Abstract
Human carbonic anhydrase (CA) is a well-studied, robust, mononuclear Zn-containing metalloprotein that serves as an excellent biological ligand system to study the thermodynamics associated with metal ion coordination chemistry in aqueous solution. The apo-form of human carbonic anhydrase II (CA) binds two equivalents of copper(II) with high affinity. The Cu2+ ions bind independently forming two non-coupled type-II copper centers in CA (CuA and CuB). However, the location and coordination mode of the CuA site in solution is unclear, compared to the CuB site that has been well characterized. Using paramagnetic NMR techniques and X-ray absorption spectroscopy we have identified an N-terminal Cu2+ binding location and collected information on the coordination mode of the CuA site in CA, which is consistent with a four to five coordinate N-terminal Cu2+ binding site reminiscent to a number of N-terminal copper(II) binding sites including the copper(II)-ATCUN and copper(II)-beta-amyloid complexes. Additionally, we report a more detailed analysis of the thermodynamics associated with copper(II) binding to CA. Although we are still unable to fully deconvolute Cu2+ binding data to the high-affinity CuA site, we have derived pH- and buffer-independent values for the thermodynamics parameters K and ΔH associated with Cu2+ binding to the CuB site of CA to be 2 × 109 and −17.4 kcal/mol, respectively.
Keywords: Carbonic anhydrase, Isothermal Titration Calorimetry, X-ray absorption spectroscopy, Paramagnetic NMR, Copper(II) binding
Carbonic anhydrase (CA) is a zinc-dependent metalloenzyme that catalyzes the reversible hydrolysis of carbon dioxide into the bicarbonate ion.1 The active site of CA contains a mononuclear zinc(II) ion coordinated to three histidine residues (His94, His96, and His119) and one water/hydroxide ligand in a tetrahedral coordination mode. The metal center is kinetically labile, where the metal-free form of CA (apoCA) is relatively stable and easy to generate using pyridine-2,6-dicarboxylic acid (DPA) dialysis.2 This has allowed for the reconstitution of the three-histidine ((His)3) active site in CA with a range of biologically available metal ions.3 However, our recent report (Song et al. J Biol. Inorg. Chem. 2013 18 595) confirms earlier experimental data that shows that Cu2+ binding to CA is more complex than a single site binding event.3–6 There are two, thermodynamically distinctive copper binding sites in apoCA, which we have termed CuA and CuB. These binding sites have different affinities for Cu2+.6 Curiously, the lower affinity site, CuB, was found to be the native (His)3 metal binding site of CA, whereas the location and coordination geometry of the high affinity CuA site is unknown. There are several crystallographically characterized Cu-substituted CAs,3,7,8 but only 1RZC.pdb has a second Cu ion. However this second Cu2+ site is poorly resolved near the N-terminus of the protein, where two conformers within the structure complicates ligand assignments (Figure 1). Here we report our efforts using NMR and XAS techniques to characterize the Cu2+ binding sites within CA; specifically determining the relative location and coordination mode of the high affinity copper(II) binding site (CuA) in CA. Additionally we report the deconvoluted thermodynamic parameters associated with copper(II) coordination to the traditional metal binding site of CA.
Figure 1.
Copper(II) substituted carbonic anhydrase. (A) Location of the crystallographically characterized Cu2+ binding sites in Cu2CA from 1RZC.pdb. Traditional metal binding site in CA is highlighted in pink, where the novel Cu-binding site is shown in yellow. (B) The novel Cu binding site in CA, proposed to be the CuA center. His4 and His64 in the CuA site are observed to have two different conformers (shown in cyan and colored ball and stick) within 1RZC.pdb.
Materials and Methods
Sample Preparation
CA was overexpressed and purified from the pACA plasmid in E. coli BL21(DE3) cells as previously described.9 ApoCA was prepared by dialyzing native CA against 1 L of 20 mM ACES (N-(2-Acetamido)-2-aminoethanesulfonic acid) containing 50 mM DPA (pH 7.0). Extra DPA was removed with dialysis of apoCA against 2 L of 20 mM Tris (pH 7.0) buffer, and then with chromatography onto a Sephacryl S-200 size-exclusion column. The concentration of apoCA was determined by UV absorption at 280 nm (ε280 = 54,000 M−1cm−1).10 There are three forms of CA prepared for the study; reconstituted CA (ZnCA, adding 1 equivalent Zn2+ to apoCA), copper/zinc CA (Cu/ZnCA, addition of 1 equivalent Cu2+ into ZnCA), and dicopper CA (Cu/CuCA, addition of 2 equivalent Cu2+ into apoCA). For NMR experiments, 15N-labeled CA was obtained by growing E. coli BL21(DE3)[pACA] in M9 minimal media containing 1 g/L 15N-ammonium chloride as the sole nitrogen source. The 15N-labeled CA species were then purified and prepared as described above.
NMR Measurements
All NMR measurements were performed at 298 K on a Bruker 600 MHz Avance III spectrometer with cryoprobe attachment. 15N-labeled samples containing 100 µM CA were prepared in 20 mM phosphate buffer at pH 7.0 (5% v/v D2O). Three samples were prepared: an apoCA sample containing no metal cofactors, a Cu/ZnCA sample with Zn2+ bound in the active site, and a Cu/CuCA sample where Cu2+ occupies both the CuA and CuB sites. 15N-1H TROSY spectra were acquired on all samples, and assignments were found to be in excellent agreement with those determined previously.11 Ambiguous assignments were resolved using a 3D 15N-1H MT-PARE-HMQC NOESY with a 150 ms mixing time.12,13 Addition of paramagnetic Cu2+ ions introduced broadening around the CuA and CuB sites, chemical shifts were largely similar between the three samples, suggesting little to no structural changes in the protein (Figures S1–S3 and Tables S1–S3 found in the supporting information).
Paramagnetic relaxation enhancements (1H Γ2 values) were calculated from the difference in 1H TROSY R2 rates measured in samples with and without Cu2+ . Relaxation rates were measured from a series of interleaved 15N-1H TROSY experiments with an additional T2 relaxation echo introduced during the final readout period as described previously.14 The relaxation times used were 0, 6, 12, 18, 24, and 36 ms. T2 values were determined using the relaxation fitting module from Sparky, and uncertainties were determined from the spectral noise.15 The pH for samples with and without Cu2+ and Zn2+ were controlled to within 0.2 pH units, and the pH of samples were checked before and after measurement.
PRE Data Analysis
To fit the positions of paramagnetic Cu2+ ions in the structure of CA, a rigid-body approach was used. Using this approach, the x, y, and z coordinates of one or two Cu2+ ions could be optimized in the molecular frame. Proton Γ2 values were interpreted using a previously-determined high resolution crystal structure of CA (PDB ID 2CBA).16 Amide protons were added using REDUCE.17 Optimization coordinates were obtained by maximizing the agreement (minimizing χ2) between the observed and calculated Γ2 PRE values. The Solomon-Bloembergen equation was used to calculate Γ2, assuming each Cu2+ site influenced relaxation independently:18,19
| (1) |
| (2) |
Where rIS is the distance between the Cu2+ center and the amide proton nucleus, τc is the correlation time for the electron-nuclear interaction vector, and ω is the nuclei Larmor frequency in rad s−1 (i.e. 3.8 × 107 s−1 at 600 MHz). The constant K is calculated from the permeability of vacuum, μ0,the gyromagnetic ratio of the 1H nucleus, γ I, the electron spin quantum number, s, the electron g-factor, gS, and the electron Bohr magneton, μ B. The value for τc was estimated to be 7 ns from 1H T2 measurements.20 Steric overlap was avoided by applying a soft-sphere repulsive potential from the LINUS simulation package.21 Using these constraints, optimization of the x, y, and z coordinates for both Cu2+ nuclei was carried out using in-house scripts. The final coordinates were reproducible and did not change significantly when the soft-sphere repulsive potential was applied.
XAS Data Collection and Analysis
X-ray absorption spectroscopy data was collected at beamline X3B of the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory. A sagitally focusing Si(111) double crystal monochromator was used for energy selection, with a nickel-coated mirror downstream of the monochromator providing harmonic rejection. Cu-K edge XAS spectra were collected in fluorescence mode over an energy range of 8779–9689 eV, using a 31 element Canberra Ge detector. Samples were maintained at a temperature of 20 K under vacuum using a helium Displex cryostat. For internal energy calibration, a copper foil spectrum was collected concomitantly and its first inflection point was set to 8979 eV.
Two samples (Cu/ZnCA: 1.0 mM in 20 mM ACES with 30% glycerol, and Cu/CuCA: 1.0 mM in 20 mM ACES with 30% glycerol) were transferred to Lucite cuvettes covered with Kapton tape as an X-ray transparent window material and quickly frozen in liquid nitrogen. Three scans were collected and averaged for the Cu/ZnCA sample, and four scans for the Cu/CuCA sample. Samples were monitored for evidence of photoreduction, and only those scans showing no photoreduction were included in the final average. Examination of individual scans, averaging, and extraction of the χ(k) EXAFS (extended X-ray absorption fine structure) was carried out using EXAFSPAK.22 Specifically, a Gaussian function was fitted to the curved pre-edge background (8779 – 8955 eV) and this function was subtracted from the entire spectrum to remove background absorption. A 3-segment spline function having fourth order components was fit to the background subtracted data (9000 – 9639 eV) to extract χ (k). EXAFS analysis was carried out using the opt program of EXAFSPAK. Theoretical scattering paths were built from the coordinates of a model of the active site of human CA extracted from the crystal structure coordinates (PDB ID 2CBA),16 with ab initio phase and amplitude parameters for these paths calculated using FEFF 6.23 Coordination numbers (n) were varied in integer steps, while the pathlengths (r) and Debye-Waller factors (σ2) were allowed to freely float. Coordinated His ligands were simulated using a rigid body approximation,24 in which the pathlengths of single- and multiple-scattering paths were constrained to a constant difference from one another. The scale factor was fixed at 0.9 in all fits. E0, the point at which k = 0 Å−1, was defined as 9020 eV, and the edge shift parameter ΔE0 was allowed to float as a single common value for all shells. Fits to Fourier-filtered first-shell EXAFS data used the goodness-of-fit parameter F', defined as F' = [Σ (χexptl− χcalc)2]2 / (NIDP − NVAR), where NVAR is the number of floated variables in the fit, while NIDP is the number of independent data points and is defined as NIDP = 2ΔkΔr/π. In the latter equation, Δk is the k-range over which the data is fit, while Δr is the back-transformation range employed in fitting Fourier-filtered data. F' provides a useful assessment of the effect of additional shells on improving fit quality.(Riggs-Gelasco, 1995 #326) The bond-valence sum (BVS) was also used to assess appropriateness of a given first-shell fit. The BVS was calculated using Σ(exp[(r0 − r)/0.37]), where r0 is an empirically derived parameter for a given pair of atoms (r0 = 1.751 for Cu2+–N and 1.649 for Cu2+–O) and r is the actual bond length.25 For fits to unfiltered EXAFS data, the goodness of fit F factor was defined as [Σk6(χexptl− χcalc)2/Σk6χexptl2]1/2.
UV-Vis Spectroscopy and Reactivity Studies
Electronic absorption spectra of Cu/ZnCA and Cu/CuCA were recorded on the OLIS modernized HP 8452A spectrophotometer. Each spectra were collected on 200 µM protein in 20 mM HEPES buffer at pH 7.4. Para-nitrophenyl acetate (pNPA) hydrolysis assays were performed as previously described with some modification.9,26 All assays were performed in 50 mM Tris buffer at pH 7.4. Reactivity was monitored by the conversion of 10 µM pNPA to p-nitrophenol (ε404 = 17,300 M−1cm−1) and acetate with 10 µM CA. Para-nitrophenyl acetate (pNPA) hydrolysis assays were also performed with ZnCA in the presence of copper(II). Reactivity was monitored by the conversion of 10 µM pNPA to p-nitrophenol by ZnCA as described above. However prior to substrate addition aliquots of copper(II) were added to the reaction buffer and the solution was allowed to incubate for 30 seconds at 25 °C before the ignition of the reaction. Reaction rates are normalized based zinc(II) content.
Isothermal Titration Calorimetry
ITC experiments were carried out at 25 °C, unless indicated otherwise, on a MicroCal VP-ITC calorimeter. Titrations were run in three buffers with different ionization energies (BES, Bis-Tris, TES), to calculate the number of protons released. A typical experiment consisted of 70 µM apoCA titrated with 1.48 mM Cu(NO3)2 in TES buffer. All solutions were generated with exactly matched buffers and degassed under vacuum prior to running the experiments. Protein concentrations were calculated from known molar absorptivity of both apoCA and native CA.
ITC data were fit with the Origin software package provided by MicroCal and the CHASM software package. CHASM uses nonlinear least-squares fitting to optimize χ2 for an arbitrarily-specified binding model.27 Both software packages produced similar optimal values for the stoichiometry (nITC), change in enthalpy (ΔHITC), and association constant (KITC). Comparison of the goodness of fit with different models was based on the calculated χ2 value. Three or more data sets were collected for each type of titration, and the best-fit values were averaged and reported. Estimated uncertainties were calculated using the standard error of the mean. All ITC data that are presented herein are shown as the baseline-adjusted raw data and the peak integrated, concentration-normalized heat of reaction versus the molar ratio of metal ion to protein. The free energy change for the overall equilibrium of each ITC titration, ΔGITC, was determined from the equilibrium constant obtained for the best fit of the experimental data, KITC.
Results
Paramagnetic NMR spectroscopy
NMR spectroscopy was used to estimate the location of the Cu2+ binding sites within the molecular frame of CA. The spectra of apoCA and ZnCA were obtained to test the suitability of studies involving Cu2+, and assignments were found to be in good agreement with those determined previously (data not shown).11 The similarity of both spectra suggests that structure of these metal-substituted CAs experience only small structural changes when a foreign metal ion is bound at the active site. In Cu/ZnCA, the traditional metal binding site is occupied by Zn2+, allowing us to directly probe the CuA binding site, whereas the Cu/CuCA has both copper ions bound to CA. The paramagnetic Cu2+ ions manifest two physical effects in NMR spectra: First, small changes in chemical shifts are expected, corresponding to pseudocontact shifts (PCS) induced by the aniosotropy in the Cu binding site.18,28 Larger shifts are possible if dramatic structural changes occur with binding. Second, the presence of a paramagnetic electron increases the proton relaxation rate through paramagnetic relaxation enhancement (PRE).19,29 Both of these effects are observed in Cu/ZnCA, and a several residues are broadened beyond detection. Additionally, several chemical shift differences are observed, although these are relatively small (c.f. supporting Information Tables S1–3). Because chemical shift changes could reflect both structural changes as well as PCS contributions, interpreting chemical shift differences can be challenging.29 PRE rates are more straight-forward to interpret, and in this study PREs are used to localize the CuA binding site in CA, assuming a rigid-body conformation of the CA structure.
Good agreement is observed between measured PREs and the optimized coordinates of CuA (Figure 2). PREs were observed for 181 out of 260 residues, and these rates indicate that CuA is located in close proximity to the N-terminus of Cu/ZnCA (Figure 3). Coordinate optimization places the CuA site close to the site originally observed by Håkansson et al.,3 His3 and His4 seem like reasonable candidates to coordinate Cu2+ in this site. While our experiments cannot determine the precise orientation of the histidine side chains, however it seems likely that these residues can re-orient to adopt a reasonable metal binding mode. The crystallographically characterized Cu2CA shows His4 adopting two different conformations within the crystal structure (Figure 1).3
Figure 2.
1H Γ2 PRE rates in Zn/CuCA. A comparison of observed PRE rates for backbone protons (black points) to the predicted rates (red lines) using the optimal Cu2+ coordinates in Zn/CuCA. The coordinates of the optimal Cu center are shown in Fig 3. PREs were measured using the pulse program described by Anthis, et al.14 and coordinate optimization was performed using the previously-determined structure of HCA (PDB ID 2CBA).
Figure 3.
Best-fit coordinates for Cu2+ bound to the CuA site in CA. The optimized position of the paramagnetic copper(II) ion (CuA site) in both Zn/CuCA and Cu2CA is shown as a yellow spheroid. This region lacks secondary structure, and the protein was treated as a rigid body during optimization, and the size and shape of this binding site is representative of the statistical uncertainty in the data. Note the copper atom (orange) described by the previously reported crystal structure of the two copper(II) coordination (1RZC.pdb).
PREs were also measured on Cu/CuCA, with the observation that more residues are broadened beyond detection, and chemical shift differences [presumably from pseudocontact shifts (PCS)] are more significant (Supporting Information). For this construct, only 133 PREs could be measured, and because of this there were fewer constraints for determining the location of the CuA site. Nevertheless, the Cu/CuCA dataset yields a binding site consistent with the Cu/ZnCA data. In this case, the coordinates of the active site CuB binding site were fixed to the crystallographic position while the CuB binding site was optimized. The resulting optimized position was less than 2 Å away from the Cu/ZnCA coordinates, indicating a consistent location for this binding site. Importantly, the binding pocket identified by both datasets is consistent, placing the Cu2+ in close proximity to the His3 and His4 side chains.
X-ray Absorption Spectroscopy
Since the paramagnetic NMR experiments above have localized copper binding sites near the N-terminal domain of CA (CuA) and in the traditional zinc bindings center (CuB) and the structure of the native (His)3 metal CuB binding site is structurally well-characterized, our main objective is to characterize the local structure of the high affinity CuA site. Figure 4A shows the XANES spectrum of Cu/ZnCA, which is relatively featureless aside from a weak shoulder feature at 8984 eV that can be assigned to a 1s → 4p electronic transition typically seen for Cu2+.30 The pre-edge peak at 8978 eV associated with a 1s → 3d transition is nearly undetectable. However, the low-energy absorption tail through the 8983–8985 eV region supports a tetragonal Cu2+ site in the protein31 Further evidence for this notion comes from EXAFS analysis of this sample.
Figure 4.
XANES spectra (A) unfiltered k3χ(k) EXAFS spectra (B), and the Fourier transform (C, k = 2.0 − 12.0 Å−1) for Cu/ZnCA ensuring Cu2+ coordination to the CuA site. Experimental data are represented by dotted lines, while best fits (given in bold in Table S5) are shown as solid black lines.
EXAFS analysis typically provides metal-ligand distances within 0.02 Å, identify donor atoms present (Z ± 1), and an estimate of the coordination number (± 25 %).32 Cu/ZnCA exhibited well-defined k3χ(k) EXAFS modulations to k = 12 Å−1 (Figure 4B), while the Fourier transform shows a single intense peak at r’ = 1.5 Å associated with first-shell ligands and a set of weak outer-shell features distributed over r’ = 2.2−4.0 Å (Figure 4C). Analysis of the Fourier-filtered first-shell data for Cu/ZnCA indicates that the first shell is fit well by 3–4 N/O scatterers at 1.93 Å (Table S4), with a fit to 4 N/O ligands favored on the basis of bond-valence sum calculations. Splitting of this shell into two subshells with 2–3 Cu–N/O scatterers at approximately 2.00 Å and 1–2 Cu–O/N scatterers at ca. 1.90 Å produces a moderate improvement in fit quality. However, the difference in bond lengths was smaller than allowed by the 0.16 Å resolution of the data while the σ2 disorder parameter of the shorter subshell was typically large, indicating that two-shell fits cannot be justified with the available EXAFS data. Similar results were obtained for fits to unfiltered data (Table 1). The EXAFS data for Cu/ZnCA also shows evidence for the presence of rigid imidazoles, based on the double-humped feature present at k ≈ 3−5 Å−1 in the k3-weighted EXAFS as well as the shape of the outer-shell features in the FT from r' = 2.4 to 4.0 Å (Figures 4B, 4C). Multiple-scattering analysis of these features shows that they are best fit by contributions from 2 symmetrically bound His ligands (fit 10, Table 1), with fits to other numbers of His ligands producing poorer quality fits. We also carried out fits in which the His multiple scattering paths are dependent on the length of the Cu-NHis first shell scatterer.33 Once again, the best fit involves two His ligands with an average Cu-NHis bond length of 1.98 Å and two additional N/O atoms at 1.88 Å (fit 13, Table 1). While EXAFS does not provide direct insight into the origin of the two N/O scatterers at ~1.9 Å, they are likely either solvent or protein-derived ligands (vide infra). XAS data was also collected for Cu/CuCA, which is less informative as the XAS spectra are a weighted average of the structurally distinct CuA and CuB sites. Thus, precise metrical parameters for the CuB site cannot be obtained from direct fits to the EXAFS data. Nevertheless, a first-shell EXAFS analysis of Cu/CuCA shows an average Cu environment of 4–5 N/O scatters at 1.97 Å (Table S5), suggesting that the CuB metal-ligand bond lengths are moderately longer than those of the CuA site.
Table 1.
EXAFS Analysis of Unfiltered Data of Cu/Zn- CA.a
| Cu-N/O | Cu⋯C(His) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| fit | n | R | σ2 | n | r | σ2 | F | F-factor | ΔE0 |
| 1 | 3 | 1.92 | 2.9 | 254.5 | 0.550 | −13.9 | |||
| 2 | 4 | 1.92 | 4.8 | 234.4 | 0.539 | −14.1 | |||
| 3 | 5 | 1.92 | 6.4 | 242.9 | 0.548 | −14.9 | |||
| 4 | 6 | 1.92 | 8.0 | 261.4 | 0.568 | −15.6 | |||
| 5 | 7 | 1.92 | 9.4 | 286.7 | 0.595 | −16.4 | |||
| 6 | 4 | 1.92 | 4.6 | 2C | 2.86 | 2.0 | 193.2 | 0.489 | −14.0 |
| 7 | 4 | 1.92 | 4.6 | 4C | 2.86 | 5.3 | 187.0 | 0.481 | −13.9 |
| 8 | 4 | 1.92 | 4.6 | 6C | 2.87 | 8.0 | 189.3 | 0.484 | −13.6 |
| 2.86 | 1.5 | ||||||||
| 9 | 4 | 1.92 | 4.6 | 1His | 4.03 | 0.8 | 155.0 | 0.438 | −14.6 |
| 4.06 | 0.8 | ||||||||
| 2.86 | 5.2 | ||||||||
| 10 | 4 | 1.92 | 4.6 | 2His | 4.04 | 4.9 | 143.2 | 0.420 | −14.0 |
| 4.07 | 4.9 | ||||||||
| 2.87 | 8.2 | ||||||||
| 11 | 4 | 1.92 | 4.6 | 3His | 4.05 | 7.9 | 146.7 | 0.426 | −13.7 |
| 4.08 | 7.9 | ||||||||
| 2.87 | 11 | ||||||||
| 12 | 4 | 1.92 | 4.6 | 4His | 4.05 | 10 | 158.1 | 0.442 | −13.6 |
| 4.08 | 10 | ||||||||
| 13 | 2 | 1.88 | 2.9 | 2His | 1.98 | 6.0 | 129.0 | 0.394 | −12.5 |
| 2.88 | 5.9 | ||||||||
| 4.04 | 5.9 | ||||||||
| 4.07 | 5.9 | ||||||||
| 14 | 1 | 1.88 | 0.6 | 3His | 1.96 | 6.5 | 130.6 | 0.402 | −4.73 |
| 2.89 | 9.0 | ||||||||
| 4.06 | 9.0 | ||||||||
| 4.08 | 9.0 | ||||||||
r is in units of Å; σ2 is in units of 10−3 Å2; ΔE0 is in units of eV. All fits are to unfiltered EXAFS data over k = 2.0− 12.0 Å−1. In fits 9–12, the His imidazoles were treated as a rigid body, and included single scattering paths for Cu–Cα and both three and four body paths involving Cu–Cα–Cβ/Nβ (Cβ and Nβ are the more distant atoms of the imidazole). Fit 13–14 also included a three-body path for Cu–NHis–Cα. The pathlengths for the rigid imidazole were constrained to a constant difference from one another and σ2 was constrained to be identical for paths containing the same set of atoms.
UV-Vis Spectroscopy and Reactivity Studies
The UV/Vis spectra of Cu/ZnCA and Cu/CuCA are indistinguishable from each other and have no features within the visible region. Both Cu/ZnCA and Cu/CuCA show some hydrolytic activity using the classical p-nitrophenylacetate activity assay.1,34,35 Cu/ZnCA shows 72 % of the reactivity of the wild-type ZnCA, whereas the Cu/CuCA has only 28 % reactivity. This retained reactivity, especially in the Cu/ZnCA, suggests that many of the mechanistically important amino-acids are not dramatically affected by Cu2+ coordination to the CuA site. This data seems to limit the likelihood that His64 is involved in the CuA site. Copper(II) is a known inhibitor of CA,36 and the inhibition mechanism is likely due to metal substitution into the active site (CuB) where copper is apparently less efficient at activating water compared to Zn2+. Additionally, we noted no decrease in p-nitrophenylacetate activity of ZnCA with increasing amounts of copper(II) in solution under these conditions (Figure S4.)
Isothermal Titration Calorimetry
We have reported preliminary ITC results of Cu2+ binding to apoCA previously.6 However recently new data has become available that allows a further deconvolution of Cu2+ binding data,37,38 which allow for the determination of specific parameters from the equilibria associated with Cu2+ binding to apoCA. A representative isotherm for the addition of Cu2+ to apoCA in TES, the integrated enthalpy change data, and nonlinear regression fit are shown in Figure 5. The ITC titration of Cu2+ into apoCA yields a complex set of equilibria that can be readily fit to a two-site binding model. Similar experiments were performed in three different buffers yielding the experimentally measured ITC data (KITC, ΔHITC) shown in Table 2. The ΔGITC and –TΔSITC terms can be easily derived from the following equations respectively, ΔGITC = −RTlnKITC and ΔGITC = ΔHITC − TΔSITC. A high affinity CuA binding site coordinates a stoichiometric amount of Cu2+ with an equilibrium constant (KITC) of approximately 1.8 × 108. This value is nearly 1000 times higher than the KITC for the low-affinity CuB site (1.4 × 105). The two sites elucidated via Cu(NO3)2 titration into ZnCA were described previously.5 It should be noted that the KITC is the product of the Ka for metal ion binding to apoCA and the Kd of the metal-buffer complex. The KITC values measured for the two Cu2+ binding sites can be compared but require more analysis to achieve a pH and buffer independent binding constant.
Figure 5.
ITC data of copper binding to apoCA. (A) Raw data from the titration of a 1.5 mL cell containing 70 µM apoCA was titrated with 80 × 3 µL of 1.48 mM Cu(NO3)2 in 100 mM TES at pH 7.4. (B) integrated isotherm and the best associated fit for a two-site binding model. The average thermodynamic parameters associated with Cu2+ binding to apoCA are reported in Table 1.
Table 2.
Experimental values for Cu(NO3)2 Binding to apoCA from ITC experiments in 100 mM Buffer at pH 7.4
| Site | Buffer | nITC | KITC | ΔHITC (kcal/mol) |
ΔGITC (kcal/mol) |
−TΔSITC (kcal/mol) |
|---|---|---|---|---|---|---|
| A | BES | 0.9 | 2.8 (±2.0)×108 | −4.2 ± 0.1 | −11.4 ± 0.5 | −7.2 ± 0.5 |
| Bis-Tris | 1.0 | 7.2 (±2.8)×107 | −4.1 ± 0.1 | −10.7 ± 0.2 | −6.6 ± 0.2 | |
| TES | 1.0 | 1.9 (±0.5)×108 | −8.1 ± 0.1 | −11.3 ± 0.1 | −3.2 ± 0.1 | |
| B | BES | 1.1 | 2.4 (±0.8)×105 | −5.7 ± 0.1 | −7.3 ± 0.2 | −1.6 ± 0.2 |
| Bis-Tris | 1.0 | 1.7 (±0.3)×104 | −4.8 ± 0.2 | −5.8 ± 0.1 | −1.0 ± 0.2 | |
| TES | 1.0 | 1.6 (±0.3)×105 | −8.0 ± 0.1 | −7.1 ± 0.1 | 0.9 ± 0.1 | |
Similar to the KITC discussed above, the change in enthalpy measured from the ITC experiments (ΔHITC) is the sum of reaction enthalpies linked to the metal binding process. Upon metal binding, pKa values from local ionizable groups will change. Typically this is observed as a loss of proton density to the solvent, where the buffer ionization also generates added heat based ionization energy (ΔHH-Buffer). This notion is evident from the linear dependence of the addition of observed enthalpy from ITC and metal–buffer interaction enthalpy (ΔHITC + ΔHCu-Buffer) versus the change in enthalpy of ionization for each buffer (ΔHH-Buffer) as shown in Figure 6. This linear relationship can be fit by equation 3, where ΔHH–Buffer, ΔHCu–Buffer, ΔHH–CA, and ΔHCuCA are heats associated with proton and metal binding to the buffer and apoCA with nP as the number of protons released.9
| (3) |
By performing our reactions in 100 mM buffer fixed at pH 7.4, a single speciation of buffer-copper complex can be assumed. The high concentration of buffer allows for only a slight reduction in the buffering capacity from copper-buffer complex formation while still maintaining a set pH. Studying the metal binding process in multiple buffers and plotting the metal-buffer and observed enthalpy vs. the ionization enthalpy, we can estimate the proton release of the reaction, as described above. As a result, the slope of this curve suggests ~1.8 protons are displaced by Cu2+ binding from the high affinity CuA site, whereas 0.8 protons are released from the CuB site, shown in Figure 6.
Figure 6.
Plot of the addition of observed enthalpy from ITC and metal-buffer interaction enthalpy (ΔHITC + ∆HCu-Buffer) versus buffer ionization enthalpies (∆HH-Buffer) in various buffers at pH 7.4. Values for ΔHITC, ∆HCu-Buffer and ∆HH-Buffer in various buffers are listed in Table 3.3 and Table 3.4. Symbols shown as diamonds (CuA) and squares (CuB), and the linear regression values are y = 1.76 × − 0.68, R2 = 0.99, and y = 0.82 × − 6.46, R2 = 0.99, respectively.
The ITC experiment measures the total heat upon addition of titrant, however, there are competitive binding equilibriums associated with metal binding events. In this case, two competing equilibria are involved, buffer competing with protein for access to the metal as well as protons competing with metal for the protein.37 By utilizing these well understood processes the buffer- and pH-independent thermodynamic values of copper binding to apoCA could be derived from a thermodynamic cycle in a similar strategy to those previously reported.39 The overall equilibrium measured in the ITC is a summation of the dissociation of the Cu2+-buffer complex and the association of Cu2+ to the apoCA, along with the dissociation of protons from the CA and the association of protons to the bulk buffer. Although the equilibrium constants associated with a number of Cu-buffer species have been previously reported,40–42 other thermodynamic parameters where characterized using EDTA displacement experiments (Figure S5 and Table S6). To fully understand the thermodynamics associated with copper binding, the coordination environment of the copper ions needs to be structurally characterized. Without specific information regarding the coordination of the metal ion, as is the case for the CuA site, we cannot complete a full analysis of the thermodynamic parameters with the binding event.
At the traditional (His)3 CuB site, the pKa values associated with the copper-bound histidine ligands (His94, His96, and His119) are less than 6 in apoCA, thus it seems unlikely for deprotonation to occur from the side-chain residues.9 The other reasonable option would be that the protons could be released from copper bound water molecules.6,43 The pKa of copper-bound water is unknown, but we can back calculate the value from experimental data. Given there are two labile coordination sites in the CuB site that are likely filled by water,3 and 0.8 protons released are released during our experiments at pH 7.4, we can establish an average pKa of approximately 8.0 for the copper bound waters. The protons released from CA as metal ion binds will be released into the bulk solvent and interact with buffer, and the thermodynamic terms associated with the ionization of various buffers are well known.40,41,44 Thus, a release of 0.8 protons from CA is shown to change the ionization states of the buffer. Using the data collected from the complex equilibrium and the three previously described simpler equilibria, we can estimate the thermodynamic parameters associated with Cu2+ binding to apoCA with the disentanglement of the linked equilibria associated with this system.
In the traditional (His)3 metal binding site (CuB) binds Cu2+ with a higher affinity than Zn2+, which follows the Irving Williams series.9 The average condition-independent change in enthalpy (ΔH) of this binding event is calculated to be −17.4 kcal/mol, and the related change in entropy is estimated to be −13.1 cal/mol/K (where −TΔS = 3.9 kcal/mol). The Cu2+ binding event to the CuB site is highly enthalpy driven. However this process suffers a slight entropic penalty, which is likely due to increasing the order of the active site which is likely derived from the ordering of water molecules and formation of hydrogen bonding networks between second sphere residues and coordination residues, and/or the coordination of the three His residues (His94, His96, and His119). This analysis generates significantly different thermodynamic parameters than were previously reported for Cu2+ binding to apoCA,43 where the metal buffer interactions were not considered as part of their analysis.
Discussion
Carbonic anhydrase is an excellent model protein for biophysics, bioanalysis, the physical-organic chemistry of inhibitor design, and medicinal chemistry.1 This designation holds true when considering the thermodynamic data discussed here for the classic three-His metal binding site and the active site of CA (described here as the CuB site). Our analysis yields terms consistent with the strong association constant observed for Cu2+ binding to the CuB site of apoCA (K ~ 2 × 109), where the corresponding condition-independent thermodynamic terms ΔG°, ΔH, and −TΔS terms are −13.5, −17.4, and +3.9 kcal/mol, respectively. These values compare well with the thermodynamic terms associated with the enthalpy driven Zn2+ binding to apoCA reported earlier.9 The Cu2+ and Zn2+ binding data appear to follow the Irving-Williams series,45 a classical observation on the stability of transition metal complexes. In this specific example, however, there are dramatic differences in the metal coordination geometries of Cu2+ and Zn2+ in the active site of CA. Cu2+ favors a trigonal pyramidal geometry in this system, whereas Zn2+ binds in a tetrahedral geometry.
Combining the data reported above with our previously published data we can draw two conclusions regarding the CuA site in copper-substituted CA. (A) The paramagnetic NMR data shown above locates the CuA site in both Cu/ZnCA and Cu2CA in the unstructured N-terminal region of the protein. (B) The XAS data on the CuA site is fully consistent with a 4-coordinate tetragonal Cu2+ site, with two Histidine ligands. The CuA site appears to be a member of a growing number of proteins and peptides that have been found to have an N-terminal Cu2+ binding site. The previously reported EPR spectra for the CuA site and binding isotherms are strikingly comparable to those observed in proteins with N-terminal copper binding sites, such as the ATCUN (amino terminal Cu2+ and Ni2+) binding motif found in serum albumin and in a range of other small bioactive peptides,6,46–50 and the Cu-binding site found in some amyloid β-peptide complexes (A-β),51,52 as shown in Figure 7.
Figure 7.
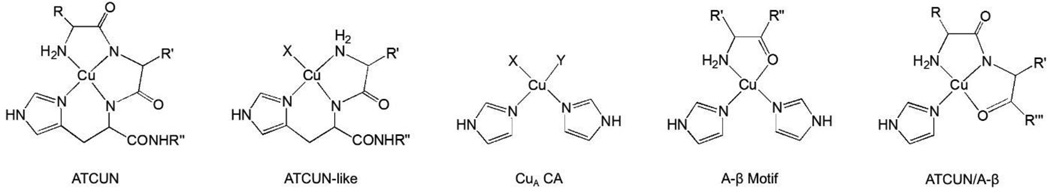
A comparison of the four possible structures of the CuA site. The ATCUN having an N-terminal binding, two backbone amides, and histidine coordinated with the Cu ion. The ATCUN-like have an N-terminal binding, one backbone amide, a histidine, and another N/O donor ligand. The expected structure of the CuA site. The A-β structure with an N-terminal binding, carboxyl oxygen coordination, and two histidines. The final possibility is the ATCUN/A-β with an N-terminal binding, backbone amide, carboxyl oxygen, and histidine coordination to the copper ion.
The ATCUN motif is traditionally thought to coordinate copper through an imino nitrogen of His side chain, two deprotonated backbone amides, and the N-terminal amine. The sequence requirement for this binding mode is an XXH sequence at the N-termini, which is highly consistent with the N-terminal domain of the recombinant CA. The ATCUN motif has a distinctive visible absorption spectrum and has been shown to release three protons as metal coordinates (where the N-termini and two back-bone amides are deprotonated.)44 This is inconsistent with our results reported here, where the CuA site has no distinctive visible spectra and we only see ~ 2 H+ released as Cu2+ binds to the CuA site in CA. Additionally, we have attempted a range of Ni2+ titrations with apoCA, which consistently show only one coordination event in apoCA (data not shown). This is consistent with the ITC data and structural characterization of the NiCA complex that has Ni2+ coordinated to the traditional (His)3 active site of CA.3,43 From these data, we hypothesize that the CuA site is not a traditional ATCUN binding domain, although some structural features of the two may still be similar.
Interestingly there is also an ATCUN-like binding mode, which was previously described for the short peptide GHK.44 Here the same imino nitrogen of a His side chain is coordinated, along with the N-terminal amine, but only one amide nitrogen is coordinated from the first three amino acids. The final coordination site is an unknown ligand, predicted to be an oxygen atom (water/buffer).53 There has been ITC analysis of Cu2+ binding to both the GHK and DAHK (ATCUN-like motif and ATCUN motif respectively), unfortunately however, the full thermodynamic analysis was never carried out for these two motifs. Upon coordination of copper(II) to the GHK peptide, two protons are released, and the proton release to the ATCUN motif was three protons. Our ITC analysis of copper(II) coordination to the CuA site shows a similar proton release to the ATCUN-like motif (Figure 7).
While the CuA site may indeed be the ATCUN-like binding motif described, there is yet another reasonable possibility for the coordination site. The β-amyloid (A-β) coordination to copper(II) shows many similarities to our CuA site. It has been reported the coordination of Cu2+ to different lengths of the β-amyloid,54 we can see more similarities in the spectra between the CuA site and the β-amyloid (A-β) coordination to copper(II), when compared with the ATCUN-like XAS reported in Hureau et al.53 The similarities between the two spectra include two His residues and two oxygen or nitrogen donors. As can be deduced, the XAS data fits with both the ATCUN-like binding motif and the A-β coordination sphere described previously. From the above spectroscopic studies, it is reasonable to say that the N-terminus region of CA has a coordination environment that is similar to the ATCUN-like and the A-β5–23 or a combination of the two.
The data is consistent with the CuA binding two His side-chain residues and an N-terminal amine. The N-terminal amine of CA is expected to have a pKa ~ 9.7,55 where the N-terminus will lose a proton to coordinate to the Cu2+ center. This is the only proton than can be assumed from the data. One option for the other 0.8 protons is the coordination of one of four His residues in close proximity to the Cu2+ with pKa values near 7.1, giving a similar structure to the A-β56,57 The next possibility is the proton coming from a backbone amide, modeling the ATCUN-like motif or ATCUN/A-β motif, or a different coordinating moiety such as a solvent molecule (water or buffer). Without a reliable structure a full thermodynamic analysis is impossible, and the calculated pH- and buffer- independent thermodynamic parameters for this site cannot be reported at this time.
It is unclear why CA has the CuA site in addition to the more traditional metal binding active site. We have found no reactivity associated with the CuA site, which could suggest that it has no accessible or labile coordination sites,6 and this could imply that the physiological role may be related to just sequestering adventitious Cu2+. Copper(II) has been reported to inhibit ZnCA previously,36,58 although these studies were performed at relatively high concentrations of Cu2+. Perhaps the high affinity CuA binding site and the relatively high cellular expression of CA could work together to limit the levels of adventitious Cu2+ during periods of copper dishomeostasis. This notion is supported by our reactivity studies that suggest there is limited effect of up to 2 equivalents of Cu2+ in solution to the ZnCA catalyzed p-nitrophenylacetate hydrolysis activity. The CuA site seems to effectively defend the native zinc(II)-(His)3 active site of CA as well as other metalloproteins from Cu2+ displacement, and in some cases inactivation. In the end it is not clear what role the CuA site is playing in vivo; however, it is clear that this is a novel high affinity Cu2+ binding domain in CA raises new questions about this old protein.
Supplementary Material
Table 3.
Thermodynamic Cycle for Cu2+ Binding to the CuB site in apoCA in 100 mM TES, pH = 7.4
Table 4.
Summary of pH- and Buffer- Independent Thermodynamic Values for Cu2+ Binding to ApoCA.
| CuB[(His)3-Cu2+] | BES | 4.1 (±0.8)×109 | −17.4 ± 0.1 | −13.1 ± 0.2 | 4.3 ± 0.2 |
| Bis-Tris | 4.9 (±0.3)×1010 | −17.4 ± 0.2 | −14.6 ± 0.1 | 2.8 ± 0.2 | |
| TES | 2.7 (±0.3)×109 | −17.4 ± 0.1 | −12.9 ± 0.1 | 4.5 ± 0.1 | |
| Average | 1.9 × 1010 | −17.4 | −13.5 | 3.9 | |
Acknowledgements
The authors thank Mississippi State University for start-up funding to JPE and NCF. All XAS experiments were carried out at beamline X3B of the NSLS at Brookhaven National Laboratory. Beamline X3B is operated by the Case Center for Synchrotron Biosciences, supported by National Institutes of Health NIBIB Grant P30-EB-009998. NSLS is supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886.
Abbreviations
- ACES
N-(2-Acetamido)-2-aminoethanesulfonic acid
- apoCA
metal free CA
- ATCUN
amino terminal Cu2+ and Ni2+
- A-β
beta amyloid
- BES
N,N-Bis(2-hydroxyethyl)-2-aminoethanesulfonic acid
- Bis-Tris
2,2-Bis(hydroxymethyl)-2,2′,2″-nitrilotriethanol
- CA
human carbonic anhydrase II
- CuA
high affinity Cu2+ binding site, unknown location
- CuB
low affinity Cu2+ binding site, (His)3 center
- Cu/CuCA
CA with two bound Cu2+ ions (CuA and CuB)
- Cu/ZnCA
Zn2+ containing CA with one Cu2+ bound (CuA)
- DPA
pyridine-2,6-dicarboxylic acid
- EXAFS
extended X-ray absorption fine structure
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- (His)3
three histidine, native metal binding site in CA
- HMQC
Heteronuclear Multiple-Quantum Correlation
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- MT-PARE
mixed-time parallel evolution
- NOESY
heteronuclear Overhauser effect spectroscopy
- PCS
pseudocontact shifts
- pNPA
p-nitrophenyl acetate
- pNP
p-nitrophenol
- PRE
Paramagnetic relaxation enhancements
- TES
2-[(2-Hydroxy-1,1-bis(hydroxymethyl)ethyl)amino]ethanesulfonic acid
- TROSY
Transverse relaxation optimized spectroscopy
- XAS
X-ray absorption spectroscopy
- XANES
X-ray absorption near-edge structure
Footnotes
This information is available free of charge via the internet at http://pubs.acs.org.
The authors declare no competing financial interest.
References
- 1.Krishnamurthy VM, Kaufman GK, Urbach AR, Gitlin I, Gudiksen KL, Weibel DB, Whitesides GM. Chem Rev. 2008;108:946–1051. doi: 10.1021/cr050262p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunt JB, Rhee M-J, Storm CB. Anal Biochem. 1977;79:614–617. doi: 10.1016/0003-2697(77)90444-4. [DOI] [PubMed] [Google Scholar]
- 3.Håkansson K, Wehnert A, Liljas A. Acta Crystallogr D Biol Crystallogr. 1994;50:93–100. doi: 10.1107/S0907444993008790. [DOI] [PubMed] [Google Scholar]
- 4.Wischeler JS, Heine A, Klebe G. Curr Chem Biol. 2011;5:1–8. [Google Scholar]
- 5.Hunt JA, Ahmed M, Fierke CA. Biochemistry. 1999;38:9054–9062. doi: 10.1021/bi9900166. [DOI] [PubMed] [Google Scholar]
- 6.Song H, Weitz AC, Hendrich MP, Lewis EA, Emerson JP. J Biol Inorg Chem. 2013;18:595–598. doi: 10.1007/s00775-013-1009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox JD, Hunt JA, Compher KM, Fierke CA, Christianson DW. Biochemistry. 2000;39:13687–13694. doi: 10.1021/bi001649j. [DOI] [PubMed] [Google Scholar]
- 8.Jude KM, Banerjee AL, Haldar MK, Manokaran S, Roy B, Mallik S, Srivastava DK, Christianson DW. J Am Chem Soc. 2006;128:3011–3018. doi: 10.1021/ja057257n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song H, Wilson DL, Farquhar ER, Lewis EA, Emerson JP. Inorg Chem. 2012;51:11098–105. doi: 10.1021/ic301645j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexander RS, Kiefer LL, Fierke CA, Christianson DW. Biochemistry. 1993;32:1510–1518. doi: 10.1021/bi00057a015. [DOI] [PubMed] [Google Scholar]
- 11.Venters RA, Farmer BT, II, Fierke CA, Spicer LD. J Mol Biol. 1996;264:1101–1116. doi: 10.1006/jmbi.1996.0699. [DOI] [PubMed] [Google Scholar]
- 12.Fitzkee NC, Masse JE, Shen Y, Davies DR, Bax A. J Biol Chem. 2010;285:18072–18084. doi: 10.1074/jbc.M110.113407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ying J, Chill JH, Louis JM, Bax A. J Biolmol NMR. 2007;37:195–204. doi: 10.1007/s10858-006-9120-z. [DOI] [PubMed] [Google Scholar]
- 14.Anthis NJ, Doucleff M, Clore GM. J Am Chem Soc. 2011;133:18966–18974. doi: 10.1021/ja2082813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goddard TD, Kneller DG. SPARKY 3. University of California; [Google Scholar]
- 16.Håkansson K, Carlsson M, Svensson LA, Liljas A. J Mol Biol. 1992;227:1192–1204. doi: 10.1016/0022-2836(92)90531-n. [DOI] [PubMed] [Google Scholar]
- 17.Word JM, Lovell SC, Richardson JS, Richardson DC. J Mol Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 18.Bertini I, Luchinat C, Rosato A. Prog Biophys Molec Biol. 1996;66:43–80. doi: 10.1016/s0079-6107(96)00016-8. [DOI] [PubMed] [Google Scholar]
- 19.Solomon I, Bloembergen N. Nuclear Magnetic Interactions in the HF Molecule. 1956 [Google Scholar]
- 20.Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton NJ, Rance M. Protein NMR Spectroscopy: Principles and Practice. Imprint: Academic Press; 2006. [Google Scholar]
- 21.Srinivasan R, Fleming PJ, Rose GR. Methods in Enzymology. 2004;383:48–66. doi: 10.1016/S0076-6879(04)83003-9. [DOI] [PubMed] [Google Scholar]
- 22.George GN, Pickering IJ. "EXAFSPACK," Stanford Synchrotron Radation Laboratory. 2000 [Google Scholar]
- 23.Zabinsky SI, Rehr JJ, Ankudinov A, Albers RC, Eller MJ. Phys Rev B. 1995;52:2995–3009. doi: 10.1103/physrevb.52.2995. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Lee MH, Hausinger RP, Clark PA, Wilcox DE, Scott RA. Inorg Chem. 1994;33:1589–1593. [Google Scholar]
- 25.Liu W, Thorpe HH. Inorg Chem. 1993;32:4102–4105. [Google Scholar]
- 26.Okrasa K, Kazlauskas RJ. Chem Eur J. 2006;12:1587–1596. doi: 10.1002/chem.200501413. [DOI] [PubMed] [Google Scholar]
- 27.Le VH, Buscaglia R, Chaires JB, Lewis EA. Anal Biochem. 2013;434:233–241. doi: 10.1016/j.ab.2012.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertini I, Fragai M, Lee Y-M, Luchinat C, Terni B. Angew Chem Int Ed. 2004;43:2254–2256. doi: 10.1002/anie.200353453. [DOI] [PubMed] [Google Scholar]
- 29.Arnesano F, Banci L, Bertini I, Felli IC, Luchinat C, Thompsett AR. J Am Chem Soc. 2003;125:7200–7208. doi: 10.1021/ja034112c. [DOI] [PubMed] [Google Scholar]
- 30.Smith SR, Bencze KZ, Wasiukanis K, Stemmler TL, Benore-Parsons M. Open Inorg Chem. 2008;2:22–24. doi: 10.2174/1874098700802010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kau LS, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO, Solomon EI. J Am Chem Soc. 1987;109:6433–6442. [Google Scholar]
- 32.Leitch S, Bradley MJ, Rowe JL, Chivers PT, Maroney MJ. J Am Chem Soc. 2007;129:5085–5095. doi: 10.1021/ja068505y. [DOI] [PubMed] [Google Scholar]
- 33.Periyannan GR, Costello AL, Tierney DL, Yang K-W, Bennett B, Crowder MW. Biochemistry. 2005;45:1313–1320. doi: 10.1021/bi051105n. [DOI] [PubMed] [Google Scholar]
- 34.Vallee BL, Auld DS. Proc Natl Acad Sci USA. 1990;87:220–224. doi: 10.1073/pnas.87.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JH, Meriwether BP, Clodfelder P, Cunningham LW. J Biol Chem. 1961;236:136–141. [PubMed] [Google Scholar]
- 36.Tu C, Wynns GC, Silverman DN. J Biol Chem. 1961;256:9468–9470. [PubMed] [Google Scholar]
- 37.Grossoehme NE, Spuches AM, Wilcox DE. J Biol Inorg Chem. 2010;15:1183–1191. doi: 10.1007/s00775-010-0693-3. [DOI] [PubMed] [Google Scholar]
- 38.Hong L, Bush WD, Hatcher LQ, Simon J. J Phys Chem B. 2008;112:604–611. doi: 10.1021/jp075747r. [DOI] [PubMed] [Google Scholar]
- 39.Quinn CF. A Thermodynamic Study of Essential and Toxic Metals Binding to Proteins, Peptides and Small Molecules involved in Detoxification, Storage, and Gene Regulation. Dartmouth University; 2009. [Google Scholar]
- 40.NIST Standard Reference Datebase 46. Technology, National Institute of Standards and Technology; Gaithersburg, VA: 2003. [Google Scholar]
- 41.Nagaj J, Stokowa-Sołtys K, Kurowska E, Frączyk T, Jeżowska-Bojczuk M, Bal W. Inorg Chem. 2013;52:13927–13933. doi: 10.1021/ic401451s. [DOI] [PubMed] [Google Scholar]
- 42.Zawisza I, Rózga M, Poznański J, Bal W. J Inorg Biochem. 2013;129:58–61. doi: 10.1016/j.jinorgbio.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 43.DiTusa CA, Christensen T, McCall KA, Fierke CA, Toone EJ. Biochemistry. 2001;40:5338–5344. doi: 10.1021/bi001731e. [DOI] [PubMed] [Google Scholar]
- 44.Tarapaidze A, Hureau C, Bal W, Winterhalter M, Faller P. J Biol Inorg Chem. 2012;17:37–47. doi: 10.1007/s00775-011-0824-5. [DOI] [PubMed] [Google Scholar]
- 45.Irving H, Williams RJP. Order of Stability of Metal Complexes. Nature. 1948;162:746–747. [Google Scholar]
- 46.Zhang Y, Wilcox DE. J Biol Inorg Chem. 2002;7:327–337. doi: 10.1007/s00775-001-0302-6. [DOI] [PubMed] [Google Scholar]
- 47.Sankararamakrishnan R, Verma S, Kumar S. Proteins. 2005;58:211–221. doi: 10.1002/prot.20265. [DOI] [PubMed] [Google Scholar]
- 48.Harford C, Sarkar B. Biochem Biophys Res Commun. 1995;209:877–882. doi: 10.1006/bbrc.1995.1580. [DOI] [PubMed] [Google Scholar]
- 49.Gogineni DP, Spuches AM, Burns CS. Inorg Chem. 2014;54:441–447. doi: 10.1021/ic502014x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sacco C, Skowronsky RA, Gade S, Kenney JM, Spuches AM. J Biol Inorg Chem. 2012;17:531–541. doi: 10.1007/s00775-012-0874-3. [DOI] [PubMed] [Google Scholar]
- 51.Hong L, Carducci TM, Bush WD, Dudzik CG, Millhauser GL, Simon JD. J Phys Chem B. 2010;114:11261–11271. doi: 10.1021/jp103272v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ginotra YP, Ramteke SN, Srikanth R, Kulkarni PP. Inorg Chem. 2012;51:7960–7962. doi: 10.1021/ic301244x. [DOI] [PubMed] [Google Scholar]
- 53.Hureau C, Eury H, Guillot R, Bijani C, Sayen S, Solari P-L, Guillon E, Faller P, Dorlet P. Chem Eur J. 2011;17:10151–10160. doi: 10.1002/chem.201100751. [DOI] [PubMed] [Google Scholar]
- 54.Minicozzi V, Stellato F, Comai M, Serra MD, Potrich C, Meyer-Klaucke W, Morante S. J Biol Chem. 2008;283:10784. doi: 10.1074/jbc.M707109200. [DOI] [PubMed] [Google Scholar]
- 55.King RW, Roberts GCK. Biochemistry. 1971;10:558–565. doi: 10.1021/bi00780a003. [DOI] [PubMed] [Google Scholar]
- 56.Dehareng D, Dive G. Int J Mol Sci. 2004;5:301–332. [Google Scholar]
- 57.Bhattacharya S, Lecomte JTJ. Biophys J. 1997;73:3241–3256. doi: 10.1016/S0006-3495(97)78349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bertini I, Canti G, Luchinat C, Scozzafava A. J Chem Soc Dalton Trans. 1978 [Google Scholar]
- 59.Fukada H, Takahashi K. Proteins. 1998;33:159–166. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








