Abstract
Mathematical and computational models have become indispensable tools for integrating and interpreting heterogeneous biological data, understanding fundamental principles of biological system functions, generating reliable testable hypotheses, and identifying potential diagnostic markers and therapeutic targets. Thus, such tools are now routinely used in the theoretical and experimental systematic investigation of biological system dynamics. Here, we discuss model building as an essential part of the theoretical and experimental analysis of biomolecular network dynamics. Specifically, we describe a procedure for defining kinetic equations and parameters of biomolecular processes, and we illustrate the use of fractional activity functions for modeling gene expression regulation by single and multiple regulators. We further discuss the evaluation of model complexity and the selection of an optimal model based on information criteria. Finally, we discuss the critical roles of sensitivity, robustness analysis, and optimal experiment design in the model building cycle.
Keywords: Biomolecular network, Differential equation, Dynamical system, Inverse problem, Mathematical model, Systems biology
1. Introduction
Molecular biology research has been profoundly impacted by the development of high-throughput measurement technologies such as next-generation sequencing, DNA microarrays, and mass spec-trometry, and by the application of these technologies in large-scale functional genomic, proteomic, metabolomic, protein–protein interaction, and protein–DNA interaction assays. These advances are enabling researchers to comprehensively map the molecular components, processes, networks, and functions that underlie biological processes and human diseases. A systems biology approach integrating these heterogeneous molecular data in quantitative network models promises to facilitate the comprehensive understanding of biological systems, the identification of novel diagnostic markers and drug targets, and rational intervention into disease processes.
The dynamic biomolecular network model is a powerful framework for integrating heterogeneous experimental data (1, 2). It allows us to interpret the behavior of complex biological systems both quantitatively and mechanistically, and to generate targeted experimentally testable hypotheses. Thus, modeling is an essentially component of the systems biology cycle – “experiment– model–hypothesis–experiment” (Fig. 1). To construct models with predictive utility, it is critical to describe cellular mechanisms with the appropriate level of detail. In the predictive kinetic biomolecular models we describe below, we seek to compile information on the system process details including reaction rates and concentrations of molecular constituents. Estimating reaction rates and other parameters is often challenging due to the heterogeneity of experimental data (e.g., different source origins, setups, cell types, experimental conditions, etc.) and the difficulty of obtaining data relevant to the specific in vivo conditions.
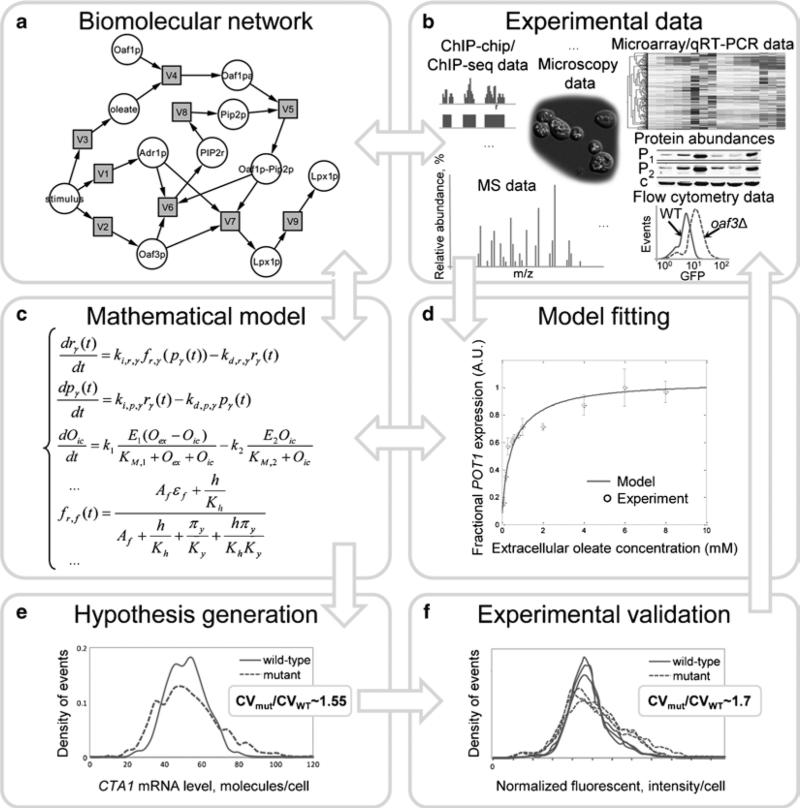
Fig. 1.
Workflow of the theoretical and experimental analysis of biomolecular network dynamics (the model building cycle). Arrows denote information flow, illustrating how the model can lead to the formulation of hypotheses that can be experimentally tested. The results from experimental validation of the model are used to improve the model, and simulations from the refined model can lead to new predictions. (a) Diagram showing regulatory network controlling oleate response in yeast. (b) Types of data used in systems biology. (c) Equations showing the structure of the ordinary differential equation (ODE) model of the oleate response network. (d) Predicted and measured steady-state dose-response of the oleate-inducible target gene, POT1. (e) Predicted cell population distributions of the mRNA level of an oleate-inducible gene in the wild-type oleate network and in a perturbed (“mutant”) network. (f) Measured cell population distributions of the level of a GFP-tagged, oleate-inducible protein in wild-type yeast and in a yeast strain bearing a mutation corresponding to the network perturbation in the “mutant” model.
Despite the tremendous data-generating capabilities of new high-throughput technologies, the compendium of available measurements of many cellular component levels and processes in space and time remains sparse. Often, the available data are not sufficient to infer the detailed molecular mechanisms underlying a given cellular process. Furthermore, current knowledge of molecular mechanisms is highly nonuniform, varying from the well supported and very detailed to the hypothetical and poorly described. Thus, it is impossible to describe all processes in the model equally comprehensively. The ideal modeling method should allow rational selection of the most appropriate level of detail in the model based on prior knowledge of relevant pathways and the resolution of relevant measurements (3, 4).
In engineering terminology, one could describe the problem of inferring regulatory mechanisms within biological systems from prior pathway/interaction knowledge and measurements of system behavior as a “structural inverse problem with prior information.” Numerous theoretical and experimental approaches have been developed to attempt to solve this biological inverse problem (5–12), and the investigation of integrative methods to identify biomolecular interactions and mechanisms from heterogeneous data remains an active area of research (13). This problem naturally divides into two components: mapping the network of molecular components and processes relevant to the system, and constructing a kinetic and quantitative model of the network behavior. It is the latter component that is the specific area of focus of this chapter.
Broadly speaking, mathematical approaches to modeling biological networks can be grouped by the type of mathematical abstraction used to represent the state vector of the system, such as discrete-valued (14–16), continuous-valued (17–19), stochastic (20–22), and various combinations of the three types (hybrid models) (18, 19, 23–26). In this chapter, we focus primarily on a continuous-valued approach for modeling biomolecular network dynamics using nonlinear ordinary or delay differential equations and the possible combination with the discrete approach for modeling such events as transitions from different environments, cell divisions, etc. We use the fatty-acid (FA)-induced transcriptional regulatory network model (27) as a running example to illustrate different steps of the workflow of the theoretical and experimental analysis of the molecular network dynamics.
2. Methods
2.1. Construction of a Mathematical Model
Construction of a mathematical model of a biomolecular network of interest (the model building cycle) starts from stating the purpose and objectives of the model (see Note 1). For example, the main purpose of the FA-responsive transcriptional regulatory network model is to understand the carbon source-sensing mechanism of the core regulatory network as well as the interplay between transcription factors (TFs) that combinatorially regulate of the expression of target genes. Ideally, the FA-responsive network model should help us understand how the variations in the input stimulus (FA concentration) and the molecular system parameters (e.g., the FA transport efficiency, the ligand–sensor affinity, TF activities, tran scription and translation initiation rates, component degradation rates, etc.) affect the network response.
Collect/gather all available information on the structural and functional organization of the biomolecular network of interest. Define the list of processes and variables (molecular components: genes, RNAs, proteins, small molecules, intermediate complexes, etc.) that should be included in the model. For example, the core FA-responsive transcriptional network consists of carbon source-sensing transcription factors that regulate key target genes through an overlapping feed-forward network motif. The model describes the transport of extracellular oleate into the cell, which activates a molecular network governing peroxisome protein production and organelle proliferation. The model describes the oleate-dependent expression and function of four TF genes (ADR1, PIP2, OAF1, and OAF3), as well as the expression of three oleate-inducible reporter genes that are downstream targets of these TFs (POT1, CTA1, and LPX1). For each gene in the model, both the gene-specific mRNA and total protein are accounted for. The model takes into account the synthesis and degradation of each species of mRNA and protein (27).
The structural organization of the biomolecular network of interest can be illustrated as a bipartite graph (Fig. 1a), which presents two independent disjoint sets of vertices. Within the graph, circular nodes represent variables of the model, i.e., levels of molecular species (RNA and protein components). Square nodes represent molecular processes that together define the dynamic relationships among physical entities (e.g., translation of RNA to protein). Edges of the graph define the relationships between the participants in each process of the model. The bipartite graph of the model at initial steps helps to reveal the model structure, its inputs and end points and to detect possible gaps and incompleteness, which is particularly important for large-scale models. For example, the graph of the oleate response network (Fig. 1a) shows that transcription factors regulate oleate-responsive genes in a feed-forward network topology, which has important implications for the stability and robustness of control of oleate-responsive genes (27).
Collect/gather all available quantitative and qualitative experimental data for molecular components and processes of the model (concentrations of molecular components at different states (e.g., basal, induced, repressed, relative concentrations in different conditions); kinetics of molecular components during transitions between different conditions/environments; synthesis and degradation rates of molecular components of the model; interaction affinities; dependencies of process rates on the level of regulator(s), any available kinetic constants and their estimates of the processes of interest, etc.) (Fig. 1b). Examples of experimental techniques and quantitative measurements for kinetic models are listed in Table 1.
Define possible mechanisms and formally describe each process in the model. For example, gene expression in the FA-induced transcription model (hereafter, FAIT model) is represented using a fractional activity function for each gene, which is a function of the protein concentrations in the model (for a description of how to derive a fractional activity function from the mechanistic details of the molecular process, see Subheading 2.2). Protein synthesis is modeled as proportional to mRNA concentration. Often, mechanisms that occur rapidly relative to the time scales of interest in the model can be simplified using the assumption of quasi-steady-state (28). For example, activation of the oleate-sensing transcription factor Oaf1p is assumed to occur rapidly so that activation and complex dissociation are at quasi-steady-state with respect to the slowly varying total Oaf1p concentration (moreover, the extracellular oleate concentration is taken to be constant, and depletion of oleate from the media is not modeled). The transport of FA across the plasma membrane and subsequent esterification with coenzyme A (CoA) are modeled by assuming a hyperbolic saturating function of extracellular oleate concentration for the rate of transport and Michaelis–Menten kinetics for fatty acyl-CoA synthesis. Turnover is modeled using first-order kinetics (27).
- Each component of the system might participate simultaneously in several processes with nonzero stoichiometry. For example, the kinetics of changes in the abundance of the ith mRNA species can be described based on its time and state-dependent overall synthesis and degradation rates as the following differential equation
In the more specific case, when the synthesis of the ith mRNA is described in terms of a fractional activity fi (see Subheading 2.2) and the mRNA degradation is proportional to the mRNA concentration in the cell, the kinetics of the mRNA changes can be described as follows:(1)
where is the maximum rate constant for the mRNA synthesis and is the mRNA degradation rate constant.(2) -
For each dynamic species in the model, a kinetic equation governing its time rate of change is derived by combining contributions from all relevant dynamic processes (from step 5), and taking into account the stoichiometry of the species in each process. When completed, this yields a system of differential equations
where xi is the ith dynamic species of the kinetic model; is a rate of the nth process in which xi is produced and is a rate of the mth process in which xi is consumed, where .(3) Examples of differential equations of the oleate model are shown in Fig. 1c.
-
Once the equations governing the model's dynamics (Eq. 3) are defined, the model is then specified by determining initial species concentrations and rate parameter values. Usually, some model parameter values are known from the literature or have been measured in direct assays and, therefore, can be fixed in the model. The values of other parameters may only be known to fall within a certain range. Parameters or species concentrations whose values are not known or adequately constrained by previous measurements need to be estimated (within physiologically relevant ranges), using the model, from the available measurements of whole-system response (see Note 2). Formally, the parameter estimation procedure can be described as solving the parametric inverse problem. In brief, a cost function representing the total deviation of the model predictions from the experimental data is defined, and the model parameters are then varied to identify the parameter set that minimizes the cost function. For example, the cost function (D) can be calculated as a sum of the squares of the model prediction error defined as a deviation of the quantitative characteristics calculated by the model (e.g., concentration of molecular components in different conditions) (ypredicted) from the relevant experimental measurements (ymeasured)
where is the variance in the ith measurement.(4) Numerous optimization algorithms have been developed to solve the parametric inverse problem in many fields of science (29). The gradient-based optimization algorithms are often used for the model parameter optimization. These algorithms are well suited to the task of finding a local minimum of an optimization function. Nonlinear mathematical models usually have multiple local minima and applying only gradient-based optimization methods is not effective for finding the global minimum of the cost function or the globally optimal solution. To overcome this problem, one can combine the traditional gradient methods with Monte Carlo methods (i.e., methods with random sampling). Evolutionary and genetic algorithms are often used as Monte Carlo heuristic methods. These algorithms incorporate into the optimization procedure basic principles of evolution such as mutation, recombination, selection, inheritance, etc. There are also other Monte Carlo optimization methods such as simulated annealing, stochastic optimization, etc. (see Notes 3 and 4).
In the case of the FAIT model, optimization was carried out using the constrained optimizers ga (genetic algorithm) and fmincon (constrained nonlinear optimization) in MATLAB. The undetermined model parameters were optimized within the physiologically relevant constraints to minimize the sum-squared of the model prediction error for the time-course and steady-state gene expression measurements. A comparison of the optimized FAIT model characteristics with experimental data is shown in Fig. 1d.
Steps 5 to 8 can be repeated for the models with different mechanistic representations of the processes in the model. For example, in the model of transcriptional induction of the yeast galactose metabolic pathway in yeast, two different model scenarios were studied. These two scenarios represented two different hypotheses regarding the mechanism of galactose-dependent de-repression of the GAL regulon by Gal3p, namely, direct nuclear binding of Gal3p and indirect derepression. Comparison of the fitness of the two model scenarios favored the indirect mechanism (30, as well as unpublished data).
-
Analyze/estimate the model complexity and/or choose the optimal model. Ideally, the model complexity should be commensurate with the amount and granularity of available experimental data. For example, given a body of highly fine-grained time-course data and data from a diverse set of system perturbations, a very simplistic model might be expected to poorly recapitulate the body of data. On the contrary, an inappropriately complex model with many undetermined parameters will likely have poor performance recapitulating experimental measurements that were held out from the body of measurements used to train the model, a problem known as overfitting. Usually, in both cases the model would be said to have a low predictive power (see Note 5). To compare two or more models with differing levels of parametric complexity, one can apply the Bayesian or Akaike information criteria (BIC (31) and AIC (32), respectively). These criteria quantify the trade-off relationship between model prediction performance and the number of the model parameters. For example, the potential for overfitting in the FAIT model was estimated by quantifying the bias-versus-variance trade-off. The small-sample-corrected Akaike Information Criterion (c-AIC) (33) was used for this purpose. The Akaike penalty term was computed using the following formula:
where k is a number of model parameters that were determined by parameter estimation and N is the number of experimental data points. The variance was computed as the sum-squared model prediction error using Eq. 4.(5) The Penalty/D ratio for the model was found to have a value of 1.4. Given that c-AIC is a conservative estimate of model fitness (i.e., the c-AIC involves a quite strong penalty for complexity), this penalty/error ratio in conjunction with results of the sensitivity analysis (see step 12) suggested that the available number and diversity of measurements used for model training are adequate to discern the best-fit model in the space of parameter values, within the model class necessitated by prior biological knowledge of the underlying molecular interactions.
At this point, we have an optimized model that can be used as a tool for exploring quantitative and qualitative characteristics of the molecular system of interest.
-
For both uncertainty analysis of model predictions and identifying potential targets for perturbation analysis in the biological system, a sensitivity analysis of the model output predictions can be beneficial. For example, it is often useful to perform a sensitivity analysis of the model output characteristics to variable inputs (stimuli). In general, sensitivity analysis allows one to explore quantitatively the relationship between input and output characteristics and detect the ranges of the input signal variations to which the model responses are most sensitive as well as the ranges to which the system is not responsive or saturated (see Note 6).
The sensitivity matrix (Sio) of the model response changes to input signal variations is defined as follows:
For example, the model of the FAIT regulatory network was used to analyze whether the Oaf3p regulator acts to buffer the induced genetic switch against variations in the level of FA in the environment (27). The expression kinetics of the target gene (POT1) were investigated in WT and oaf3Δ model strains exposed to an oscillating oleate concentration. The kinetic model predicted that the transcriptional repressor Oaf3p modulates the amplitude of variation of expression levels of oleate-responsive element (ORE)-driven genes in a fluctuating environment. The target gene undergoes larger-amplitude variations in the oaf3Δ model than in the WT model, indicating that the loss of Oaf3p destabilizes the genetic switch with respect to transient oleate variations (27). The dependence of the target gene expression amplitudes on the oleate variation frequency and amplitude was also systematically explored (27). The model simulations show that the POT1 amplitude difference between the oaf3Δ model and WT model increases with decreasing frequency of an oleate pulse, suggesting that the oaf3Δ strain is less able than the WT, to filter out oleate variations on a time scale of >40 min. Varying both the amplitude and period of the oleate concentration oscillations reveals a nonlinear relationship between amplitude and period. The maximal differences between oaf3Δ and WT strains shift toward greater period as the amplitude increases (27), which illustrates the potential of mathematical models of complex biomolecular networks to reveal nontrivial system-level properties.(6) Perform parameter sensitivity analysis (PSA). PSA can be used to systematically investigate how perturbations/changes in the model parameters impact on the system outcomes. PSA is very useful for analyzing the robustness of the behavior of the system with respect to perturbations of its components (e.g., pharmacological inhibition, knockouts, etc.), as well as for identifying the limiting parameters for system functioning. This analysis is also helpful for estimating the required precision levels of the parameter values that are necessary to achieve a given precision level in a prediction of system output/behavior (see Note 6 and 7). There are two major types of PSA: local and global.
Table 1.
Examples of experimental techniques and quantitative measurements for kinetic models
| Quantity in the kinetic model | Experimental technique |
|---|---|
| Concentrations of molecular components | qRT-PCR microarray; quantitative northern blot; fluorescence microscopy; quantitative mass spectrometry; flow cytometry; etc. |
| Enzymatic activity (initial rate, transient and relaxation kinetics, etc.) | Enzyme assays (spectrophotometric, fluorometric, calorimetric, chemiluminescent, light scattering, chromatographic and other assays) |
| Kinetics of molecular interactions | In vitro measurements; time-course measurements of protein activation; etc. |
| Time-course of protein-DNA binding | DNA-binding protein immunoprecipitation combined with microarray (ChIP-chip), sequencing (ChIP-seq) or PCR in vitro binding assays for purified interacting partners; etc. |
| Time-course of protein-RNA binding | RNA-binding protein immunoprecipitation combined with microarray, sequencing or PCR; in vitro binding assays for purified interacting partners; etc. |
| Kinetics of protein-protein binding | Affinity electrophoresis; co-immunoprecipitation combined with quantitative mass spectrometry; quantitative immunoprecipitation combined with knock-downs; fluorescence correlation spectroscopy (FCS); surface plasmon resonance (SPR); static and dynamic light scattering; etc. |
| Kinetics of protein-ligand binding | Ligand-binding assays; radiolabeling, affinity electrophoresis; SPR; light scattering; etc. |
| Kinetics of molecular transport | Imaging; fluorescence microscopy; radiolabeling, pulse chase; etc. |
| Synthesis and degradation rates | Time-course abundance data (during induction or deactivation of system, or after inhibition of transcription); pulse chase; etc. |
-
(a)Local PSA: The sensitivity matrix (Sp) of the model response to changes to the model parameters can be defined as follows
Local PSA allows exploring the model response changes to one parameter variation at a time.(7) -
(b)
Global PSA allows varying multiple model parameters simultaneously to analyze their effect on the model outputs. Global PSA methods usually apply random sampling techniques because it is impossible to explore systematically the influence of all possible combinations of the model parameter changes of the model outputs. PSA is extensively described and applied in a number of publications (34–39).
As an example application of PSA, the relative model prediction error for alternative values of the 14 fitted parameters was estimated in the FAIT regulatory network. The relative model prediction error (i.e., the optimization cost function) was calculated using Eq. 4. The model error was analyzed as a function of varying a single model parameter over a range of approximately eightfold up or down relative to the best-fit parameter value. In the variation of all 14 parameters except KM,s, the Michaelis constant for the activation of transcriptional response of ADR1 and OAF3 by FA (27), a significant increase in the cost function value (i.e., decrease in the model fitness) was seen over the range of alternative parameter values. In the case of KM,s, the relative fitness-insensitivity to the parameter value may indicate that the expression data, in conjunction with the model, are most consistent with FA-dependent activation of Adr1p and Oaf3p being essentially on-or-off (27).
-
13
Optimal experiment design. The systematic analysis of the mathematical model allows one to detect the most sensitive processes/parameters to a particular environmental/experimental condition that is relevant to a particular function of the system. For example, in the case of the FAIT model, the role of the TF Adr1p in reducing the cell population heterogeneity of expression of the target genes (i.e., CTA1, LPX1, etc.) has been investigated. The model was modified to simulate a hypothetical Adr1p-independent mutant strain in which Adr1p regulates neither PIP2 nor downstream target targets, but for which these genes fully induce in the presence of oleate (which might be envisioned as an adr1Δ strain with elevated ORE-binding affinity). To compare the cell population heterogeneity of expression (arising from the intrinsic stochasticity of gene expression) of the target gene CTA1 in the presence of oleate in the two models, the steady-state stochastic dynamics in both models were simulated using the Gibson–Bruck stochastic chemical kinetic algorithm (21). The simulations revealed that the histogram of CTA1 transcript levels has a broader distribution in the mutant model than in the WT model (Fig. 1e), indicating a greater heterogeneity of gene expression (approximately 1.5-fold, as measured by coefficient of variation (CV)) (27), which provides an explanation for the network structure and suggests experiments to test the predictions.
To investigate the FAIT model prediction that Adr1p serves as a noise reducer in the oleate-responsive network, the variability of expression of LPX1, was examined experimentally. The level of LPX1 gene expression was measured in WT and adr1Δ yeast strains in the presence of oleate using flow cytometry and a GFP-tagged Lpx1p reporter. Consistent with simulation results, the CV of Lpx1-GFP in adr1Δ cells was 1.7-fold higher than in WT cells (Fig. 1f).
2.2. Formal Description of Fractional Activity Functions of Molecular Processes
The purpose of this section is to illustrate how to derive the so-called fractional activity functions for a formal description of the dependence of quasi steady-state rates of molecular processes on component concentrations of the model (e.g., TFs or other types of regulators). To give a flavor how to describe formally the fractional activity functions of molecular processes, which would, therefore, form the right parts of differential equations of the model (see Subheading 2.1, steps 6 and 7), we consider below some cases of a gene expression regulation by single and multiple TFs. The approach described here borrows from methods originally developed for modeling enzyme kinetics, and for this reason, it is helpful to have some familiarity with the King–Altman method (40, 41) and the generalized Hill function approach (this method is very flexible and does not require prior knowledge of the detailed mechanism of the molecular process of interest) (4). The reader is also referred to (17–19), which provide additional relevant background. Here, we outline the procedure for deriving the fractional activity function for a complex biological process (e.g., transcription) controlled by multiple regulators:
-
Constructing a fractional activity function for the rate of initiation of transcription of a gene y (hereafter written Ty) regulated by a single TF x requires initially answering a few questions. First, is x an activator or a repressor in the context of the model (sometimes the regulator may have a dual role)? What is the constitutive level of the target gene expression? At which concentration of x does Ty reach one half of its maximum level? Is the regulation of Ty by x linear, or does x act in a cooperative fashion as it regulates Ty? Nonlinear regulation of Ty by x can be captured by a “Hill coefficient” – see below.
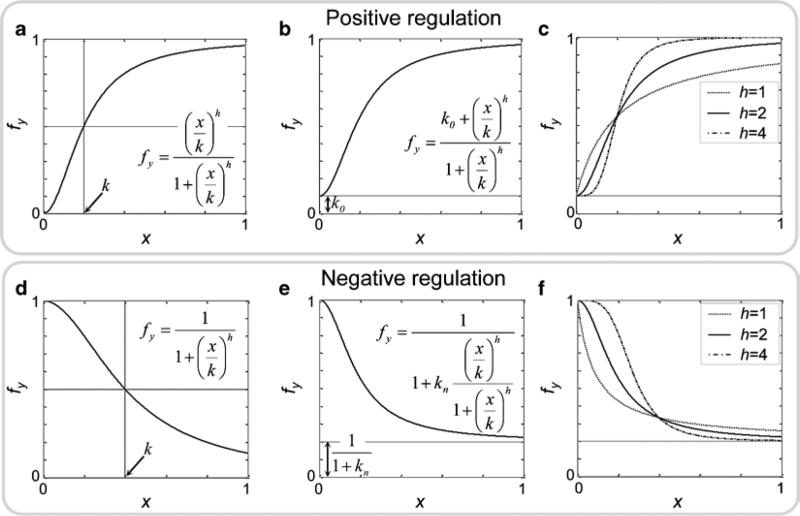
The fractional activity function of transcriptional initiation for a gene y, denoted by fy, that is upregulated by x can be generally described as follows
where x is a concentration of the regulator, k0 is the basal expression level of y; k1 and k2 are efficiency constants for the regulator x, and h1 and h2 are Hill coefficients of the regulator x. Examples of the fractional activity function (Eq. 8) dependence on the concentration of x with different parameter values are illustrated in Fig. 2a–c.(8) The fractional activity function for transcription of a gene y that is downregulated by x can be modeled as follows
The partial inhibition of y by x can be described as follows(9)
where 1 / (1 + kn) represents the level to which the expression of y can be downregulated by x when x →∞. Examples of how the fractional activity functions (Eqs. 9 and 10) depend on the concentration of x with different parameter values are illustrated in Fig. 2d–f.(10) -
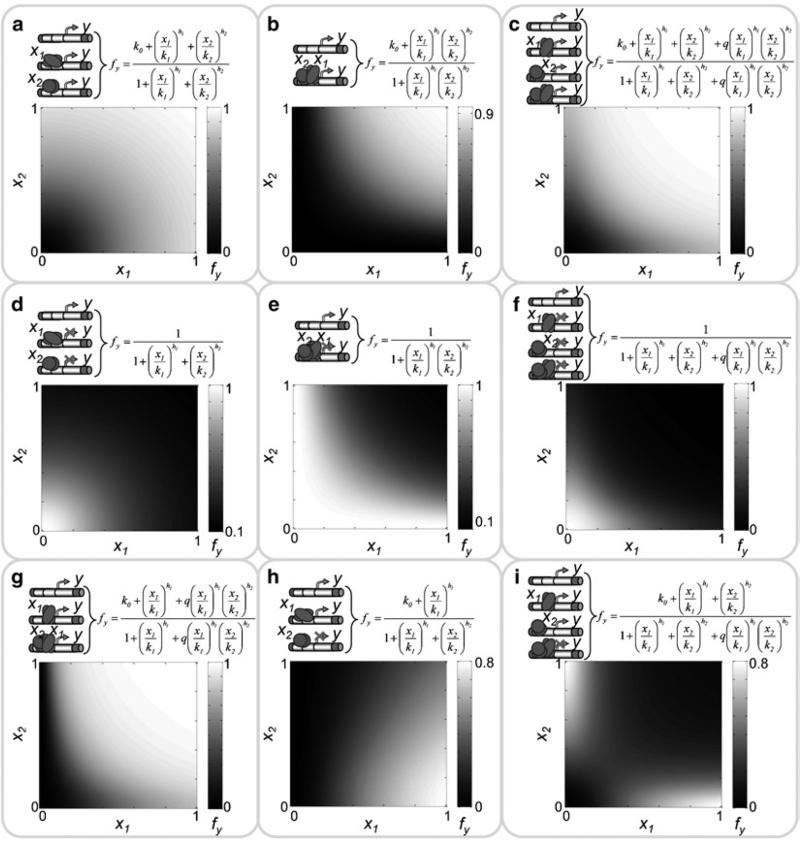
The expression level of a gene y upregulated by two TFs (x1 and x2) acting combinatorially can be described as follows
where x1 and x2 are concentrations of corresponding regulators, k0 is the basal expression level of y, k1 and k2 are efficiency constants for x1 and x2, respectively, h1 and h2 are Hill coefficients for x1 and x2, respectively, and q is a cooperativity/synergistic constant for the joint regulation of y by x1 and x2. An example of the fractional activity function shown in Eq. 11 as a function of concentrations of x1 and x2 is illustrated in Fig. 3c.(11) Equation 11 can be modified for different mechanisms of combinatorial up- down- and mixed regulation (e.g., some terms in numerator and/or in denominator can be set to zero). Examples of using such modified functions for a competitive activation of y by x1 and x2 and the upregulation of y only when both factors are presented (e.g., x1 and x2 bind the promoter of y as a heterocomplex (x1 / x2) to regulate the transcription initiation of gene y) are illustrated in Fig. 3a–b, respectively. Other examples are presented in Fig. 3d–i.
- Equation 11 can be easily generalized for an arbitrary number of TFs with mixed types and mechanisms of the regulation, as follows
where xi is a concentration of the ith regulator in the system, ki is an efficiency constant for xi, CA,m is a number of different combinations of independently acting upregulators ; and CI,A,n is a number of different combinations of independently acting up- and downregulators . In the more general case, ki and hi parameter values can vary with time and depend on the regulator levels as well as other environmental and intracellular factors (4).(12)
Fig. 2.
Examples of regulation of transcription of a gene y by a single regulator x and their formal descriptions using a fractional activity function fy(x). Plots show the simulated dependence of the fractional activity on the level of regulator x, for different regulatory scenarios (positive and negative regulation) and for different kinetic parameter values. (a) Gene y has no basal level of expression. (b) Gene y has a constitutive level of expression (k0). (c) Dependence of fy on the level of activator (x) with the different cooperative activities (Hill coefficients, h). (d) Simplest formal description of negative regulation. (e) Partial negative regulation (kn). (f) Dependence of fy on the level of the inhibitor (x) with different cooperative activities. The following parameters were used for the simulations: k0 = 0.1; k = 0.2 (a–c); k = 0.4 (d–f); kn = 4; h = 2.
Fig. 3.
Examples of combinatorial regulation of the expression level (initiation of transcription) of gene y (fy) by two regulators (x1 and x2) and their formal descriptions. (a) Competitive activation of y by x1 and x2. (b) Upregulation of y only when both factors bind the promoter (e.g., by x1 / x2 heterocomplex). (c) Independent activation of y by x1 and x2 with the synergistic effect (q) when both factors bind the promoter. (d) Competitive inhibition of y by x1 and x2. (e) Downregulation of y when only both factors bind the promoter (e.g., by x1 / x2 heterocomplex). (f) Independent inhibition of y by x1 and x2 with synergistic (q) downregulation when both factors bind the promoter. (g) Sequential activation of y by x1 and x2 when x1 binds the promoter first and both factors synergistically upregulate the target gene expression (h) Competitive activation and inhibition of y by x1 and x2, respectively. (i) Activation of y by x1 and x2 when they bind the promoter separately and synergistic inhibition when they bind the promoter together. The following parameters were used for the simulations: k0 = 0; k1 = k2 = 0.5; q = 10; h1 = h2 = 2.
Acknowledgments
This work was supported by the National Institutes of Health through grants from the National Institute of General Medical Sciences (R01-GM075152 and P50-GM076547), the National Technology Centers for Networks and Pathways (U54-RR022220), and the National Heart, Lung, and Blood Institute (K25-HL098807 to S.A.R).
Footnotes
To better formulate the purpose and objectives of the model, one should first state a set of questions/problems about the behavior of the system of interest which the model may help to answer.
Ideally, the available experimental data should be divided into at least two subsets, with one subset used for parameter estimation and the other subset used for the model cross-validation or the estimation of the model prediction power.
An important common feature of stochastic algorithms is that for complex cost functions, they are, in general, not guaranteed to find the globally optimal solution; instead, they will identify a solution that is very close to globally optimal, where the degree of difference is controlled by the criteria used to cease the optimization process (stopping criteria; e.g., number of generations, time limits, fitness limits, etc.) and other parameters of the optimization (42).
Occasionally, to solve the parametric inverse problem it is necessary to divide the model into small parts or submodels and fit each submodel to an appropriate set of experimental data. Such subdivisions/submodels and the corresponding experimental data represent a set of scenarios that might describe different experimental setups, conditions, etc. For instance, the framework of modeling in terms of scenarios is helpful for an optimal experiment design, hypothesis generation, etc.
Generally, the potential for overfitting in the model depends not only on the number of parameters but also on the model structure that may inappropriately reduce the model prediction error in comparison with the noise level in the experimental data.
The sensitivity analysis of the model input/output characteristics (see Subheading 2.1, step 11) can be repeatedly performed in the context of the parameter sensitivity analysis (see Subheading 2.1, step 12). This approach allows for the systematic investigation and quantitative comparative analysis of the biomolecular network dynamics relative to system parameters.
The sensitivity analysis can also precede the parameter estimation procedure (solving the inverse problem) and be used to rank parameters based on a sensitivity measure. The low-rank parameters can be roughly estimated or even eliminated from the models by changing relevant parts of the model (e.g., equations for fractional activities, etc.), whereas high-rank parameters must be estimated with an appropriate accuracy/precision.
References
- 1.Bornholdt S. Systems biology. Less is more in modeling large genetic networks. Science. 2005;310:449–451. doi: 10.1126/science.1119959. [DOI] [PubMed] [Google Scholar]
- 2.Kim HD, Shay T, O'Shea EK, Regev A. Transcriptional regulatory circuits: predicting numbers from alphabets. Science. 2009;325:429–432. doi: 10.1126/science.1171347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Riel NA. Dynamic modelling and analysis of biochemical networks: mechanism-based models and model-based experiments. Briefings in Bioinformatics. 2006;7:364–374. doi: 10.1093/bib/bbl040. [DOI] [PubMed] [Google Scholar]
- 4.Likhoshvai V, Ratushny A. Generalized Hill function method for modeling molecular processes. J Bioinform Comput Biol. 2007;5:521–531. doi: 10.1142/s0219720007002837. [DOI] [PubMed] [Google Scholar]
- 5.Wagner A. How to reconstruct a large genetic network from n gene perturbations in fewer than n(2) easy steps. Bioinformatics. 2001;17:1183–1197. doi: 10.1093/bioinformatics/17.12.1183. [DOI] [PubMed] [Google Scholar]
- 6.Liao JC, Boscolo R, Yang YL, Tran LM, Sabatti C, Roychowdhury VP. Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci USA. 2003;100:15522–15527. doi: 10.1073/pnas.2136632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrgard MJ, Covert MW, Palsson BO. Reconstruction of microbial transcriptional regulatory networks. Current opinion in bio-technology. 2004;15:70–77. doi: 10.1016/j.copbio.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 10.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 12.Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, Schubert LA, Birditt B, Shay T, Goren A, Zhang X, Smith Z, Deering R, McDonald RC, Cabili M, Bernstein BE, Rinn JL, Meissner A, Root DE, Hacohen N, Regev A. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marbach D, Prill RJ, Schaffter T, Mattiussi C, Floreano D, Stolovitzky G. Revealing strengths and weaknesses of methods for gene network inference. Proc Natl Acad Sci USA. 2010;107:6286–6291. doi: 10.1073/pnas.0913357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol. 1969;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- 15.Thomas R. Boolean formalization of genetic control circuits. J Theor Biol. 1973;42:563–585. doi: 10.1016/0022-5193(73)90247-6. [DOI] [PubMed] [Google Scholar]
- 16.Kauffman SA. The Origins of Order: Self-Organization and Selection in Evolution. Oxford Univ. Press; New York: 1993. [Google Scholar]
- 17.Edelstein-Keshet L. Mathematical Models in Biology. SIAM: Society for Industrial and Applied Mathematics; New York: 2005. [Google Scholar]
- 18.Bolouri H. Computational Modelling Of Gene Regulatory Networks - A Primer. Imperial College Press; London: 2008. [Google Scholar]
- 19.Klipp E, Liebermeister W, Wierling C, Kowald A, Lehrach H, Herwig R. Systems Biology: A Textbook. WILEY-VCH; Weinheim: 2009. [Google Scholar]
- 20.Gillespie DT. A General Method for Numerically Simulating the Stochastic Time Evolution of Coupled Chemical Reactions. Journal of Computational Physics. 1976;22:403–434. [Google Scholar]
- 21.Gibson MA, Bruck J. Efficient exact stochastic simulation of chemical systems with many species and many channels. J Phys Chem A. 2000;104:1876–1889. [Google Scholar]
- 22.Wilkinson DJ. Stochastic Modelling for Systems Biology. Chapman & Hall/CRC; Boca Raton, FL.: 2006. [Google Scholar]
- 23.McAdams HH, Shapiro L. Circuit simulation of genetic networks. Science. 1995;269:650–656. doi: 10.1126/science.7624793. [DOI] [PubMed] [Google Scholar]
- 24.Likhoshvai VA, Matushkin IuG, Ratushnyi AV, Anan'ko EA, Ignat'eva EV, Podkolodnaia OA. [A generalized chemical-kinetic method for modeling gene networks] Molekuliarnaia Biologiia. 2001;35:1072–1079. [PubMed] [Google Scholar]
- 25.Shmulevich I, Dougherty ER, Kim S, Zhang W. Probabilistic Boolean Networks: a rule-based uncertainty model for gene regulatory networks. Bioinformatics. 2002;18:261–274. doi: 10.1093/bioinformatics/18.2.261. [DOI] [PubMed] [Google Scholar]
- 26.de Jong H. Modeling and simulation of genetic regulatory systems: a literature review. J Comput Biol. 2002;9:67–103. doi: 10.1089/10665270252833208. [DOI] [PubMed] [Google Scholar]
- 27.Ratushny AV, Ramsey SA, Roda O, Wan Y, Smith JJ, Aitchison JD. Control of transcriptional variability by overlapping feed-forward regulatory motifs. Biophysical Journal. 2008;95:3715–3723. doi: 10.1529/biophysj.108.134064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao CV, Arkin AP. Stochastic chemical kinetics and the quasi-steady-state assumption: Application to the Gillespie algorithm. J Chem Phys. 2003;118:4999–5010. [Google Scholar]
- 29.Banga JR. Optimization in computational systems biology. BMC systems biology. 2008;2:47. doi: 10.1186/1752-0509-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Atauri P, Orrell D, Ramsey S, Bolouri H. Evolution of “design” principles in biochemical networks. IET Sys Biol. 2004;1:28–40. doi: 10.1049/sb:20045013. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz G. Estimating the Dimension of a Model. The Annals of Statistics. 1978;6:461–464. [Google Scholar]
- 32.Akaike H. A New Look at the Statistical Model Identfication. IEEE Trans. Automatic Control. 1974;19:716–723. [Google Scholar]
- 33.Burnham KP, Anderson DR. Model Selection and Multi-Model Inference. Springer-Verlag New York, LLC.; 2002. [Google Scholar]
- 34.Rabitz H. Systems analysis at the molecular scale. Science. 1989;246:221–226. doi: 10.1126/science.246.4927.221. [DOI] [PubMed] [Google Scholar]
- 35.Ratushny AV, Likhoshvai VA, Ignatieva EV, Goryanin II, Kolchanov NA. Resilience of Cholesterol Concentration to a Wide Range of Mutations in the Cell. Complexus. 2003;1:142–148. [Google Scholar]
- 36.Ratushnyi AV, Likhoshvai VA, Ignat'eva EV, Matushkin YG, Goryanin II, Kolchanov NA. A computer model of the gene network of the cholesterol biosynthesis regulation in the cell: analysis of the effect of mutations. Doklady Biochemistry and Biophysics. 2003;389:90–93. doi: 10.1023/a:1023679921536. [DOI] [PubMed] [Google Scholar]
- 37.Stites EC, Trampont PC, Ma Z, Ravichandran KS. Network analysis of oncogenic Ras activation in cancer. Science. 2007;318:463–467. doi: 10.1126/science.1144642. [DOI] [PubMed] [Google Scholar]
- 38.Saltelli A, Ratto M, Andres T, Campolongo F, Cariboni J, Gatelli D, Saisana M, Tarantola S. Global sensitivity analysis: the primer. John Wiley & Sons Ltd; Chichester, UK.: 2008. [Google Scholar]
- 39.Chen WW, Schoeberl B, Jasper PJ, Niepel M, Nielsen UB, Lauffenburger DA, Sorger PK. Input-output behavior of ErbB signaling pathways as revealed by a mass action model trained against dynamic data. Mol Syst Biol. 2009;5:239. doi: 10.1038/msb.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King EL, Altman C. A Schematic method of deriving the rate laws for enzyme-catalyzed reactions. The Journal of physical chemistry. 1956;60:1375–1378. [Google Scholar]
- 41.Cornish-Bowden A. An automatic method for deriving steady-state rate equations. Biochem J. 1977;165:55–59. doi: 10.1042/bj1650055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes with Source Code CD-ROM 3rd Edition: The Art of Scientific Computing. 3rd edn. Cambridge University Press; New York: 2007. [Google Scholar]