (Wang et al. 2010). Among the three variations, only the 968 (C/T) led to an amino acid change, which may have caused the phenotypic difference (Wang et al. 2010, Xu et al. 2010).
Here, we analyzed the erucic acid content of 1981 B. rapa landraces and found that over 90% of the samples had more than 40% erucic acid. An LEA landrace was isolated and FAE1 was analyzed. We found that the variation in the CDS of FAE1 did not cause the variable erucic acid content of B. rapa, and the LEA B. rapa formation may be attributable to the decrease in FAE1 expression. The 24-base AT-rich region deletion in the FAE1 promoter of the LEA B. rapa may be responsible for altering the expression of FAE1. A molecular marker was developed based on the deletions, and the genotype with the deletions co-segregated with the LEA trait in the segregating population. In the study, the formation of LEA in B. rapa is explained in a novel way. The discovery of the LEA landrace could be used in LEA breeding.
Materials and Methods
Plant materials
The seeds of most of the B. rapa accessions were obtained from the Chinese Crop Germplasms Information System (CGRIS, a germplasm repository for collecting worldwide genetic resources of oilseed crops). In this study, 1981 B. rapa landraces from China were used to reveal the erucic acid variation among landraces and to screen for zero erucic acid landraces. Twenty-four inbred B. rapa accessions (11 landraces selected from the 1981 landraces and 13 modern elite cultivars including 4 foreign cultivars) were selected to clone the CDS of FAE1 (Supplemental Table 1). The GenBank accession numbers for the nucleotide sequences range from KF999615 to KF999639. All of the self-pollinated seeds were harvested for determination of the erucic acid content.
To establish an F2 population segregating for different alleles at the FAE1 promoter locus, Sanjiecaizi (an LEA type containing zero erucic acid, e/e) was crossed with Nanhualinggongdacaizi (an HEA type containing 55% erucic acid, E/E) (Supplemental Table 1). The effect of the deletions on erucic acid formation was examined using 118 F2 seeds.
Fatty-acid analysis
Approximately thirty mature oven-dried seeds were first ground in tubes and then ground to a fine powder using small pestles. After dissolution in 1 mL petroleum ether, each sample was treated using ultrasonic irradiation (using ultrasonic power, i.e., 100 W, at 20°C) for 20 min. After saponification in 3 mL of 0.4 N methanolic-KOH, the samples were sonicated for 20 min and centrifuged, and the supernatant was then collected. The supernatant samples were added to 3 mL of H2O and vortexed for 15 min. After centrifugation, the 2-μl upper layer was loaded into a gas chromatographic analyzer (GC, Agilent 6890N). The GC conditions were set according to Hu et al. (2009). The erucic acid content was determined by measuring the area of the peak.
Fifty seeds from each sample with an erucic acid content less than 20% were analyzed using the half-seed method (one cotyledon was analyzed using the GC, and the other was sown and harvested by self-crossing) to isolate zero erucic acid materials. Fatty-acid analysis of the half-seed method was the same as the method described above except one-tenth of the reagents was used. The half-seed method was also used for erucic acid determination of the F2 seeds (Gupta et al. 2004).
Cloning the FAE1 coding and promoter sequences
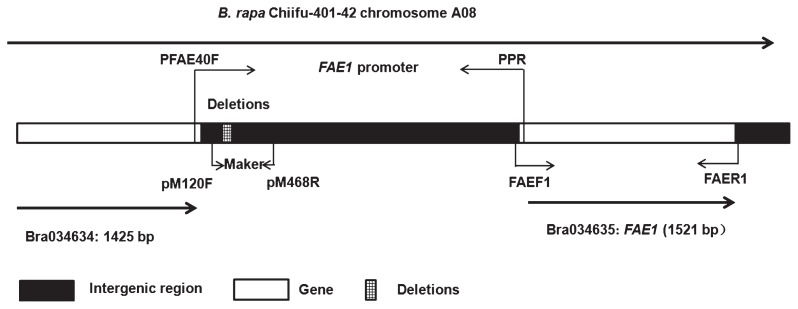
Based on the FAE1 sequences from BRAD Bra034635.1 (Wang et al. 2011, http://www.brassicadb.org), the forward primer (FAEF1: 5′-ATTCTCCGACACACACACTG-3′) and the reverse primer (FAER1: 5′-AGAGAAACATCGTAGCCATCA-3′) were designed to isolate the CDS of FAE1. To obtain the full length of the upstream sequence of FAE1, one pair of specific primers (PFAE40F: 5′-TGCATCCATAGATATCCTGT-3′; PPR: 5′-AACGGAAAGAAGCAAAGGT-3′) was designed within the 3′ and 5′ sequences of Bra034634 and FAE1. The primers of pM120F (5′-TCGGTAAAAGAAAAATCA-3′) and pM468R (5′-CTCATCTAAACTATATTAAGTG-3′) were designed based on the deletions of the LEA promoter for genotyping the segregating population (Fig. 1).
Fig. 1.
Schematic representation of the position of FAE1 and its promoter in the B. rapa chromosome.
Total DNA was extracted from leaves according to the methods described by Murray and Thompson (1980). Genomic DNA was used as a template for PCR, which was carried out using a KOD-plus kit (Toyobo, Japan) and the primers mentioned above. The reactions were prepared in a total volume of 50 μl containing 1 μl of genome DNA, 1 μl of each 10 μM primer, 5 μl of 10× buffer, 2 μl of Mg2+, 5 μl of dNTPs, 1 μl of KOD-plus and 34 μl of ddH2O. The PCR was carried out in a PTC-200 Peltier Thermal Cycler (Bio-Rad, USA) using the following program: 5 min of initial denaturation at 94°C; 35 cycles of 1 min at 94°C, 1 min at 56°C and 1 min 50 sec at 68°C for FAE1; and a final extension of 10 min at 68°C. The annealing temperature was 54°C for FAE1 promoter amplification.
A nucleotide of adenine was added to the end of the PCR products and they were purified using a PCR purification kit. The FAE1 genes from different sources were cloned into the pEASY-T1 vector using a pEASY-T1 Cloning Kit (Transgen Biotech, China) and sequenced using the M13 forward and reverse primers (Shanghai Sangon, China). Approximately 5 clones bearing PCR products from each cultivar were sequenced. The sequence alignment was carried out using the Vector NTI suite 9.0 software package.
Determination of FAE1 transcript levels
The transcript levels of FAE1 in the developing seeds of the inbred LEA Sanjiecaizi, HJa 96368 and HEA Nanhualinggongdacaizi were analyzed using quantitative reverse transcription (qRT-PCR). Total RNA was extracted according to the protocol described for Arabidopsis seeds (Vicient and Delseny 1999). RNA pellets were dissolved in DEPC-treated water, quantified by absorbance at 260 nm and checked for quality using agarose gel electrophoresis. Total RNA samples were reverse transcribed to first-strand cDNA using a ReverTra Ace-α-TM qPCR RT kit (Toyobo) and stored at −80°C. The expression of the FAE1 gene was normalized to ACTIN using the 2−ΔΔCT method (Livak and Schmittgen 2001). The FAE1 and ACTIN primers and the amplification conditions have been described previously (Hu et al. 2009).
Real-time PCR reactions were performed in triplicate using the 2× SYBR Green qPCR Master Mixes (Toyobo). The iCycler iQ5 (Bio-Rad) was used for all amplifications. For each gene, triplicate sets of the PCR reaction samples were prepared and run in an 8-tube strip (Bio-Rad). The PCR experiments were repeated for each plate to ensure that similar results were obtained. The PCR protocol consisted of an initial denaturation step at 94°C for 2 min, followed by 40 repeats of denaturation at 94°C for 10 sec and a combined primer annealing/elongation step at 58°C for 20 sec. A melting curve cycle followed the amplification cycle to confirm PCR product specificity: 94°C for 1 min, 58°C for 1 min, and acquisition of a melting curve from 55°C to 95°C with temperature change values of 0.5°C and a dwell time of 30 s.
Statistical analysis
Box plots of erucic acid contents in F2 individuals grouped by FAE1 genotypes were drawn and the differences in the erucic acid contents among different genotypes were tested using R software Version 3.11 for Windows (http://www.r-project.org/). P < 0.01 was considered statistically significant.
A Chi-square test was applied to determine the segregation of the genotypes in the F2 population. The expected ratio of the homozygous (Sanjiecaizi, e/e) to heterozygous (E/e) to homozygous (Nanhualinggongdacaizi, E/E) genotypes was 1 : 2 : 1.
Results
Survey of erucic acid content variation in Chinese landraces of B. rapa
The phenotypic distribution of the erucic acid content in the 1981 B. rapa lines is shown in Table 1. There were no lines that contained less than 10% erucic acid. There were five classes of phenotypes (10–20%, 20–30%, 30–40%, 40–50% and 50–60%). Among the samples, 92.07% (1824 accessions) had a high erucic acid content, i.e., more than 40%. Of these, 52.70% had an erucic acid content of 40–50%, and 39.37% had an erucic acid content up to 50–60%. From the perspective of geography, we found that the winter B. rapa from the Yangtze valley (comprised of Jiangsu, Anhui, Jiangxi, Hunan, Hubei, Sichuan and Yunnan and Xizang provinces) had an erucic acid content of 40–60%, whereas spring B. rapa from Qinghai, Gansu and Xinjiang had an erucic acid content of 30–40%. In 18 accessions, the erucic acid contents were between 20% and 30%. Only 17 landraces had 10–20% erucic acid; these were distributed across Anhui, Sichuan, Guizhou, and so on. The 17 landraces with 10–20% erucic acid in the seeds were then used to isolate zero-EAC individuals.
Table 1.
Survey of erucic acid content variation in Chinese landraces of B. rapa
| Origin | Number of accessions with different erucic acid contents | Total | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 0–10% | 10–20% | 20–30% | 30–40% | 40–50% | 50–60% | ||
| Jiangsu | 10 | 37 | 47 | ||||
| Zhejiang | 2 | 29 | 48 | 79 | |||
| Anhui | 3 | 70 | 138 | 211 | |||
| Jiangxi | 1 | 117 | 10 | 128 | |||
| Hunan | 64 | 25 | 89 | ||||
| Hubei | 3 | 95 | 131 | 229 | |||
| Sichuan | 4 | 2 | 75 | 84 | 165 | ||
| Guizhou | 3 | 7 | 19 | 134 | 149 | 312 | |
| Yunnan | 37 | 84 | 121 | ||||
| Shaanxi | 6 | 168 | 10 | 184 | |||
| Henan | 5 | 30 | 9 | 44 | |||
| Shanxi | 1 | 1 | 59 | 23 | 84 | ||
| Guangdong | 13 | 4 | 17 | ||||
| HongKong | 1 | 1 | |||||
| Taiwan | 2 | 1 | 3 | ||||
| Fujian | 14 | 10 | 24 | ||||
| Gansu | 2 | 35 | 8 | 1 | 46 | ||
| Qinghai | 3 | 40 | 13 | 2 | 58 | ||
| Xinjiang | 4 | 3 | 1 | 8 | |||
| Xizang | 1 | 2 | 8 | 103 | 14 | 128 | |
| Mongolia | 2 | 1 | 3 | ||||
| Total | 0 | 17 | 18 | 122 | 1044 | 780 | 1981 |
| Proportion | 0 | 0.86% | 0.91% | 6.16% | 52.70% | 39.37% | 100% |
Isolation of zero erucic acid B. rapa
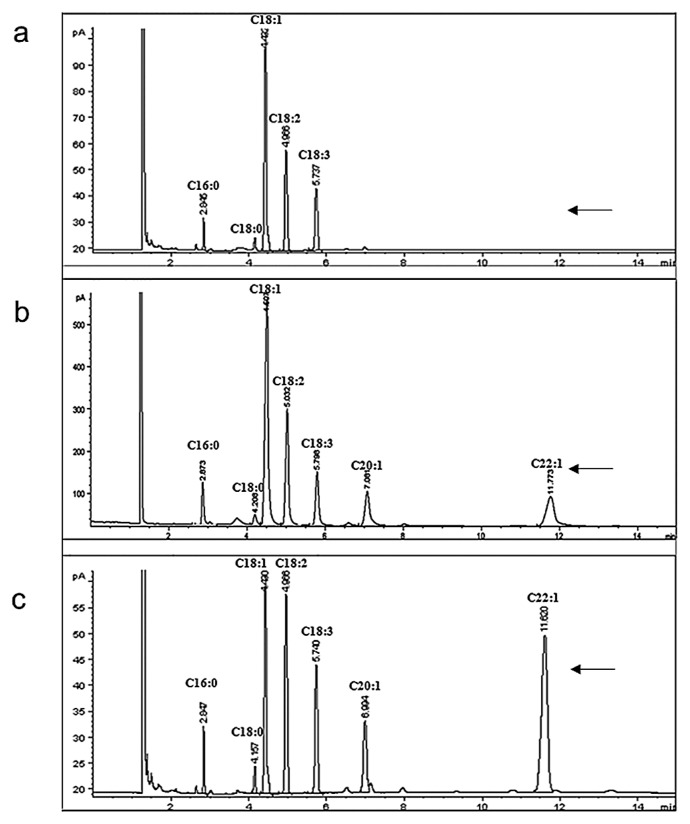
Half-seed GC analysis of the 17 landraces with 10–20% erucic acid showed that their erucic acid contents varied from 0% to 36.6% (data not shown). The Sanjiecaizi individuals had the largest variation of erucic acid content (varying from 0 to 34%) among the 17 landraces, and the erucic acid contents of the other 16 landraces varied from 11.0% to 36.6%. Among the fifty seeds of the Sanjiecaizi analyzed, in 26 seeds (52%), the erucic acid contents were zero; in three seeds (6%), between 10% and 20%; in thirteen seeds (26%), between 20% and 30%; and in eight seeds (16%), between 30% and 40% (Fig. 2, Supplemental Table 2). The Sanjiecaizi individuals with zero erucic acid were planted and self-pollinated, and we ultimately obtained stable zero erucic acid B. rapa lines through several self-pollinations.
Fig. 2.
GC analysis showing fatty acid profiles of different individuals of Sanjiecaizi. a: Sanjiecaizi-1 (erucic acid content: 0); b: Sanjiecaizi-28 (erucic acid content: 13.84%); c: Sanjiecaizi-49 (erucic acid content: 33.22%).
FAE1 polymorphisms in the 24 B. rapa with different erucic acid contents
The 24 accessions of B. rapa whose erucic acid contents were between 0 and 55% (with a normal distribution) were collected for analysis of the CDS of FAE1, and the materials contained three zero erucic acid B. rapa accessions: HJa96368 (a cultivar from Finland), Sanjiecaizi (a landrace from China), and Qingyou 11 (a cultivar from China). PCR amplification of each of the 24 lines generated a band at 1.5 kb that was cloned into the pEASY-T vectors. The corresponding clones were designated by the name of the lines. Alignment of the FAE1 CDS revealed 26 SNPs along the CDS, with a similarity of up to 98–100% (Supplemental Table 1). The SNPs led to 13 mutations in the deduced amino acid sequences. Most samples had three variable sites (at positions 122, 591 and 735) (Supplemental Table 1). However, no sites of the FAE1 CDS were distinguishable between the HEA and LEA accessions. The results indicate that the variation in the FAE1 CDS did not contribute to the variable erucic acid phenotype. Moreover, the CDS of the LEA cultivars HJa 96368 from Finland and Qingyou 11 from China shared 100% identity. However, the LEA landrace Sanjiecaizi had three variations (at positions 591, 735 and 968) compared with the CDS of HJa 96368 and Qingyou 11.
FAE1 gene expression in LEA and HEA B. rapa
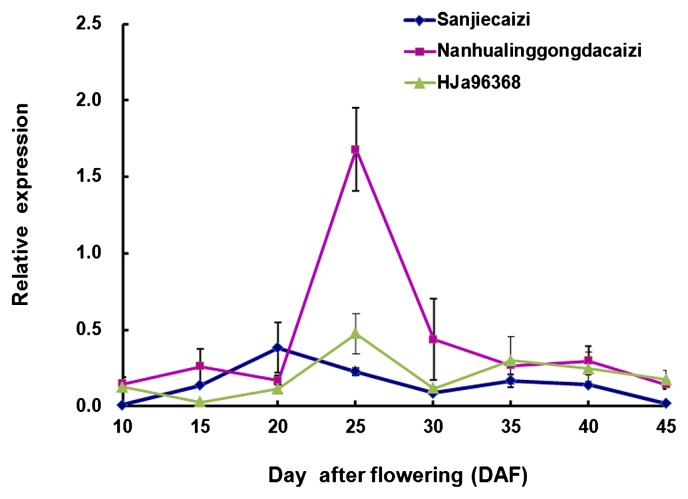
To confirm whether the FAE1 gene expression affected the erucic acid content of B. rapa, we carried out transcriptional analysis in the developing seeds of inbred landraces LEA Sanjiecaizi, HJa 96368 and HEA Nanhualinggongdacaizi. The mRNA developmental profiles during seed development differed between the HEA and LEA seeds (Fig. 3). In the HEA accession, the FAE1 expression could be detected 10 days after flowering (DAF). The expression decreased at 15–20 DAF and increased significantly at 20–25 DAF. The maximal expression levels were reached at 25 DAF. The expression decreased sharply at 25–30 DAF and then steadily declined at 30–45 DAF. In contrast, in LEA B. rapa, the FAE1 gene expression remained stable at lower levels compared with HEA B. rapa at most developmental stages. The maximal expression level was measured at 20 and 25 DAF in LEA Sanjiecaizi and HJa 96368, respectively. In the Sanjiecaizi individuals, the FAE1 gene transcripts decreased at 20–30 DAF and increased during 30–35 DAF. After 35 DAF, the transcript levels continued to decrease until no transcription was detected during the final stage of seed development. In HJa 96368, the FAE1 gene transcripts increased at 15–25 DAF, decreased during 25–30 DAF and increased during 30–35 DAF. After 35 DAF, the transcript levels continued to decrease. A comparison of the peak transcript levels indicated that the FAE1 gene transcript levels in HEA were 4.4-fold and 3.5-fold those measured in LEA B. rapa Sanjiecaizi and HJa 96368, respectively. Generally speaking, the FAE1 gene transcript was more abundant in the HEA than in the LEA accession during seed development.
Fig. 3.
The expression profile of the FAE1 gene during seed development in HEA B. rapa Nanhualinggongdacaizi and LEA B. rapa Sanjiecaizi and HJa 96368.
FAE1 promoter analysis in LEA and HEA B. rapa
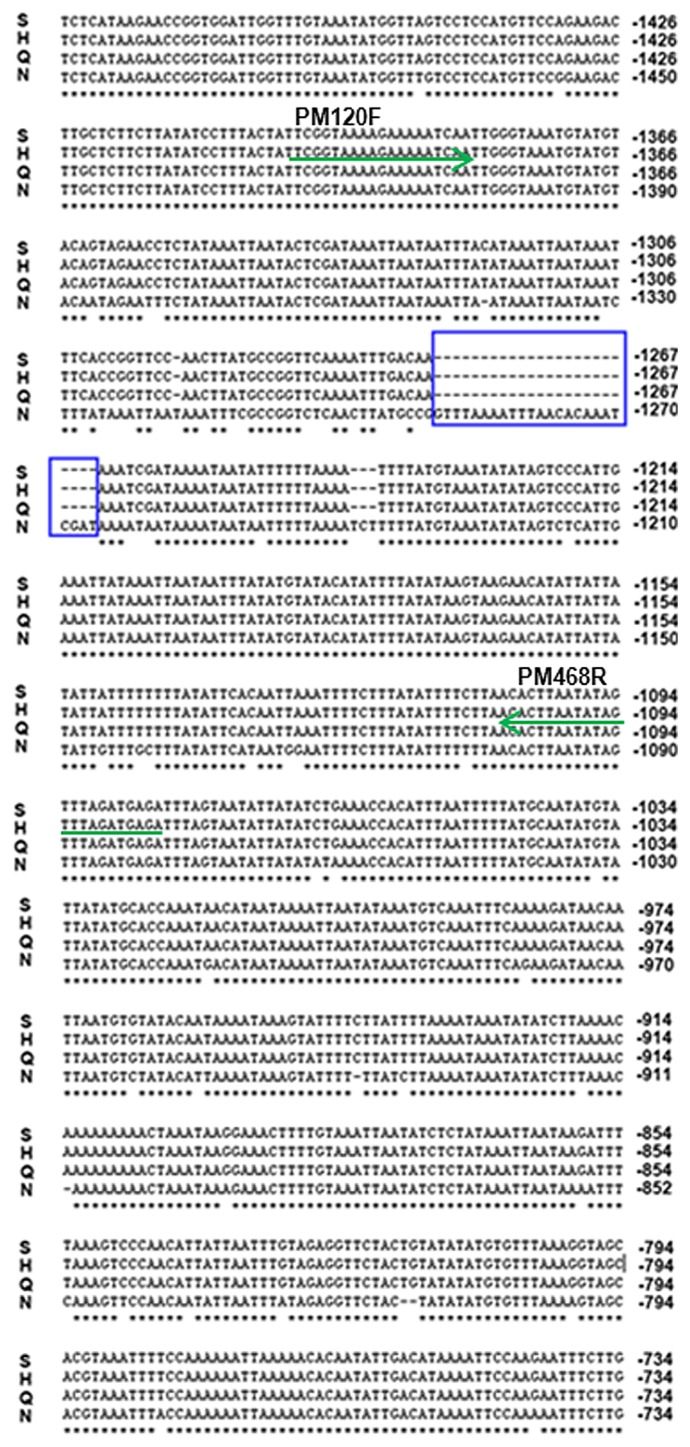
The promoter regions of the FAE1 gene from the LEA B. rapa Sanjiecaizi and Qingyou 11 and HJa 96368, and the HEA Nanhualinggongdacaizi were isolated based on the Chiifu-401-42 sequence (Wang et al. 2011) (Fig. 1). The alignment results showed that the three LEA promoter sequences shared 95% similarity with the HEA sequences; however, the sequence of the landrace LEA Sanjiecaizi was 100% similar to the LEA cultivar Qingyou 11 and the foreign cultivar HJa 96368. There were 56 SNPs and 37 INDELs between the HEA and LEA promoter sequences. Among the INDELs, a total of 28 bases were deleted at a position approximately 1300 bp from the translation initiation site (ATG) of the LEA allele promoter (Fig. 4). The deletions caused the LEA sequence to lose a 24-base AT-rich region. The variations were mainly located in the region shown in Fig. 4.
Fig. 4.
The sequences alignment results of the HEA and LEA FAE1 promoters. N: HEA B. rapa Nanhualinggongdacaizi (accession number: KF999632). S: LEA B. rapa Sanjiecaizi (accession number: KF999615). H: LEA B. rapa HJa 96368 (accession number: KF999623). Q: LEA B. rapa Qingyou 11 (accession number: KP718763). ▭ The A/T-rich sequence deleted from LEA FAE1 promoters. The promoter regions do not contain CpG islands. The sequence data presented here have been submitted to GenBank. → The positions of pM120F and pM468R.
Association between the erucic acid content and the 28 bases deletions in the promoter region
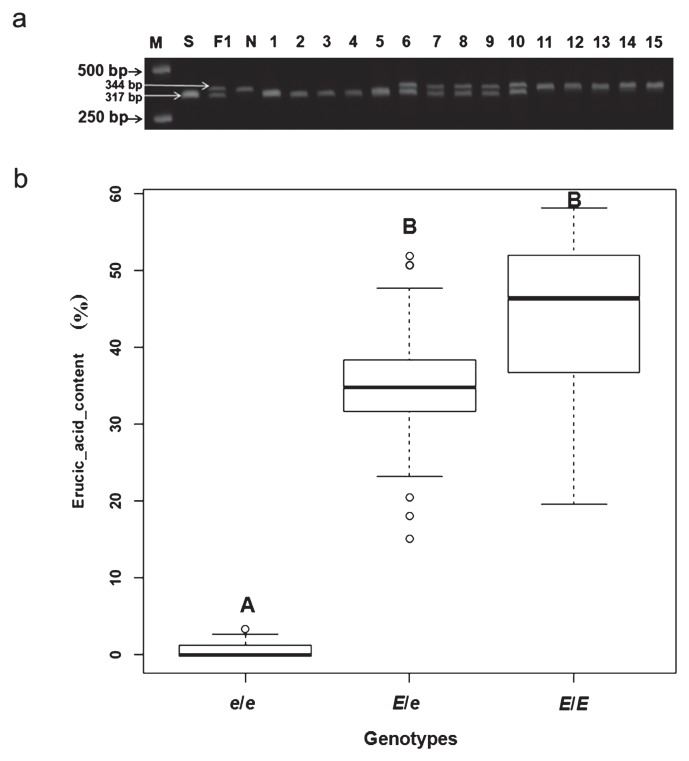
The primer pair pM120F/pM468R was specifically designed for the region carrying the 28 bases deletions in the promoter of FAE1 to reveal the relationship between the phenotypes and the deletions (as shown in Fig. 4). The amplicons could be accurately scored by size difference on 2.5% agarose gels. The primers produced a 317-bp fragment in the zero erucic acid Sanjiecaizi, whereas a 344-bp fragment was produced in the HEA Nanhualinggongdacaizi; 317-bp and 344-bp fragments were produced in the heterozygous F1 individuals (with an erucic acid content of 30.36 ± 5.02%), as predicted (Fig. 5a). In 118 F2 plants from the Sanjiecaizi (e/e) × Nanhualinggongdacaizi (E/E) cross, 30 plants with amplicons resembling the zero erucic acid parent Sanjiecaizi were LEA lines (0.75% ± 1.32%); 38 plants with amplicons resembling the HEA parent Nanhualinggongdacaizi had an erucic acid content of 43.64 ± 10.60%, and 50 heterozygous individuals had an erucic acid content of 35.12 ± 7.94% (Fig. 5a, 5b). The segregation ratio of the three genotypes in the F2 population agreed perfectly with the expected ratio of 1 : 2 : 1 (χ2 = 3.83, P = 0.147). The result showed that all LEA plants were e/e homozygous and the erucic acid content of the e/e genotype was highly significantly different from that of the E/E and E/e genotypes (p < 0.01) (Fig. 5b). Therefore, the homozygosity with the 28 bases deletions co-segregated with the LEA phenotypes.
Fig. 5.
The amplicons amplified by the primer pair pM120F and pM468R in the F2 population of Sanjiecaizi (e/e) × Nanhualinggongdacaizi (E/E). The amplicon contained one insertion in addition to the 28 deletions (as shown in Fig. 4). Therefore, the fragments amplified from the e/e genotype were 27 bases shorter than those from the E/E genotype, which was confirmed using sequencing. (a) M, DNA ladder; S, Sanjiecaizi (e/e), zero erucic line, 317 bp; F1 plants with a erucic acid content of 30.36 ± 5.02%, Sanjiecaizi × Nanhualinggongdacaizi, 344 bp and 317 bp; N, Nanhualinggongdacaizi (E/E), high erucic line, 344 bp; 1 to 5, F2 plants with a low erucic acid content (average 0.75% ± 1.32%); 6 to 10, F2 plants with a erucic acid content of 35.12 ± 7.94%; 11 to 15, F2 plants with a high erucic acid content (43.64% ± 10.60%). (b) Box plot showing the erucic acid content range and median for the three genotypes. e/e: homozygous genotype resembling the zero erucic acid parent Sanjiecaizi. E/e: heterozygous genotype containing both alleles of the two parents. E/E: homozygous genotype resembling HEA Nanhualinggongdacaizi. Note: Capital letters indicate significant differences at the 0.01 level.
Discussion
Because of the effect of erucic acid on human health, there is a maximal allowed level of erucic acid in food oil in most countries (5% in the EU). Downey (1964) bred SPAN, the first LEA B. rapa cultivar in the world. The first zero erucic acid B. rapa cultivar Torch was registered based on SPAN in 1973. In 1989, Tian bred the first LEA B. rapa cultivar named Qingyou 11 in China using the introduced LEA genetic resource from SPAN (Liu 1985). To identify the LEA genetic resources in the landraces, we analyzed the erucic acid contents of 1981 B. rapa landraces from China. There were no lines that contained less than 10% erucic acid. At least 92.07% (1824) of the B. rapa lines had erucic acid levels up to 40%. Another 17 B. rapa landraces had 10–20% erucic acid content, which indicates it may be possible to isolate LEA from these lines. However, there was no LEA resource in the collection. We deduced that highly self-sterile, cross-pollinating plants make the LEA accessions hard to maintain. Therefore, there are very few erucic acid-free winter B. rapa cultivars registered. By generation selection, we ultimately isolated the LEA landrace Sanjiecaizi.
The sequence alignment of the FAE1 CDS indicates that the LEA landrace Sanjiecaizi had three variations compared with the LEA HJa 96368 and Qingyou 11 cultivars. In addition, there were 26 SNPs among the varieties, and these SNPs led to 13 amino acid changes. The 591 (G/A), 735 (C/T) and 968 (C/T) variations that had been reported to be associated with erucic acid content in B. rapa (Wang et al. 2010) appeared simultaneously in HEA and LEA B. rapa. The 845 (C/T) variation detected in FAE1 of the LEA B. napus A genome (Wu et al. 2008) did not occur between HEA and LEA B. rapa. Thus, it may be concluded that the LEA genetic resources in the A genomes differed between B. rapa and B. napus (Wang et al. 2010). In addition, the other 19 variations at other positions were not correlated with the erucic acid content of B. rapa through statistical analysis (data not shown). Based on the analysis above, we demonstrated that these altered DNA and amino acid sequences are not responsible for LEA.
A previous study showed that FAE1 gene(s) expression was found to be restricted to the embryo and was temporally regulated during seed development. In addition, the highest transcript levels were found at approximately 24–30 DAF, concomitant with the accumulation of 22 : 1 in rapeseed oil (Han et al. 2001). Our results show that the FAE1 gene was transcribed normally in both HEA and LEA accessions. The peak transcript levels of the LEA Sanjiecaizi, HJa 96368 and the HEA Nanhualinggongdacaizi occurred at 20, 25 and 25 DAF, respectively, which is basically identical to the results of previous reports. However, the transcription level in HEA B. rapa cultivar Nanhualinggongdacaizi was much higher than that detected in LEA Sanjiecaizi and HJa 96368. It is clear that the formation of LEA B. rapa is due to the decreased expression level of FAE1. In B. napus, though the FAE1 expression in the LEA cultivar is much higher than in the HEA cultivar (Hu et al. 2009, Wu et al. 2008), there is no relationship between the expression and the erucic acid types. The LEA trait in B. napus is determined by a four-base pair deletion in the CDS of the FAE1 gene that leads to a frameshift mutation (Wu et al. 2008). Similarly, in B. juncea, SNPs in FAE1.1 and FAE1.2 were identified for distinguishing the low from the high erucic types (Gupta et al. 2004). In Sinapis alba, the SNPs in the FAE1 CDS did not affect enzyme functionality; however, the transposable element insertion of the promoter and epigenetic modification decreased the expression of FAE1 sharply, which caused LEA formation (Zeng and Cheng 2014). Similarly, we found that LEA B. rapa is attributed to the expression level, not the variation of the FAE1 CDS, and the regulation mechanism for LEA in B. rapa is different from that in B. napus and B. juncea.
To determine why the expression of FAE1 decreased in LEA B. rapa, we analyzed the promoter region of FAE1. The alignment of the promoter region of the FAE1 gene indicated that a total of 28 bases were deleted at a position approximately 1300 bp from the translation initiation site (ATG) of the LEA promoter. The deletions contained a 24-base A/T-rich region. The A/T-rich sequences are able to act as quantitative, non-tissue-specific enhancer elements in higher plants (Chen et al. 1988, Sandhu et al. 1998). A 33-bp double-stranded oligonucleotide homologous to AT-rich sequences can increase anaerobic stress-induced transcription of the maize Adhl promoter (Czarnecka et al. 1992). A previous study suggested that a 31-bp A/T-rich sequence and a 26-bp random A/T sequence were able to enhance GUS expression, and the enhancer activity was correlated with the number of copies of the A/T-rich sequence (Sandhu et al. 1998). In our study, the deletions in the LEA promoter also occurred at an A/T-rich region. A PCR-based marker was developed based on the deletions and was detected in the F2 population. The segregation ratio of the three genotypes agreed perfectly with the Mendelian model of a single major gene that controls the trait, which was consistent with the report of the segregation of erucic acid phenotypes of the F2 seeds (Dorrell and Downey 1964). Most importantly, the homozygosity with the 28 bases deletions co-segregated with the LEA phenotypes. Therefore, we propose that the divergent promoter may be associated with the differential transcription of FAE1 in the HEA and LEA accessions. Our next step is to reveal the relationship between the expression and the deletions in the promoters using a transgenic approach in which the only difference between the HEA and LEA lines will be the 28 bases deletions. Other transcription factors may be simultaneously involved in the regulation of FAE1.
Supplementary Material
Acknowledgments
We would like to thank American Journal Experts for providing language editing and the National Center for Biotechnology Information for the GenBank accessions. This study was funded by the National Natural Science Foundation of China (31100911), the 973 Projects (2011CB109302), and the Natural Science Foundation of Hubei Province (2011CDB353).
Literature Cited
- Badawy, I.H., Atta, B. and Ahmed, W.M. (1994) Biochemical and toxicological studies on the effect of high and low erucic acid rapeseed oil on rats. Nahrung 38: 402–411. [DOI] [PubMed] [Google Scholar]
- Barret, P., Delourme, R., Renard, M., Domergue, F., Lessire, R., Delseny, M. and Roscoe, T.J. (1998) A rapeseed FAE1 gene is linked to the E1 locus associated with variation in the content of erucic acid. Theor. Appl. Genet. 96: 177–186. [Google Scholar]
- Beare-Rogers, J.L. (1971) Liver phospholipids of rats fed a choline-deficient diet supplemented with choline or methionine. Lipids 6: 649–651. [DOI] [PubMed] [Google Scholar]
- Chen, Z.L., Pan, N.S. and Beachy, R.N. (1988) A DNA sequence element that confers seed-specific enhancement to a constitutive promoter. EMBO J. 7: 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecka, E., Ingersoll, J.C. and Gurley, W.B. (1992) AT-rich promoter elements of soybean heat shock gene Gmhsp 17.5E bind two sets of nuclear proteins in vitro. Plant Mol. Biol. 19: 985–1000. [DOI] [PubMed] [Google Scholar]
- Das, S., Roscoe, T.J., Delseny, M., Srivastava, P.S. and Lakshmikumaran, M. (2002) Cloning and molecular characterization of the Fatty Acid Elongase 1 (FAE 1) gene from high and low erucic acid lines of Brassica campestris and Brassica oleracea. Plant Sci. 162: 245–250. [Google Scholar]
- Davies, C., Heath, R.J., White, S.W. and Rock, C. (2000) The 1.8 A crystal structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from Escherichia coli. Structure 8: 185–195. [DOI] [PubMed] [Google Scholar]
- Dorrell, D.G. and Downey, R.K. (1964) The inheritance of erucic acid content in rapeseed (Brassica campestris). Can. J. Plant Sci. 44: 499–504. [Google Scholar]
- Downey, R.K. (1964) A selection of Brassica campestris L. containing no erucic acid in its seed oil. Can. J. Plant Sci. 44: 295–298. [Google Scholar]
- Downey, R.K. and Craig, B.M. (1964) Genetic control of fatty acid biosynthesis in rapeseed (Brassica napus L.). J. Am. Oil Chem. Soc. 41: 475–478. [Google Scholar]
- Fourmann, M., Barret, P., Renard, M., Pelletier, G., Delourme, R. and Brunel, D. (1998) The two genes homologous to Arabidopsis FAE1 co-segregate with the two loci governing erucic acid content in Brassica napus. Theor. Appl. Genet. 96: 852–858. [Google Scholar]
- Ghanevati, M. and Jaworski, J.G. (2001) Active-site residues of a plant membrane-bound fatty acid elongase β-ketoacyl-CoA synthase, FAE1 KCS. Biochim. Biophys. Acta 1530: 77–85. [DOI] [PubMed] [Google Scholar]
- Ghanevati, M. and Jaworski, J.G. (2002) Engineering and mechanistic studies of the Arabidopsis FAE1 β-ketoacyl-CoA synthase, FAE1 KCS. Eur. J. Biochem. 269: 3531–3539. [DOI] [PubMed] [Google Scholar]
- Gopalan, C., Krisnamurthy, D., Shenolikar, I.S. and Krisnamurthy, K.A.V.R. (1974) Myocardial changes in monkeys fed on mustard oil. Nutr. Metab. 16: 352–365. [DOI] [PubMed] [Google Scholar]
- Gupta, V., Mukhopadhyay, A., Arumugam, N., Sodhi, Y.S., Pental, D. and Pradhan, A.K. (2004) Molecular tagging of erucic acid trait in oilseed mustard (Brassica juncea) by QTL mapping and single nucleotide polymorphisms in FAE1 gene. Theor. Appl. Genet. 108: 743–749. [DOI] [PubMed] [Google Scholar]
- Han, J., Lühs, W., Sonntag, K., Zähringer, U., Borchardt, D., Wolter, F., Heinz, E. and Frentzen, M. (2001) Functional characterization of β-ketoacyl-CoA synthase genes from Brassica napus L. Plant Mol. Biol. 46: 229–239. [DOI] [PubMed] [Google Scholar]
- Harvey, B.L. and Downey, R.K. (1964) The inheritance of erucic acid content in rapeseed (Brassica napus). Can. J. Plant Sci. 44: 104–111. [Google Scholar]
- Hu, Y., Wu, G., Cao, Y., Wu, Y., Xiao, L., Li, X. and Lu, C. (2009) Breeding response of transcript profiling in developing seeds of Brassica napus. BMC Mol. Biol. 10: 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Jia, J., Edwards, P., Dehesh, K., Schneider, G. and Lindqvist, Y. (1998) Crystal structure of β-ketoacyl-acyl carrier protein synthase II from E. coli reveals the molecular architecture of condensing enzymes. EMBO J. 17: 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, D.W., Lim, E., Keller, J., Plooy, I., Ralston, E. and Dooner, H.K. (1995) Directed tagging of the Arabidopsis fatty acid elongation 1 (FAE1) gene with the maize transposon activator. Plant Cell 7: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jez, J., Ferrer, J., Bowman, M., Dixon, R. and Noel, J. (2000) Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39: 890–902. [DOI] [PubMed] [Google Scholar]
- Jourdren, C., Barret, P., Horvais, R., Foisset, N., Delourme, R. and Renard, M. (1996) Identification of RAPD markers linked to the loci controlling erucic acid level in rapeseed. Mol. Breed. 2: 61–71. [Google Scholar]
- Katavic, V., Mietkiewska, E., Barton, D.L., Giblin, E.M., Reed, D.W. and Taylor, D.C. (2002) Restoring enzyme activity in nonfunctional low erucic acid Brassica napus fatty acid elongase l by a single amino acid substitution. Eur. J. Biochem. 269: 5625–5631. [DOI] [PubMed] [Google Scholar]
- Katavic, V., Barton, D.L., Giblin, E.M., Reed, D.W., Kumar, A. and Taylor, D.C. (2004) Gaining insight into the role of serine 282 in B. napus FAE1 condensing enzyme. FEBS Lett. 562: 118–124. [DOI] [PubMed] [Google Scholar]
- Kirk, J.T.O. and Oram, R.N. (1978) Mustard as possible oil and protein crops for Australia. J. Aust. Inst. Agric. Sci. 44: 143–156. [Google Scholar]
- Liu, H.. (1985) Rapeseed genetics and breeding. Shanghai Science and Technology Press, Shanghai. [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe, T.J., Lessireb, R., Puyaubertb, J., Renardc, M. and Delsenya, M. (2001) Mutations in the fatty acid elongation 1 gene are associated with a loss of L-ketoacyl-CoA synthase activity in low erucic acid rapeseed. FEBS Lett. 492: 107–111. [DOI] [PubMed] [Google Scholar]
- Sandhu, J.S., Webster, C.I. and Gray, J.C. (1998) A/T-rich sequences act as quantitative enhancers of gene expression in transgenic tobacco and potato plants. Plant Mol. Biol. 37: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient, C.M. and Delseny, M. (1999) Isolation of total RNA from Arabidopsis thaliana seeds. Anal. Biochem. 268: 412–413. [DOI] [PubMed] [Google Scholar]
- Wang, N., Shi, L., Tian, F., Ning, H., Wu, X., Long, Y. and Meng, J. (2010) Assessment of FAE1 polymorphisms in three Brassica species using EcoTILLING and their association with differences in seed erucic acid contents. BMC Plant Biol. 10: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., Bai, Y., Mun, J.H., Bancroft, I., Cheng, F.et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Wu, G., Wu, Y., Xiao, L., Li, X. and Lu, C. (2008) Zero erucic acid trait of rapeseed (Brassica napus L.) results from a deletion of four base pairs in the fatty acid elongase 1 gene. Theor. Appl. Genet. 116: 491–499. [DOI] [PubMed] [Google Scholar]
- Xu, A., Huang, Z., Ma, C., Xiao, E., Zhang, X., Tu, J. and Fu, T. (2010) FAE1 sequence characteristics and its relationship with erucic acid content in Brassica juncea. Acta Agron. Sin. 36: 794–800. [Google Scholar]
- Zeng, F. and Cheng, B. (2014) Transposable element insertion and epigenetic modification cause the multiallelic variation in the expression of FAE1 in Sinapis alba. Plant Cell 26: 2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.