Abstract
Viruses are intracellular parasites that can only replicate and spread in cells of susceptible hosts. Alpha herpesviruses (α-HVs) contain double stranded DNA genomes of at least 120 kb, encoding for 70 or more genes. The viral genome is contained in an icosahedral capsid that is surrounded by a proteinaceous tegument layer and a lipid envelope. Infection starts in epithelial cells and spreads to the peripheral nervous system (PNS). In the natural host, α-HVs establish a chronic latent infection that can be reactivated, rarely spread to the central nervous system (CNS). In the non-natural host, viral infection will in most cases spread to the CNS with often fatal outcome. The host response plays a crucial role in the outcome of viral infection. α-HVs do not encode all the genes required for viral replication and spread. They need a variety of host gene products including RNA polymerase, ribosomes, dynein, and kinesin. As a result, the infected cell is dramatically different from the uninfected cell revealing a complex and dynamic interplay of viral and host components required to complete the virus life cycle. In this review, we describe the pivotal contribution of mass spectrometry-based proteomics (MSBP) studies over the past 15 years to understanding the complicated life cycle and pathogenesis of four α-HV species from the alphaherpesvirinae subfamily: Herpes simplex virus-1 (HSV-1), varicella zoster virus (VZV), pseudorabies virus (PRV) and bovine herpes virus (BHV-1). We describe the viral proteome dynamics during host infection and the host proteomic response to counteract such pathogens.
Keywords: alpha herpesvirus, HSV-1, PRV, BHV-1, VZV, Proteomics
1 INTRODUCTION
The Alphaherpesvirinae subfamily of the family Herpesviridae includes the genera simplexvirus and varicellovirus [1]. These two genera contain several pathogenic species infecting a broad variety of mammals, which for the purpose of this review will be noted as alpha herpesvirus (α-HV). The large double-stranded DNA genome, virion size and structure (icosahedral capsid, tegument and glycosylated lipid envelope), and the latency-reactivation cycle are common features of all α-HVs [2, 3]. These viruses are pantropic and neuroinvasive pathogens that establish a persistent or latent infection in the nervous systems of the natural hosts [4]. Upon reactivation, α-HVs cause diverse effects that vary from mild epithelial lesions, to life threatening necrotizing brain infections and death [5, 6]. The complex and dynamic interplay between virus and host components is an active area of research. The interactions between viral-viral and viral-cell proteins are constantly modified during productive and latent infection. Moreover, the function and localization of viral and cell proteins are regulated at transcriptional, translational and posttranslational (PTM) levels. Spread of infection in the highly connected nervous system, is regulated by directional movement of viral proteins in axons [7]. Spread in the retrograde direction occurs during primary infection (from infected epithelial cells to the innervating neuron), and in the anterograde direction after reactivation (from infected PNS neurons to epithelial cells). This pattern of infection moving into and out of the PNS is a hallmark of α-HVs. The viral and host mechanisms that control viral sorting, transport and egress from axons and dendrites remains partially understood and are an active area of research [8, 9].
In this review, we focus on four α-HV species (see Table 1) where innovative mass spectrometry-based proteomics (MSBP) methods have contributed towards understanding basic and clinical aspects of these pathogens [10]. We review two main topics: 1. The viral and host proteins required to complete the replication cycle; and 2. The cellular response to viral infection and the co-evolution of virus and host (see Fig. 1).
Table 1.
Viral species from the alphaherpesvirinae subfamily included in the present review. For each virus, the acronym is shown on the left column and a brief description of the susceptible host, pathogenesis and life cycle is described on the right column.
| BHV-1 | Bovine herpes virus-1 infects cattle causing rhinotracheitis symptoms that predispose animals to secondary bacterial infections resulting in ‘shipping fever’. BHV-1 is also associated with conjunctivitis, reproductive tract lesions, encephalitis and fetal infections. Economic losses result mainly from shipping fever, milk drop, loss of body weight, early pregnancy termination and death. BHV-1 can establish latency within the natural host [103]. |
| PRV | Pseudorabies virus is a pathogen of swine that causes important economic losses worldwide. It is also known by its taxonomic name, suid herpesvirus 1, or by its original name, Aujeszky’s disease virus. The disease was originally described as “mad itch” because of the intense itching symptoms. The natural host is the swine, although PRV can infect a broad range of vertebrates including cattle, sheep, dogs, cats, goats, chickens, raccoons, possums, skunks, rodents, rabbits, and guinea pigs. PRV is tropic for both respiratory and nervous system tissues of swine. Young swine are severely affected by PRV infection of the central nervous system. Older swine show symptoms of respiratory disease. Several live attenuated vaccine versions are used in PRV eradication programs [41]. PRV is commonly use by neuroscientists to study neuronal connectivity and function in animal models [21, 104]. |
| VZV | Varicella-zoster virus (VZV) is a ubiquitous human pathogen that spreads from mucosal epithelial cells, where initial acute infection occurs, to the skin via a T cell-associated viremia, causing varicella (chicken pox) to young age patients. Viremia and cutaneous infection allow transfer of VZV to sensory nerve ganglia, where a latent infection is established in neurons. Zoster (shingles) is caused by VZV reactivation from latently infected neurons producing a painful vesicular rash (dermatome). Live attenuated VZV vaccines are effective against varicella and zoster [105]. |
| HSV-1 | Herpes simplex virus-1 (HSV-1) is a widespread human pathogen that establishes a life long reactivateable infection in neurons of the peripheral nervous system of the human host. Upon reactivation, HSV-1 particles travel in axons back to epithelial tissue causing oropharyngeal lesions. Occasionally, the infection will spread to the cornea and cause keratitis, or to the central nervous system to cause herpes simplex encephalitis (HSE) with high mortality rates [2]. Currently, no effective vaccine exists and the only treatment is based on antiviral drugs that inhibit viral replication. HSV-1 is widely used to study neuronal connectivity and function in animal models [40]. Ongoing human clinical trials are testing the efficacy of modified HSV-1 strains as oncolytic vectors [106]. |
Figure 1. Use of MSBP in understanding alpha herpesvirus biology.
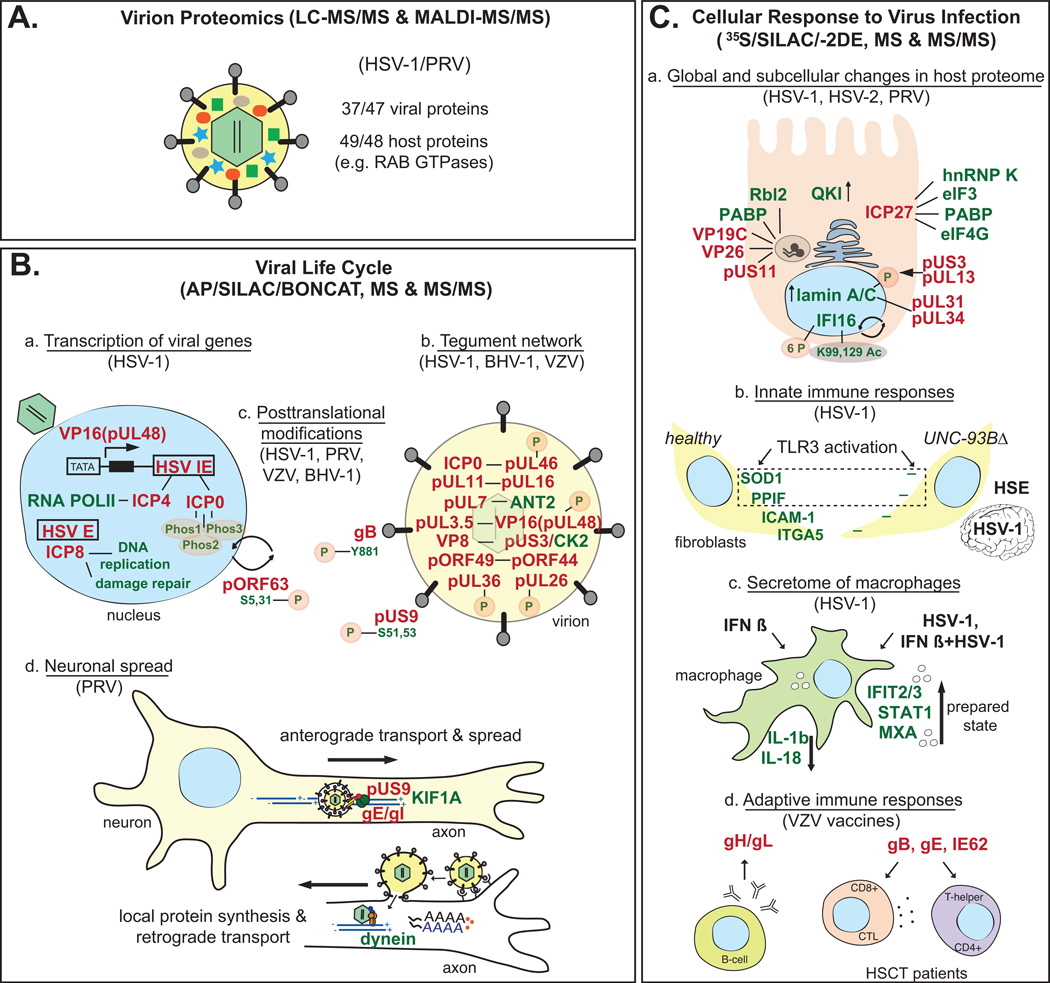
(A) Understanding the complex structure of α-HV virions. HSV-1 and PRV virions are comprised of several viral and cellular proteins. (B) Understanding the viral life cycle. (a) Upon α-HV infection and entry, viral IE genes are transcribed in the nucleus with the action of tegument protein VP16 (pUL48) and its cellular interacting partners. IE proteins (e.g. ICP0 and ICP4) interact with several cellular and viral proteins. Cellular interacting partners of ICP4 and E protein ICP8 were identified by MSBP. (b) MSBP has also contributed to understand the complex tegument network. Several viral and cellular proteins were found to interact with each other and some of them (e.g. pUL26, pUL36, pUL46, pUL48) contained novel PTM’s. (c) Two viral envelope proteins (PRV pUS9 and VZV gB) were phosphorylated (P), this PTM is important for the protein function. PTM analyses revealed 3 phosphorylation regions (Phos1-3) in HSV-1 ICP0, which is a multifunctional nucleocytoplasmic shuttling protein (depicted by arrows). Similarly, VZV protein pORF63 was modified by phosphorylation, which possibly regulates its subcellular regulation. (d) How newly made PRV virions are sorted into axons during neuronal infection was clarified with the identification of KIF1A and pUS9 interaction by MSBP. This interaction is stabilized by two other glycoproteins, gE and gI. The rapid response after PRV entry into axons was also characterized by MSBP, and several host proteins were found to be locally synthesized in axons. Some of these proteins aid retrograde transport of viral particles. (C) Characterizing the changes in the host cell proteome after α-HV infection. (a) Proteins regulating translation (e.g. PABP, hnRNP K, eIF3, eIF4G, QKI), transcription (e.g. Rbl2) and nuclear proteins (e.g. lamin A/C) enabling capsid assembly and egress were characterized by MSBP. Innate immune responses involving DNA sensor IFI16 were identified upon HSV-1 infection by MSBP. (b) Differences in innate immune responses between healthy individuals and HSE patients with UNC-93B mutations were studied detecting differential regulation of SOD1, PPIF, ICAM-1 and ITGA5 in fibroblasts. (c) IFNβ treatment with or without HSV-1 infection in macrophages showed that IFNβ alarms cells by increasing the secretion of proteins such as IFIT2 and 3, STAT1 and MxA (prepared state). HSV-1 proteins counteract these anti-viral responses by decreasing the secretion of cytokines such as IL-1b and IL-18. (c) MSBP was used to characterize the adaptive immune responses to the with VZV vaccine Zostavax, and to identify immunogenic proteins to develop subunit vaccines for HSCT patients. Viral and host proteins are shown in red and green, respectively. Interactions between viral and cellular proteins are depicted as a horizontal line. Increase/decrease in protein levels are shown with arrows. All other abbreviations and references are detailed in the text.
2 ALPHA HERPESVIRUS INFECTION AND INTERACTION WITH HOST PROTEINS
2.1 The composition of alpha herpesvirus virions
Newly assembled herpesvirus particles egress from infected cells using the host secretory pathway that is repurposed by viral components. Mature viral particles travel to the plasma membrane inside acidified secretory vesicles to sites of exocytosis where vesicles fuse with the plasma membrane and release mature virions [11]. The precise global composition of PRV and HSV-1 mature virions has been well studied by MSBP (Fig. 1A) [12–14]. Both PRV and HSV-1 virions have a conserved capsid structure with many common components [15]. In the HSV-1 study, Loret et al. used highly purified virions and performed in-gel digestion, followed by liquid chromatography and tandem mass spectrometry (nLC-MS/MS). They identified 37 of the 40 previously identified virion components, and correctly confirmed the absence of 21 non-structural proteins [14]. Additionally, 4 novel virion components were identified (pUL7, pUL23, pUL50, and pUL55). 54% of all the peptides identified, correspond to tegument proteins. In the PRV study [12], Kramer et al. also used highly purified virions and combined two complementary approaches: First they did in-gel digestion and LTQ Orbitrap XL MS, configured with a matrix-assisted laser desorption/ionization (MALDI) source; Second they performed in-solution digestion by filter-aided sample preparation (FASP) and nLC-MS/MS analyses to compensate for sample loss in the first approach. They identified 47 PRV virion proteins, including seven novel viral components (pUL8, pUL20, pUL32, pUL40, pUL42, pUL50, and Rsp40/ICP22). They used proteinase K digestion to exclude non-specific proteins that could adhere to virions and compared the results with uninfected extracts. Additionally, the PRV study identified phosphorylation sites in virion proteins pUL26, pUL36, pUL46, and pUL48. Strikingly, both the HSV-1 and PRV studies identified as many as 49 host proteins that were incorporated into virions, including several Rab GTPases that are relevant for viral trafficking and egress from infected cells (Fig. 1A). The role of these low copy number host proteins in virions is a subject of research and debate. These proteins could function immediately after viral entry into cells, perhaps facilitating establishment of the infection. For example, the presence of host translation initiation and elongation factor in virions could jump start the production of new viral components [13]. Another idea is that virions may incorporate host proteins from one cell type that are not present in a different cell type perhaps allowing a wider infectious range.
Studies on the virion composition of BHV-1 are ongoing (http://vivo.usda.gov/display/NIFA-1001199-PROJ).
2.2 Viral and host protein interactions during viral transcription, translation and replication
After fusion-mediated cell entry, α-HV capsids and some tegument proteins are actively transported by dynein to the cell nucleus where viral DNA is delivered and transcribed. Viral genes are transcribed in three kinetic groups: immediate early (IE), early (E) and late (L). The timing of gene expression is regulated at transcriptional and posttranslational levels by viral and host proteins. The HSV-1 genome encodes five IE genes, called infected-cell polypeptides or ICP’s. ICP0, ICP4, ICP22, ICP27 and ICP47 proteins have multiple regulatory and immunomodulatory functions and are essential for a productive infection [16]. IE genes are transcribed efficiently before viral protein synthesis or viral replication primarily because the incoming virion carries the tegument protein VP16 (UL48), which is a transcription activator (Fig. 1B, panel a). After synthesis, the IE proteins modulate the transcription of early viral genes (E), which are required for viral DNA replication, transcription of late viral genes (L), and assembly of progeny virions. IE proteins can either activate or repress transcription of most viral genes. To do so, the IE proteins must interact with the host transcription machinery [17]. Since α-HVs do not encode their own RNA polymerase, the host RNA polymerase II complex (RNA pol II) must be repurposed for transcription of viral genes. Several host general transcription factors (GTF) along with RNA pol II and the viral IE protein ICP4 assemble on viral DNA promoters to form the preinitiation complex (PIC). ICP4 is a 350 KDa homodimeric phosphoprotein containing a DNA binding domain, a nuclear localization signal, and regions that modulate the transcription of viral and host genes including its own repression. ICP4 stabilizes the formation of PICs on viral promoters and interacts with GTFs [18]. Affinity purification (AP) of ICP4 followed by MSBP enabled identification of the proteins involved in this process. Forty-six proteins that co-purify with ICP4 were identified by LC-MS/MS, including 11 GTF components of the TFIID complex and 4 components of the Mediator complex. TFIID is a protein complex that includes TATA binding proteins (TBP) that engage essentially all HSV promoters. Mediator is a large multicomponent complex that can facilitate the entry of RNA pol II into the PIC. Mediator interacts with VP16, further demonstrating the relevance of these interactions to recruit all the necessary components to transcribe IE viral genes. Determining the precise number of cell proteins required for efficient cell and viral replication is an area of active research [19, 20].
Viral IE genes not only play a pivotal role in the precise timing of transcription of E and L viral genes, they also have multiple functions in regulating both productive and latent infections. Most IE genes are promiscuous transactivators and modulate transcription and translation of host genes, occasionally leading to cytotoxic effects [21]. ICP27 is an HSV-1 IE gene product that co-purifies with translation initiation factors such as poly A binding protein (PABP), eukaryotic initiation factor 3 (eIF3), and eukaryotic initiation factor 4G (eIF4G). In these complexes, ICP27 stimulates or inhibits viral and host mRNA translation (Fig. 1C, panel a) [22]. Viral protein functions, including those of the viral IE proteins are also regulated by posttranslational modifications (PTMs) [23]. PTM sites in viral proteins are readily identified by MSBP (See section 2.5). The ICP0 IE protein contains multiple phosphorylation sites. MS studies enabled the identification of three regions within or adjacent to transactivation domains of the protein. In total, 11 phosphorylation sites were detected in regions Phos 1, Phos 2 and Phos 3 (Fig. 1B, panel a). These sites were mutated to alanine by site directed mutagenesis to determine their relevance in ICP0 function after viral infection. Mutations in the phosphorylation sites of ICP0 affected its subcellular localization. The most dramatic case was observed when residues in Phos 3 were mutated and ICP0 localization switched from being almost exclusively nuclear to localize both in the nucleus and cytoplasm. Interestingly only mutations in region Phos 1 disabled the ability of ICP0 to complement ICP0 null viral mutants [24].
E genes are expressed only after the IE proteins engage the host transcription machinery. E genes have several functions including the reduction of IE gene expression, DNA replication and recombination, and activation of L gene transcription. The viral replication proteins include pUL30 (DNA polymerase), pUL29 and pUL42 (DNA binding proteins), ORI binding proteins (pUL9) and the helicase/primase complex (pUL5, pUL8 and pUL52). MSBP studies on ICP8 (encoded by UL29 gene) revealed an extensive list of interacting cellular partners that may be relevant for viral replication. ICP8 is a 128 KDa non-structural multifunctional zinc metalloprotein that binds single stranded DNA and localizes to viral replication compartments in the infected cell nucleus. ICP8 is involved not only in viral DNA replication, but also in repression of transcription of parental viral genomes, and stimulating transcription of L genes. A study from Taylor and Knipe [25] identified 49 host proteins and 6 viral proteins that interact with ICP8. Most of the host proteins identified are involved in DNA replication and damage repair, nonhomologous and homologous recombination, and chromatin remodeling (Fig. 1B, panel a). The authors showed that the cellular nonhomologous end-joining machinery (NHEJ) inhibits viral replication as tested in a mutant murine fibroblast line with a disrupted NHEJ pathway. Some of these cell proteins are recruited by ICP8 to viral replication compartments [25, 26]. After translation of the last wave of viral L proteins, all the players required for viral assembly of viral particles are available. The many viral and host components required for virion assembly need to be in the right place at the right time and in the precise stoichiometry to complete this elaborate process. Precise targeting of virion components to the diverse cell compartments required for α−HV assembly is only now being understood.
2.3 Alpha herpesvirus tegument proteins and their relevance in infection
As discussed previously, the most abundant proteins in the virion are found in the tegument. It is a proteinaceous complex that fills the space between the nucleocapsid and viral envelope membrane Such complex tegument is unique to herpesviruses and is comprised of a variety of viral and host proteins, all involved in many viral processes, either immediately after infection or during assembly of the virions. It is not well understood how the tegument components assemble into newly synthesized virions. MSBP however, has been essential to define and analyze this unusual collection of proteins (Fig. 1B, panel b). Michael et al. showed that deletion of the PRV tegument protein pUL21 resulted in a drastic decrease in the incorporation of three tegument proteins, pUL46, pUL49, and pUS3, into mature virions [27]. The attenuated vaccine strain, PRV Bartha, contains missense mutations in pUL21 and in the unique short (US) region and also failed to package pUL46, pUL49, and pUS3 in virions. In a similar study, the authors used stable isotope labeling with amino acids in culture (SILAC) and MSBP, to show that the composition of the virion changes when tegument proteins pUS3, pUL47, pUL49, or glycoprotein E (gE) are not expressed. When the gE was not expressed, mutant virions had significant reductions in the pUL46, pUL48 and pUL49 tegument proteins. The authors also observed an increase in the amount of a smaller isoform of pUL48 and host actin in the gE null virions [28]. In HSV-1, the tegument protein pUL46 modulates the transactivating activity of VP16 (pUL48), however it was not known if other cell or viral proteins interact with pUL46, or if it has additional roles early in infection. Lin et al. showed that HSV-1 pUL46 is phosphorylated in at least 23 sites and specifically interacts with the viral protein ICP0, an E3 ubiquitin ligase [29]. They also demonstrated that pUL46 is partially degraded in a proteasome-mediated manner by ICP0 (Fig. 1B, panel b). This finding is the first report of a viral protein being targeted for degradation by another viral protein during HSV-1 infection. Interestingly, pUL46 was found in a phosphorylated form in mature PRV virions [12].
pUL11 is an HSV-1 tegument protein comprised of 96 amino acids and localizes to the cytoplasmic faces of nuclear and Golgi derived membranes in infected cells, suggesting a role in virion secondary envelopment. MSBP studies with UL11 mutants revealed that the protein is myristylated and palmitylated near the N terminus and these modifications stabilize membrane-binding and Golgi-targeting signaling. pUL11 localizes only to the Golgi membranes but not to nuclear membranes in transfected cells, strongly suggesting that that pUL11 interacts with additional viral proteins upon infection. Loomis et al. demonstrated that pUL11 interacts with pUL16, a protein involved in nucleocapsid assembly, and that this interaction is independent of other viral proteins (Fig. 1B, panel b) [30].
Tegument protein networks involving HSV-1 pUL7 and the cell mitochondrial protein adenine nucleotide translocator 2 (ANT2) have been identified (Fig. 1B, panel b). UL7 mutants are able to grow in tissue culture but have cell-cell spread and replication defects. ANT2 is part of the permeability transposition pore (PT) complex and exchanges ATP and ADP on the inner mitochondrial membrane. In apoptotic conditions ANT2 can form a nonspecific pore. The relevance of this interaction is yet to be determined [31].
The pUL3.5 tegument protein is not conserved in all α-HVs: PRV, VZV and BHV-1 genomes contain UL3.5 homologs while HSV-1 and HSV-2 genomes do not. The roles of the pUL3.5 protein in replication may not be conserved: PRV pUL3.5 is required for virion egress, but VZV mutants lacking pUL3.5 (pORF57), grow well in cell culture with normal kinetics. On the other hand, BHV-1 pUL3.5 protein can complement the egress defect of a PRV UL3.5 mutant, suggesting that they have similar functions in egress. Lam and Letchworth, identified the BHV-1 UL48 gene product (BHV-1 α-transinducing factor; αBTIF), as a pUL3.5 interacting partner (Fig. 1B, panel b) [32]. αBTIF is the HSV-1 VP16 homolog, a transactivator protein that is incorporated into the virion and promotes transcription of IE genes. Although the authors did not demonstrate the transactivation activity of αBTIF, they showed pUL3.5 and α-BTIF co-localization of in the Golgi of infected cells and that this depends on 20 amino acids (aa) from the N-terminus of pUL3.5.
Another BHV-1 tegument protein studied by MSBP is the major tegument protein VP8. The homolog of VP8 in HSV-1 and PRV is VP13/14 (UL47). Labiuk et al. demonstrated that VP8 is phosphorylated by the viral kinase pUS3 and cellular kinase CK2 and that they co-localize to the nucleus of infected cells (Fig. 1B, panel b) [33].
In VZV, the non-essential tegument protein pORF49, functions as a cell tropic factor in cell culture. ORF49 null mutants, produce small plaques, and have low infectious virus production. The interaction between pORF49 and pORF44 is eliminated by mutation of a single amino acid of pORF44. The authors identified pORF49 residues 41–44 as the pORF44 binding interface. The phenotype of ORF49 delta 41–44 was comparable to that of ORF49 null mutants, suggesting that the pORF49-44 interaction is important for VZV infection and efficient production of progeny virus [34]. The homolog of pORF49 in HSV-1 is the non-essential protein pUL11 involved in secondary envelopment of virions. In HSV-1, the pORF44 homolog is pUL16 that functions in viral entry and nuclear and cytoplasmic egress. MSBP has been essential to unveil not only the complex composition of α-HV tegument but also the plethora of functions that tegument proteins perform during infection. Tegument proteins include all three kinetic groups of proteins (IE, E, L), suggesting their essential role in every step of infection (Fig. 1B, panel b).
2.4 Post-translational modifications during alpha herpesvirus infection
MSBP is a powerful approach to globally identify PTMs in complex settings like an infected cell. Here we provide some examples illustrating the relevance of PTMs in α-HV infection. The HSV-1 genome encodes more than 80 proteins and most of them are modified after translation. So far, the most abundant PTMs detected on viral proteins are phosphorylation and glycosylation. Viral infection also affects cell protein PTM [35]. A recent study of the HSV-1 proteome identified 10 novel ubiquitylation and 95 new phosphorylation sites in a macrophage cell line lysate 8 hrs after infection [23]. Kramer et al. detected phosphorylation of PRV proteins pUL26, pUL36, pUL46 and pUL48 in purified mature virions (Fig. 1B, panel b). Although it is still unclear if this plays a biological role in viral infectious cycle, authors speculate that might activate viral proteins for specific functions in very early stages of infection [12]. When infected cells are treated with inhibitors of DNA replication, both the protein abundance and degree of PTMs change [23]. The essential IE gene product ICP0 contains 11 phosphorylation sites across three regions Phos 1, Phos 2 and Phos 3 as detailed in section 2.2 [24]. When the phosphorylated residues in Phos 1 are mutated to alanine, ICP0 is unable to complement an ICP0 HSV-1 mutant (Fig. 1B, panel a). PTMs are also important for efficient axonal transport of PRV in infected neurons. As mentioned in section 2.4, the viral phosphoprotein pUS9 contains two serine phosphorylation sites (S51 and S53) (Fig. 1B, panel c). When these residues are mutated to alanine, anterograde neuronal transport of PRV particles is impaired both in vitro and in vivo [36].
In VZV, Oliver et al. identified a novel immunoreceptor tyrosine-based inhibition motif (ITIM), in the cytoplasmic domain of viral glycoprotein B (gB) [37]. gB is a conserved protein across α-HVs and is essential for fusion-mediated viral entry into susceptible cells. The authors showed that the tyrosine in position 881 of gB (Y881) is phosphorylated and that this PTM is essential for gB function (Fig. 1B, panel c). When VZV gB Y881 is mutated to alanine, cell–cell spread is reduced in vitro, and skin infection is severely impaired in vivo. The Y881 phosphorylation-dependent cell-cell fusion mechanism may involve novel intracellular signaling through the gB cytoplasmic motif that is essential for skin pathogenesis. This study also provides evidence that ITIM’s have functions different from their known role in immune modulation. Another example of VZV PTM function involves pORF63. pORF63 is an IE protein of 278 aa important for maintenance of viral latency that shares homology with HSV-1 IE protein ICP22. Mueller et al. identified 6 phosphorylation sites in pORF63 including novel sites Serine5 and Serine31 (Fig. 1B, panel c). The authors did not analyze the contribution of these PTMs to VZV infection, but suggested based on bioinformatics analysis, that a novel cellular kinase could be phosphorylating these residues [38]. PTMs add another layer of regulation for interactions of viral and host proteins. They can affect protein function, subcellular localization, and interaction with other proteins and stability.
2.5 Neuroinvasion by alpha herpesvirus
After acute infection of epithelial tissue, newly made α-HV particles invade peripheral nervous system (PNS) axons innervating such tissues and undergo fast retrograde transport towards the distant cell body. To accomplish this long distance transport, viral nucleocapsids must engage microtubule dependent motors to move from axons to neuron cell bodies (retrograde motion). After reactivation from a latent infection, particles must reverse direction and move into axons for transport back to the peripheral tissues (anterograde motion) [7, 39–41]. Understanding how efficient invasion of axons is stimulated and how viral particles engage different motors for directional transport has been a challenge. However, recent work with MSBP has made significant contributions to identify the complicated interaction between viral and host components required for viral spread within and between neurons. Koyuncu et al. used non-canonical amino acid tagging (BONCAT) and LC-MS/MS to show that PRV infection of axon induces rapid, local protein synthesis, including proteins involved in cytoskeletal remodeling, intracellular trafficking, signaling, and metabolism [42]. The rapid translation of axonal mRNAs appears to be required for efficient PRV retrograde transport and infection of cell bodies (Fig. 1B, panel d). Strikingly, the axonal damage response in uninfected neurons also induces local synthesis of similar proteins. There is evidence that damage signals and viral particles compete for retrograde transport. The hypothesis is that a preexistent host response is repurposed for efficient PRV neuroinvasion.
α-HV genomes do not encode motor proteins and rely on the transport machinery of their hosts to spread efficiently in cells. The motors used for both retrograde and anterograde transport of viruses in neurons and other polarized cells have been identified [43, 44]. Affinity purification and MSBP studies revealed that newly made PRV virions in transport vesicles engage the microtubule motor protein Kinesin-3 (KIF1A) for sorting into axons and subsequent anterograde transport (Fig. 1B, panel d). Kinesin-3 interacts with the small viral membrane phosphoprotein pUS9, to move PRV particles that are contained in a transport vesicle [45]. pUS9 is essential for efficient in vivo axonal sorting and anterograde transport of PRV and BHV-1 [46, 47]. Two additional viral glycoproteins, gE and gI, are needed to facilitate or stabilize the interaction between Kinesin-3 and PRV US9 (Fig. 1B, panel d) [48]. The role that HSV-1 pUS9, gE and gI play in anterograde transport of viral particles is under study [49, 50].
3 CELL RESPONSE TO VIRAL INFECTION
3.1 Global proteomic profiling of alpha herpesvirus infected cells
Viral infection alters the proteomic profile of host cells. Some responses reflect the cell autonomous, intrinsic early warning signals in response to infection (danger signals), while others reflect the directed interactions of viral proteins to produce a productive environment for viral replication. How many of these responses are general and how many are specific, both to virus and to cell type? Systematic studies of viral transcription profiles are useful but they cannot report on protein composition, stability, and modifications (see for example [35]). Early studies compared the mock-infected and HSV-1-infected cell proteome using 35S pulse-labeling and two-dimensional electrophoresis (2-DE) [51]. While HSV-1 infection strongly inhibited host protein synthesis, the synthesis of a number of acidic proteins and basic ribosome proteins was maintained or stimulated. The specific identity and function of most of these dynamic proteins was not known. Now, mass spectrometry and protein sequence databases enable the analysis of the complex virus-host interactions.
Antrobus et al. combined 2-DE with LC-MS to perform the first global proteomic analysis of HSV-1 infected cells [52]. Uninfected and infected Hep-2 whole cell extracts were analyzed with 2-DE, identifying 103 protein spot changes. Subsequent LC-MS/MS of these spots, identified a total of 144 individual proteins involved in a variety of biological processes including DNA replication, chromatin remodeling, pre-mRNA splicing, mRNA stability and the ER stress response [52]. Similar whole-cell proteomic analysis of PRV infected bovine kidney cells has been reported using SILAC mass spectrometry methods [53]. Proteins from infected and uninfected cell extracts were fractionated by affinity solid-phase extraction prior to 2-DE and MALDI-MS/MS. SILAC allowed differentiation and quantification of isotopically modified peptides that are chemically identical, while pre-fractionation of whole cell extracts enhanced yield as well as resolution of proteins. Changes were observed for 103 protein spots, representing 55 individual proteins that are representatives of stress response proteins, constituents of the nuclear lamina, heterogeneous nuclear ribonuclear proteins (hnRNPs), ribosome proteins, proteins involved in trafficking and transport as well as other biological processes [53]. Only a small portion of host cell proteins was altered during early stages of infection by either HSV-1 or PRV. This finding is consistent with the earlier study using only 2-DE analysis [51]. Both studies showed that the expression levels of lamin A/C and hnRNP K was affected by infection. The HSV-1 pUS3 [54–56] and HSV-2 pUL13 [57] protein kinases can phosphorylate lamin A/C, while HSV-1 pUL31 and pUL34 affect sub-cellular localization of lamin A/C (Fig. 1C, panel a) [54, 58]. These changes may be involved in regulating viral genome replication and transcription [59], targeting pUL31 and pUL34 to lamina, and regulating viral capsid nuclear egress [58]. The shift of lamin A/C to more acidic isoforms during PRV infection indicates that HSV-1 and PRV infection affects common biological processes [53]. Other changes after HSV-1 infection involve hnRNP K, which forms a complex with ICP27 and host p32 and CK2 proteins (Fig. 1C, panel a). CK2 then phosphorylates the other three complex components in an ICP27-dependent manner. The formation of this complex may disrupt or modulate pre-mRNA splicing, facilitating splicing-independent nuclear export of viral transcripts [60–62]. hnRNP K also modulates HSV-1 egress from infected cells [63]. It is striking that no other proteins are shared among the lists of proteome changes for either PRV or HSV-1. While technical differences may explain this lack of congruence, it may be that cells sense the two α-HVs differently or that these distantly related viruses have evolved different interactions for efficient replication. Global proteomic studies have provided us with the fundamental tools to examine and address such specific differences between viruses and infected cells.
3.2 Subcellular proteomic profiling of alpha herpesvirus infected cells
Monitoring the global proteomic profile in host cells often obscures subtle alterations in subcellular fractions such as the cytoplasm, ribosomes, and nucleus [51–53]. Studies utilizing fractionation methods have expanded our knowledge of the host response to alpha herpesvirus infection.
Cytosolic, microsomal and nuclear proteomic changes were analyzed separately in HSV-1 infected human hepatoma cells. In one study biochemically fractionated cytosolic and microsomal samples were analyzed by 2D-differential in-gel electrophoresis (2D–DIGE) and nano LC-MS. In this method, proteins from mock infected and infected cells were differentially labeled with cyanine dye Cy3 or Cy5, and both samples were fractionated in the same gel. The expression levels of 12 cytosolic and 6 microsomal proteins involved in apoptosis, signal transduction and endoplasmic reticulum associated degradation pathway, were altered [64]. In another study, Sanchez-Quiles et al. analyzed nuclear isolates from HSV-1 infected hepatoma cells by two separate approaches, 2D-DIGE and tandem mass tag isobaric labeling (TMT). The two approaches identified 24 and 38 proteins, respectively, with significant changes. These two approaches complemented each other in that 2D-DIGE provided distinct information about the different isoforms and PTMs of the same proteins, while the TMT approach enabled better resolution and detection. Many of the identified proteins play roles in cell cycle regulation and RNA homeostasis. One protein, quaking (QKI), was further validated to promote viral protein synthesis [65]. Only one protein, RuvB-like 2 (RBL2), was common in both studies showing increased expression in the microsomal fraction at 4hpi, and reduced expression in the nucleus at 8hpi. RBL2 is an antiapoptotic ATPase and putative DNA helicase (Fig. 1C, panel a) [66] whose synthesis is increased after HIV infection [67]. RBL2 expression also interferes with influenza replication [68]. It is not clear what RBL2 is doing during α-HV infection. Despite the fact that different HSV-1 strains and cells are used, the sampling times are not uniform, and the proteomic methods are not identical, global and subcellular proteomic profiling gave similar results [52, 64, 65]. The expression levels of four nuclear proteins (lamin A/C, cofilin-1, hnRNP C1/C2 and NonO/P54) and three cytosolic proteins (hnRNP K, UPF0160 protein MYG1, and FK506 binding protein) were consistently altered after HSV-1 infection.
The proteome of fractionated ribosomes changes after HSV-1 infection. Greco et al. identified ribosomal and viral proteins using N-terminal sequencing and MS [69]. The viral proteins VP19C, VP26 and multiple phosphorylated forms of pUS11 as well as a host protein poly(A)-binding protein 1 (PABP), were the only non-ribosomal proteins consistently associated with the ribosome after infection (Fig. 1C, panel a). Subsequent studies also showed that HSV-1 IE protein ICP27, interacted with PABP and eIF3, eIF4G [22]. Together, these studies demonstrate the value of MSBP in determining the proteins that regulate protein synthesis and selective translation of proteins after HSV-1 infection.
Fractionating cellular compartments has been extended to the axons of neurons infected with α−HVs. The efficient invasion by α−HVs of peripheral axons after infection of epithelial surfaces is poorly understood. Most viruses that infect epithelial surfaces do not infect axons that innervate that tissue. It is well known that early in development, axons can autonomously respond to various guidance stimuli as the early nervous system is established. In addition, when damaged, axons can respond quickly by synthesizing new proteins. Indeed, a subset of mRNAs and the complete protein synthesis machinery are localized to axons in uninfected neurons [70]. Local translation in axons, not in the distant cell body, regulates growth cone navigation and integrity during development and regeneration. Such retrograde communication of axons with the cell body in adult neurons plays a key role in axonal damage signaling [71–76]. Using primary neurons cultured in chambers to physically separate axons from cell bodies, Koyuncu et al. reported that PRV infection of isolated axons also induces rapid and local protein synthesis. They identified the newly synthesized axonal proteins using BONCAT followed by tandem MS (Fig. 1B, panel d). These nascent proteins include participants in cytoskeleton remodeling, intracellular trafficking, signaling, and energy metabolism pathways. Interestingly, this de novo protein synthesis is a requisite for transport of virus particles in axons [42]. The two levels of fractionation - physically collecting only axons and BONCAT to purify and enrich for newly made proteins - combined with highly sensitive detection of MS, enabled analysis in molecular detail. While proteomic profiling of subcellular compartments provides insight of local changes within cells after α−HV infection, additional comparative studies between different subcellular compartments and cell types are needed. Comparative analyses of cells infected with different α−HV species are also missing.
3.3 Host immune response after alpha herpesvirus infection
An effective host immune response will contain and clear most viral infections. The first line of defense is the cell autonomous, intrinsic defensive response within the first infected cell where pathogen sensing through multiple pathways converge in the production of inflammatory cytokines such as type I and type II interferon (IFN). The endocrine and paracrine signaling of inflammatory cytokines activates the expression of a plethora of cellular genes that function in not only controlling the virus infection in the currently infected cell, but also in protecting surrounding uninfected cells from infection. The inflammatory cytokines also serve as important regulators of the adaptive immune response where T-lymphocytes and B-lymphocytes elicit specific antiviral mechanisms through cellular and humoral immune responses, respectively [77]. Many investigators have studied the role of immune responses in controlling productive α−HV infection or promoting a quiescent latent infection. Here we review the studies where MSBP has provided insight into the immune mechanisms that regulate α-HV infection.
3.3.1 Early sensing of alpha herpesvirus DNA in infected cells
The first and foremost step of mounting an appropriate immune response is sensing the invading pathogen through multiple pattern recognition receptors. Knowledge of how and where the nucleic acid of DNA viruses such as herpesvirus and poxvirus is detected within the cell was limited until the discovery of DNA sensors over the past decade. The DNA sensor IFI16 was first revealed during a proteomic search of vaccinia virus (VACV) DNA binding partners. IFI16 also detects HSV-1 DNA both in the nucleus and the cytoplasm. This interaction leads to expression of pro-inflammatory cytokines such as IFNβ, CXCL10, IL-6 and TNF [78]. While herpesvirus genomes replicate in the nucleus, poxvirus DNA is replicated in the cytoplasm. Therefore, IFI16 must be localized in both the nucleus and the cytoplasm. Indeed, Li et al. discovered that while the induction of IFNβ production by HSV-1 relies on a nuclear localization sequence (NLS) in the IFI16 protein, this NLS is not required for detection of VACV DNA. Combining immunoaffinity purification with MS, Li and colleagues further identified 6 phosphorylation and 9 acetylation (Ac) sites in IFI16 protein sequence (Fig. 1C, panel a). Strikingly, acetylation of two lysines (K99 and K129), within the NLS, inhibited nuclear localization of IFI16 [79]. These analyses were the first to suggest that localization of a DNA sensor (IFI16) could be regulated by posttranslational modification. It will be important to know what host and viral gene products aid in regulating precise intracellular localization. Thus further MSBP studies are required to reveal these specific mechanisms.
3.3.2 The innate immune system controls herpes simplex encephalitis
Herpes simplex virus encephalitis (HSE), a disease caused by the rare infection of the central nervous system (CNS) by HSV, may result from deficiency of type I and II IFN production in response to TLR3 activation during HSV-1 infection [80]. This conclusion follows from five genetic etiologies of HSE (UNC-93B, TLR3, TRAF3, TRIF, TBK1) that are all involved in TLR3 signaling [80]. The story is more complicated because not all HSE patients harbor deficiencies in these five genes. Moreover, 30% of HSE patients do not have deficiencies in TLR3 induced type I and III IFN production. These observations prompted Pérez de Diego et al. to use MSBP to search for other factors regulating the TLR3 signaling pathway or other independent pathways that may be associated with HSE susceptibility. The proteomic changes in fibroblasts from healthy and HSE patients were compared after TLR3 activation. The HSE fibroblasts were obtained from patients with UNC-93B deficiency or an unknown deficiency and analyzed with SILAC/MS (Fig. 1C, panel b). The presence of superoxide dismutase 2 (SOD2) and peptidyl-prolyl cis-trans isomerase F (PPIF) was detected in fibroblasts from both healthy individuals and those from the patient with unknown deficiency, but not in fibroblasts from the patient with UNC-93B deficiency. SOD2 is an antioxidant enzyme strongly increased after TLR3 activation in macrophages and HSV-1 infection in neurons. It may be involved in protecting these cells from oxidative stress and apoptosis [81, 82]. PPIF is part of the mitochondrial permeability transition pore in the inner mitochondrial membrane and a key protein regulating neuronal cell death [83–85]. These genes might be the downstream responses in the TLR3 pathway that are defective in genetic predisposition to HSE. Furthermore, the expression of ICAM-1 (an intercellular adhesion molecule) and ITGA5 (integrin alpha 5) were significantly increased only in healthy individuals and not in any HSE patients, suggesting their role in TLR3-independent pathways that affect HSE susceptibility [86] (Fig. 1C, panel b). Variation on a population scale has posed a formidable barrier for the analysis of this data because large variations are observed even among healthy individuals. In the simplest case, we do not have a precise definition of what it means to be “healthy”. Proteomic analysis of large cohort studies may help to address these challenges [80].
3.3.3 The alpha herpesvirus secretome after infection of macrophages
The complex set of macromolecules secreted by cells after infection (the secretome) affect local and global responses. For instance, pro-inflammatory cytokines produced and secreted from infected cells, will not only inform and prepare uninfected neighboring cells, they also will attract innate and adaptive immune cells to the site of infection. Traditional methods of examining the secreted components of an infected cell have been limited in scope and sensitivity. Miettienen et al. did a global profiling of the secretome from IFNβ treated and HSV-1 infected human primary macrophages. They used 4-plex labeling of primary amino groups in intact proteins using isobaric tags for relative and absolute quantitation (iTRAQ) combined with LC-MS/MS analysis. They found that IFNβ treatment, HSV-1 infection, and IFNβ priming prior to HSV-1 infection, increased the secretion of 46, 161 and 399 human proteins, respectively [87]. These observations indicate that cells exposed to IFNβ, were probably in a “prepared” state rather than a fully responsive state (Fig. 1C, panel c). Once these “prepared” cells were infected, there was a massive increase in the amount and type of secreted proteins that are likely to control the subsequent spread of infection to other cells. Another interesting finding was that 80% of the secreted proteins were known exosomal proteins. This non-classical vesicle-mediated protein secretion system appears to be activated by IFNβ priming or HSV-1 infection. Proteins related to immune and inflammatory responses, IFN-induced proteins, and endogenous danger signal proteins (such as IFIT2, IFIT3, STAT1, and MxA) were secreted. They also noted a depletion of interleukin-1β (IL-1β) and IL-18 in HSV-1-infected macrophages, suggesting that inflammasome function may be specifically antagonized (Fig. 1C, panel c) [87]. Secretome profiling has been done for cells infected by other viral families [88, 89]. It will be interesting to compare the secretome of different types of cells under various infectious conditions to define the cell, tissue, and system level responses in both natural and non-natural hosts.
3.3.4 Adaptive immune responses to alpha herpesvirus infection
The activation of adaptive immune responses is essential for effective control of α-HV during both productive and latent infection [90]. Although most α-HV infections induce a robust humoral immune response, the role of this response in controlling infections is not clear. Many studies point out specific antibody interactions with essential viral process that may contribute to effective control of infection. For example, a specific antibody reconstructed from a Zostavax-immunized male patient, targeted the VZV gH/gL complex, important in cell-cell fusion [91]. Since cell to cell spread of VZV relies in part on activities of glycoproteins gB, gH and gL on the surface of infected cells, this antibody from an immune patient suggests that it might function in limiting the spread of infection (Fig. 1C, panel d). The efficacy of antibodies is limited by their avidity. Pasman et al. showed enhanced potency of an antibody against BHV-1, by creating a single-chain variable antibody, covalently linking the variable regions (antigen recognizing regions) of the light and heavy chain. Q-TOF MS analysis of the single-chain variable antibody suggested that it formed a multimer, which was later shown to increase the potency of this antibody [92]. Such antibodies hold promise for direct transfer of antibodies to infected individuals (passive immunity).
Cellular immune responses mediated by T cells limit viral spread and are thought to be required to maintain a latent infection in immune competent individuals. Immune-deficient or compromised individuals are more prone to α-HV reactivation. For example, VZV reactivates in 25–40% in patients after allogeneic hematopoietic stem cell transplantation (HSCT) [93]. Boosting cellular immunity by the current live VZV vaccine is not appropriate for these immunocompromised patients. A possible solution arises from MSBP study by Keelmann et al. Using HPLC and MS, they identified VZV proteins gB, gE and IE62, as potent activators of both CD4+ and CD8+ T cells from HSCT patients with onset of VZV reactivation (Fig. 1C, panel d) [93]. These viral components may be used to activate specific T cells for passive transfer or for use as subunit vaccines suitable for immune-compromised patients.
Single cell mass cytometry is a powerful method. This technology is similar to conventional flow cytometry, but allows simultaneous labeling of over 40 proteins per cell by metal-isotope labeled antibodies. It is possible to determine individual cell states and phenotypes based on single cell mass differences [94]. This method was used to study human tonsil T cells infected with VZV. These cells spread VZV to the skin of infected hosts to cause varicella. In contrast to the idea that VZV has a tropism for skin tropic T cells, these authors found that VZV infection induces or enhances these properties in a variety of T cells by increasing the expression of relevant surface proteins regardless of the original profile of the cell. The increased expression of such surface proteins was triggered by Zap70 and Akt signaling pathways in a T cell receptor (TCR)-independent manner [95]. The use of single cell mass cytometry changes our view of proteomic data from a population level to single-cell level, providing important information about the heterogeneity within the infected tissue.
4. NON-PROTEOMIC USE OF MASS SPECTROMETRY IN ALPHA HERPESVIRUS RESEARCH
Mass spectrometry is used to identify molecules other than proteins involved in α−HV infections. For example, direct comparison of MALDI-TOF spectra generated from uninfected, HSV-2 or VZV infected cells, showed distinct differences in many mass species that can be used for detection and specific identification of infected cells [96]. In addition, PCR coupled with MS to identify viral nucleic acids from cerebrospinal fluid samples has proven to be a valuable tool for early diagnosis of neurological disorders correlated with infection by neurotropic viruses such as HSV-1 and VZV [97]. MS facilitated the analysis of the chemical composition of Thymus capitata extracts, which have a potential non-cytotoxic antiviral to BHV-1 [98].
Maybe one of the more challenging and useful non-proteomic MS technologies is to measure changes in small molecule metabolites before and after virus infection. Vastag et al. discovered marked differences in the metabolome after HCMV or HSV-1 infection of primary fibroblasts. HCMV infection increased components driving the tricarboxylic acid cycle (TCA) and fatty acid biosynthesis, while HSV-1 infection promotes pyrimidine biosynthesis [99]. Such studies provide novel targets for antiviral drug discovery because when these metabolic pathways were altered by inhibitors, infectious virus production was affected.
MS techniques also can be used to identify complex oligosaccharides. Liu et al. used MS techniques to characterize the structure of a heparin sulfate octasaccharide that binds to gD [100]. Viral glycoprotein gD uses cell surface heparin sulfate as the cell receptor to infect susceptible cells. The authors used MS to analyze a cell heptasulfated octasaccharide that binds gD with an affinity of 18 µM.
5 CONCLUSIONS AND PERSPECTIVES
We reviewed the state of the art of proteomics methods used in the last 15 years to study α-HVs. The literature is limited to studies done with HSV-1, PRV, VZV and BHV-1 (Table 1). We reviewed work done to identify the viral and host proteins important for transcription of viral genes and the recruitment of host RNA polymerase II. We also reported studies defining the proteome of extracellular mature HSV-1 and PRV virions. A curious finding is that the number of host proteins in virus particles exceeds that of the viral components. The function of these host proteins is not well understood. We reviewed reports that analyzed the proteome of neurons and axons infected with different α-HVs to understand the mechanisms of retrograde and anterograde transport. Finally, we discussed how posttranslational modification of host and viral proteins change after infection (Fig. 1A and B).
We also reviewed the literature that used global MSBP analyses of infected cells as well as subcellular organelles after α−HV infection. In particular, we examined work describing the immediate intrinsic defense responses of cells. MSBP has contributed extensively to understanding how cells sense and react to incoming α-HV DNA, how innate immune signaling may affect HSE pathogenesis, how the secretome components of an infected cell change, and how antibodies and T-cell activation may limit viral infection (Fig. 1C).
Many of the reports described new interacting partners of a specific viral protein. The most common technique was to isolate the protein of interest (and interacting partners) by affinity purification, followed by LC-MS/MS. Affinity purification of fluorescently labeled (GFP) fusion proteins with a GFP antibody, is a powerful procedure to preserve protein complexes and minimize nonspecific interactions prior to analyses. GFP tagged proteins can be readily visualized in living cells, enabling determination of the relationship of phenotype to localization. Using GFP antibodies broadens the range of protein targets that can be purified when specific antibodies against the protein of interest are not available [101]. This technology also is useful when purifying proteins from virus infected cells where the GC content is low as found for VZV [102]. We note that many of the papers reviewed here study the composition of a protein interacting network without detailed validation of the function of the interactions. A list of viral interactors is a strong start, but without functional investigation by other techniques (e.g. viral or cell mutants, RNAi, shRNA, qPCR, microarrays, sequencing, among others), the significance is uncertain.
The vast majority of published MS studied with α-HVs have used epithelial cells and fibroblasts. Considering that neurons play significant roles in the life cycles of most α-HVs, it is important that more proteomic studies of infected neurons in vitro, in vivo or ex vivo be completed. Only a few studies use MSBP analysis of α-HV infection of neurons [42, 45, 48]. These studies all focused on productive infection. Analyzing samples obtained from latently infected and reactivating neurons will be revealing.
In summary, MSBP is a relatively new field of study that has already contributed to the understanding of α-HVs. As the technology related to MS and subcellular fractionation evolves and improves, we expect to see more MSBP studies that will capture the changing composition of organelles and subcellular structures in α-HV infected cells and tissues. MSBP studies of single infected cells will enable the analysis of spatiotemporal changes after viral infection with higher resolution. Finally, these studies produce an enormous amount of information, so it is important that proteomics data is made available in public databases. The data will then enable comparison of results by different laboratories, accelerating data analysis, validation, and fostering collaborations.
ACKNOWLEDGEMENTS
Members of the Enquist laboratory provided helpful comments and suggestions. This work was supported by National Institutes of Health grants to L.W.E. (R01 NS33506, R01 NS060699, and P40 RR18604 and Pew Latin American Fellowship in the Biomedical Sciences (2010-000225-002) to E.A.E. We apologize to those many investigators whose interesting work could not be cited because of page limits.
Footnotes
The authors declare no conflict of interest
REFERENCES
- 1.Davison AJ, Eberle R, Ehlers B, Hayward GS, et al. The order Herpesvirales. Arch Virol. 2009;154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvin A, et al., editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge: 2007. [PubMed] [Google Scholar]
- 3.Roizman B, Whitley RJ. The nine ages of herpes simplex virus. Herpes. 2001;8:23–27. [PubMed] [Google Scholar]
- 4.Whitley RJ, Roizman B. Herpes simplex virus infections. Lancet. 2001;357:1513–1518. doi: 10.1016/S0140-6736(00)04638-9. [DOI] [PubMed] [Google Scholar]
- 5.Roizman B, Taddeo B. The strategy of herpes simplex virus replication and takeover of the host cell. In: Arvin A, et al., editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge: 2007. [PubMed] [Google Scholar]
- 6.Zerboni L, Sen N, Oliver SL, Arvin AM. Molecular mechanisms of varicella zoster virus pathogenesis. Nat Rev Microbiol. 2014;12:197–210. doi: 10.1038/nrmicro3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith G. Herpesvirus transport to the nervous system and back again. Annu Rev Microbiol. 2012;66:153–176. doi: 10.1146/annurev-micro-092611-150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koyuncu OO, Hogue IB, Enquist LW. Virus infections in the nervous system. Cell Host Microbe. 2013;13:379–393. doi: 10.1016/j.chom.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramer T, Enquist LW. Directional spread of alphaherpesviruses in the nervous system. Viruses. 2013;5:678–707. doi: 10.3390/v5020678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greco TM, Diner BA, Cristea IM. The Impact of Mass Spectrometry-Based Proteomics on Fundamental Discoveries in Virology. Annual Review of Virology. 2014;1:581–604. doi: 10.1146/annurev-virology-031413-085527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogue IB, Bosse JB, Hu JR, Thiberge SY, et al. Cellular mechanisms of alpha herpesvirus egress: live cell fluorescence microscopy of pseudorabies virus exocytosis. PLoS Pathog. 2014;10:e1004535. doi: 10.1371/journal.ppat.1004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramer T, Greco TM, Enquist LW, Cristea IM. Proteomic characterization of pseudorabies virus extracellular virions. J Virol. 2011;85:6427–6441. doi: 10.1128/JVI.02253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lippe R. Deciphering novel host-herpesvirus interactions by virion proteomics. Front Microbiol. 2012;3:181. doi: 10.3389/fmicb.2012.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loret S, Guay G, Lippe R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol. 2008;82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Homa FL, Huffman JB, Toropova K, Lopez HR, et al. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J Mol Biol. 2013;425:3415–3428. doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe D, Brockman MA, Ndung’u T, Mathews L, et al. Properties of a herpes simplex virus multiple immediate-early gene-deleted recombinant as a vaccine vector. Virology. 2007;357:186–198. doi: 10.1016/j.virol.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 18.Carrozza MJ, DeLuca NA. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol Cell Biol. 1996;16:3085–3093. doi: 10.1128/mcb.16.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lester JT, DeLuca NA. Herpes simplex virus 1 ICP4 forms complexes with TFIID and mediator in virus-infected cells. J Virol. 2011;85:5733–5744. doi: 10.1128/JVI.00385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner LM, DeLuca NA. Temporal association of herpes simplex virus ICP4 with cellular complexes functioning at multiple steps in PolII transcription. PLoS One. 2013;8:e78242. doi: 10.1371/journal.pone.0078242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu BW, Engel EA, Enquist LW. Characterization of a Replication-Incompetent Pseudorabies Virus Mutant Lacking the Sole Immediate Early Gene IE180. MBio. 2014:5. doi: 10.1128/mBio.01850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fontaine-Rodriguez EC, Taylor TJ, Olesky M, Knipe DM. Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology. 2004;330:487–492. doi: 10.1016/j.virol.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Bell C, Desjardins M, Thibault P, Radtke K. Proteomics analysis of herpes simplex virus type 1-infected cells reveals dynamic changes of viral protein expression ubiquitylation and phosphorylation. J Proteome Res. 2013;12:1820–1829. doi: 10.1021/pr301157j. [DOI] [PubMed] [Google Scholar]
- 24.Davido DJ, von Zagorski WF, Lane WS, Schaffer PA. Phosphorylation site mutations affect herpes simplex virus type 1 ICP0 function. J Virol. 2005;79:1232–1243. doi: 10.1128/JVI.79.2.1232-1243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor TJ, Knipe DM. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J Virol. 2004;78:5856–5866. doi: 10.1128/JVI.78.11.5856-5866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karttunen H, Savas JN, McKinney C, Chen YH, et al. Co-opting the Fanconi anemia genomic stability pathway enables herpesvirus DNA synthesis and productive growth. Mol Cell. 2014;55:111–122. doi: 10.1016/j.molcel.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michael K, Klupp BG, Karger A, Mettenleiter TC. Efficient incorporation of tegument proteins pUL46, pUL49, and pUS3 into pseudorabies virus particles depends on the presence of pUL21. J Virol. 2007;81:1048–1051. doi: 10.1128/JVI.01801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michael K, Klupp BG, Mettenleiter TC, Karger A. Composition of pseudorabies virus particles lacking tegument protein US3 UL47 or UL49 or envelope glycoprotein E. J Virol. 2006;80:1332–1339. doi: 10.1128/JVI.80.3.1332-1339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin AE, Greco TM, Dohner K, Sodeik B, et al. A proteomic perspective of inbuilt viral protein regulation: pUL46 tegument protein is targeted for degradation by ICP0 during herpes simplex virus type 1 infection. Mol Cell Proteomics. 2013;12:3237–3252. doi: 10.1074/mcp.M113.030866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loomis JS, Courtney RJ, Wills JW. Binding Partners for the UL11 Tegument Protein of Herpes Simplex Virus Type 1. Journal of Virology. 2003;77:11417–11424. doi: 10.1128/JVI.77.21.11417-11424.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka M, Sata T, Kawaguchi Y. The product of the Herpes simplex virus 1 UL7 gene interacts with a mitochondrial protein, adenine nucleotide translocator 2. Virol J. 2008;5:125. doi: 10.1186/1743-422X-5-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam N, Letchworth GJ. Bovine Herpesvirus 1 UL3.5 Interacts with Bovine Herpesvirus 1 alpha -Transinducing Factor. Journal of Virology. 2000;74:2876–2884. doi: 10.1128/jvi.74.6.2876-2884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labiuk SL, Babiuk LA, van Drunen Littel-van den Hurk S. Major tegument protein VP8 of bovine herpesvirus 1 is phosphorylated by viral US3 and cellular CK2 protein kinases. J Gen Virol. 2009;90:2829–2839. doi: 10.1099/vir.0.013532-0. [DOI] [PubMed] [Google Scholar]
- 34.Sadaoka T, Serada S, Kato J, Hayashi M, et al. Varicella-zoster virus ORF49 functions in the efficient production of progeny virus through its interaction with essential tegument protein ORF44. J Virol. 2014;88:188–201. doi: 10.1128/JVI.02245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skiba M, Glowinski F, Koczan D, Mettenleiter TC, et al. Gene expression profiling of Pseudorabies virus (PrV) infected bovine cells by combination of transcript analysis and quantitative proteomic techniques. Vet Microbiol. 2010;143:14–20. doi: 10.1016/j.vetmic.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 36.Kratchmarov R, Taylor MP, Enquist LW. Role of Us9 phosphorylation in axonal sorting and anterograde transport of pseudorabies virus. PLoS One. 2013;8:e58776. doi: 10.1371/journal.pone.0058776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver SL, Brady JJ, Sommer MH, Reichelt M, et al. An immunoreceptor tyrosine-based inhibition motif in varicella-zoster virus glycoprotein B regulates cell fusion and skin pathogenesis. Proc Natl Acad Sci U S A. 2013;110:1911–1916. doi: 10.1073/pnas.1216985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller NH, Walters MS, Marcus RA, Graf LL, et al. Identification of phosphorylated residues on varicella-zoster virus immediate-early protein ORF63. J Gen Virol. 2010;91:1133–1137. doi: 10.1099/vir.0.019067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enquist LW. Five questions about viral trafficking in neurons. PLoS Pathog. 2012;8:e1002472. doi: 10.1371/journal.ppat.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wojaczynski GJ, Engel EA, Steren KE, Enquist LW, et al. The neuroinvasive profiles of H129 (herpes simplex virus type 1) recombinants with putative anterograde-only transneuronal spread properties. Brain Struct Funct. 2014 doi: 10.1007/s00429-014-0733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomeranz LE, Reynolds AE, Hengartner CJ. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev. 2005;69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koyuncu OO, Perlman DH, Enquist LW. Efficient retrograde transport of pseudorabies virus within neurons requires local protein synthesis in axons. Cell Host Microbe. 2013;13:54–66. doi: 10.1016/j.chom.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bearer EL, Breakefield XO, Schuback D, Reese TS, et al. Retrograde axonal transport of herpes simplex virus: evidence for a single mechanism and a role for tegument. Proc Natl Acad Sci U S A. 2000;97:8146–8150. doi: 10.1073/pnas.97.14.8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radtke K, Kieneke D, Wolfstein A, Michael K, et al. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 2010;6:e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kramer T, Greco TM, Taylor MP, Ambrosini AE, et al. Kinesin-3 mediates axonal sorting and directional transport of alphaherpesvirus particles in neurons. Cell Host Microbe. 2012;12:806–814. doi: 10.1016/j.chom.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brideau AD, Card JP, Enquist LW. Role of pseudorabies virus Us9, a type II membrane protein in infection of tissue culture cells and the rat nervous system. J Virol. 2000;74:834–845. doi: 10.1128/jvi.74.2.834-845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butchi NB, Jones C, Perez S, Doster A, et al. Envelope protein Us9 is required for the anterograde transport of bovine herpesvirus type 1 from trigeminal ganglia to nose and eye upon reactivation. J Neurovirol. 2007;13:384–388. doi: 10.1080/13550280701375433. [DOI] [PubMed] [Google Scholar]
- 48.Kratchmarov R, Kramer T, Greco TM, Taylor MP, et al. Glycoproteins gE and gI are required for efficient KIF1A–dependent anterograde axonal transport of alphaherpesvirus particles in neurons. J Virol. 2013;87:9431–9440. doi: 10.1128/JVI.01317-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howard PW, Howard TL, Johnson DC. Herpes simplex virus membrane proteins gE/gI and US9 act cooperatively to promote transport of capsids and glycoproteins from neuron cell bodies into initial axon segments. J Virol. 2013;87:403–414. doi: 10.1128/JVI.02465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGraw HM, Awasthi S, Wojcechowskyj JA, Friedman HM. Anterograde spread of herpes simplex virus type 1 requires glycoprotein E and glycoprotein I but not Us9. J Virol. 2009;83:8315–8326. doi: 10.1128/JVI.00633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greco A, Bausch N, Coute Y, Diaz JJ. Characterization by two-dimensional gel electrophoresis of host proteins whose synthesis is sustained or stimulated during the course of herpes simplex virus type 1 infection. Electrophoresis. 2000;21:2522–2530. doi: 10.1002/1522-2683(20000701)21:12<2522::AID-ELPS2522>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 52.Antrobus R, Grant K, Gangadharan B, Chittenden D, et al. Proteomic analysis of cells in the early stages of herpes simplex virus type-1 infection reveals widespread changes in the host cell proteome. Proteomics. 2009;9:3913–3927. doi: 10.1002/pmic.200900207. [DOI] [PubMed] [Google Scholar]
- 53.Skiba M, Mettenleiter TC, Karger A. Quantitative whole-cell proteome analysis of pseudorabies virus-infected cells. J Virol. 2008;82:9689–9699. doi: 10.1128/JVI.00995-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bjerke SL, Roller RJ. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology. 2006;347:261–276. doi: 10.1016/j.virol.2005.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mou F, Forest T, Baines JD. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J Virol. 2007;81:6459–6470. doi: 10.1128/JVI.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mou F, Wills EG, Park R, Baines JD. Effects of lamin A/C, lamin B1, and viral US3 kinase activity on viral infectivity, virion egress, and the targeting of herpes simplex virus U(L)34-encoded protein to the inner nuclear membrane. J Virol. 2008;82:8094–8104. doi: 10.1128/JVI.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cano-Monreal GL, Wylie KM, Cao F, Tavis JE, et al. Herpes simplex virus 2 UL13 protein kinase disrupts nuclear lamins. Virology. 2009;392:137–147. doi: 10.1016/j.virol.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reynolds AE, Liang L, Baines JD. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J Virol. 2004;78:5564–5575. doi: 10.1128/JVI.78.11.5564-5575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silva L, Cliffe A, Chang L, Knipe DM. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 2008;4:e1000071. doi: 10.1371/journal.ppat.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bryant HE, Matthews DA, Wadd S, Scott JE, et al. Interaction between herpes simplex virus type 1 IE63 protein and cellular protein p32. J Virol. 2000;74:11322–11328. doi: 10.1128/jvi.74.23.11322-11328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koffa MD, Kean J, Zachos G, Rice SA, et al. CK2 protein kinase is stimulated and redistributed by functional herpes simplex virus ICP27 protein. J Virol. 2003;77:4315–4325. doi: 10.1128/JVI.77.7.4315-4325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wadd S, Bryant H, Filhol O, Scott JE, et al. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J Biol Chem. 1999;274:28991–28998. doi: 10.1074/jbc.274.41.28991. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt T, Striebinger H, Haas J, Bailer SM. The heterogeneous nuclear ribonucleoprotein K is important for Herpes simplex virus-1 propagation. FEBS Lett. 2010;584:4361–4365. doi: 10.1016/j.febslet.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 64.Santamaria E, Mora MI, Potel C, Fernandez-Irigoyen J, et al. Identification of replication-competent HSV-1 Cgal+ strain signaling targets in human hepatoma cells by functional organelle proteomics. Mol Cell Proteomics. 2009;8:805–815. doi: 10.1074/mcp.M800202-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanchez-Quiles V, Mora MI, Segura V, Greco A, et al. HSV-1 Cgal+ infection promotes quaking RNA binding protein production and induces nuclear-cytoplasmic shuttling of quaking I-5 isoform in human hepatoma cells. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.009126. 009126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rousseau B, Menard L, Haurie V, Taras D, et al. Overexpression and role of the ATPase and putative DNA helicase RuvB-like 2 in human hepatocellular carcinoma. Hepatology. 2007;46:1108–1118. doi: 10.1002/hep.21770. [DOI] [PubMed] [Google Scholar]
- 67.Bora A, Mohien CU, Chaerkady R, Chang L, et al. Identification of putative biomarkers for HIV-associated neurocognitive impairment in the CSF of HIV-infected patients under cART therapy determined by mass spectrometry. J Neurovirol. 2014;20:457–465. doi: 10.1007/s13365-014-0263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kakugawa S, Shimojima M, Neumann G, Goto H, et al. RuvB-like protein 2 is a suppressor of influenza A virus polymerases. J Virol. 2009;83:6429–6434. doi: 10.1128/JVI.00293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greco A, Bienvenut W, Sanchez JC, Kindbeiter K, et al. Identification of ribosome-associated viral and cellular basic proteins during the course of infection with herpes simplex virus type 1. Proteomics. 2001;1:545–549. doi: 10.1002/1615-9861(200104)1:4<545::AID-PROT545>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 70.Vuppalanchi D, Willis DE, Twiss JL. Regulation of mRNA transport and translation in axons. Results Probl Cell Differ. 2009;48:193–224. doi: 10.1007/400_2009_16. [DOI] [PubMed] [Google Scholar]
- 71.Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ben-Yaakov K, Dagan SY, Segal-Ruder Y, Shalem O, et al. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 2012;31:1350–1363. doi: 10.1038/emboj.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, et al. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hanz S, Perlson E, Willis D, Zheng JQ, et al. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- 75.Lin AC, Holt CE. Function and regulation of local axonal translation. Curr Opin Neurobiol. 2008;18:60–68. doi: 10.1016/j.conb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holt C, Schuman E. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murphy K, Travers P, Walport M, Janeway C. Janeway’s immunobiology. 8th ed. xix. New York: Garland Science; 2012. p. 868. [Google Scholar]
- 78.Unterholzner L, Keating SE, Baran M, Horan KA, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci U S A. 2012;109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perez de Diego R, Mulvey C, Casanova JL, Godovac-Zimmermann J. Proteomics in immunity and herpes simplex encephalitis. Expert Rev Proteomics. 2014;11:21–29. doi: 10.1586/14789450.2014.864954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hill JM, Lukiw WJ, Gebhardt BM, Higaki S, et al. Gene expression analyzed by microarrays in HSV-1 latent mouse trigeminal ganglion following heat stress. Virus Genes. 2001;23:273–280. doi: 10.1023/a:1012517221937. [DOI] [PubMed] [Google Scholar]
- 82.Rakkola R, Matikainen S, Nyman TA. Proteome analysis of human macrophages reveals the upregulation of manganese-containing superoxide dismutase after toll-like receptor activation. Proteomics. 2007;7:378–384. doi: 10.1002/pmic.200600582. [DOI] [PubMed] [Google Scholar]
- 83.Baines CP, Kaiser RA, Purcell NH, Blair NS, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 84.Du H, Guo L, Fang F, Chen D, et al. Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer’s disease. Nat Med. 2008;14:1097–1105. doi: 10.1038/nm.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li V, Brustovetsky T, Brustovetsky N. Role of cyclophilin D-dependent mitochondrial permeability transition in glutamate-induced calcium deregulation and excitotoxic neuronal death. Exp Neurol. 2009;218:171–182. doi: 10.1016/j.expneurol.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perez de Diego R, Mulvey C, Crawford M, Trotter MW, et al. The proteome of Toll-like receptor 3-stimulated human immortalized fibroblasts: implications for susceptibility to herpes simplex virus encephalitis. J Allergy Clin Immunol. 2013;131:1157–1166. doi: 10.1016/j.jaci.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miettinen JJ, Matikainen S, Nyman TA. Global secretome characterization of herpes simplex virus 1-infected human primary macrophages. J Virol. 2012;86:12770–12778. doi: 10.1128/JVI.01545-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ciborowski P, Kadiu I, Rozek W, Smith L, et al. Investigating the human immunodeficiency virus type 1-infected monocyte-derived macrophage secretome. Virology. 2007;363:198–209. doi: 10.1016/j.virol.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lietzen N, Ohman T, Rintahaka J, Julkunen I, et al. Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages. PLoS Pathog. 2011;7:e1001340. doi: 10.1371/journal.ppat.1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chew T, Taylor KE, Mossman KL. Innate and adaptive immune responses to herpes simplex virus. Viruses. 2009;1:979–1002. doi: 10.3390/v1030979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Birlea M, Owens GP, Eshleman EM, Ritchie A, et al. Human anti-varicella-zoster virus (VZV) recombinant monoclonal antibody produced after Zostavax immunization recognizes the gH/gL complex and neutralizes VZV infection. J Virol. 2013;87:415–421. doi: 10.1128/JVI.02561-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pasman Y, Nagy E, Kaushik AK. Enhanced bovine herpesvirus type 1 neutralization by multimerized single-chain variable antibody fragments regardless of differential glycosylation. Clin Vaccine Immunol. 2012;19:1150–1157. doi: 10.1128/CVI.00130-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kleemann P, Distler E, Wagner EM, Thomas S, et al. Varicella-zoster virus glycoproteins B and E are major targets of CD4+ and CD8+ T cells reconstituting during zoster after allogeneic transplantation. Haematologica. 2012;97:874–882. doi: 10.3324/haematol.2011.052597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bjornson ZB, Nolan GP, Fantl WJ. Single-cell mass cytometry for analysis of immune system functional states. Curr Opin Immunol. 2013;25:484–494. doi: 10.1016/j.coi.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sen N, Mukherjee G, Sen A, Bendall SC, et al. Single-cell mass cytometry analysis of human tonsil T cell remodeling by varicella zoster virus. Cell Rep. 2014;8:633–645. doi: 10.1016/j.celrep.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Erukhimovitch V, Karpasasa M, Huleihel M. Spectroscopic detection and identification of infected cells with herpes viruses. Biopolymers. 2009;91:61–67. doi: 10.1002/bip.21082. [DOI] [PubMed] [Google Scholar]
- 97.Leveque N, Legoff J, Mengelle C, Mercier-Delarue S, et al. Virological diagnosis of central nervous system infections by use of PCR coupled with mass spectrometry analysis of cerebrospinal fluid samples. J Clin Microbiol. 2014;52:212–217. doi: 10.1128/JCM.02270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boubaker Elandalousi R, Mekni Toujani M, Kaabi B, Larbi I, et al. Non-cytotoxic Thymus capitata extracts prevent Bovine herpesvirus-1 infection in cell cultures. BMC Vet Res. 2014;10:231. doi: 10.1186/s12917-014-0231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vastag L, Koyuncu E, Grady SL, Shenk TE, et al. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7:e1002124. doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu J, Shriver Z, Pope RM, Thorp SC, et al. Characterization of a heparan sulfate octasaccharide that binds to herpes simplex virus type 1 glycoprotein D. J Biol Chem. 2002;277:33456–33467. doi: 10.1074/jbc.M202034200. [DOI] [PubMed] [Google Scholar]
- 101.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 102.Vizoso Pinto MG, Villegas JM, Peter J, Haase R, et al. LuMPIS--a modified luminescence-based mammalian interactome mapping pull-down assay for the investigation of protein-protein interactions encoded by GC-low ORFs. Proteomics. 2009;9:5303–5308. doi: 10.1002/pmic.200900298. [DOI] [PubMed] [Google Scholar]
- 103.Jones C, Chowdhury S. A review of the biology of bovine herpesvirus type 1 (BHV-1), its role as a cofactor in the bovine respiratory disease complex and development of improved vaccines. Anim Health Res Rev. 2007;8:187–205. doi: 10.1017/S146625230700134X. [DOI] [PubMed] [Google Scholar]
- 104.Card JP, Enquist LW. Transneuronal circuit analysis with pseudorabies viruses. Curr Protoc Neurosci. 2014;68 doi: 10.1002/0471142301.ns0105s68. 1 5 1-1 5 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arvin AM. Varicella-zoster virus. Clin Microbiol Rev. 1996;9:361–381. doi: 10.1128/cmr.9.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lim F, Khalique H, Ventosa M, Baldo A. Biosafety of gene therapy vectors derived from herpes simplex virus type 1. Curr Gene Ther. 2013;13:478–491. doi: 10.2174/156652321306140103224550. [DOI] [PubMed] [Google Scholar]