Abstract
Background As adoption of electronic health records continues to increase, there is an opportunity to incorporate clinical documentation as well as laboratory values and demographics into risk prediction modeling.
Objective The authors develop a risk prediction model for chronic kidney disease (CKD) progression from stage III to stage IV that includes longitudinal data and features drawn from clinical documentation.
Methods The study cohort consisted of 2908 primary-care clinic patients who had at least three visits prior to January 1, 2013 and developed CKD stage III during their documented history. Development and validation cohorts were randomly selected from this cohort and the study datasets included longitudinal inpatient and outpatient data from these populations. Time series analysis (Kalman filter) and survival analysis (Cox proportional hazards) were combined to produce a range of risk models. These models were evaluated using concordance, a discriminatory statistic.
Results A risk model incorporating longitudinal data on clinical documentation and laboratory test results (concordance 0.849) predicts progression from state III CKD to stage IV CKD more accurately when compared to a similar model without laboratory test results (concordance 0.733, P<.001), a model that only considers the most recent laboratory test results (concordance 0.819, P < .031) and a model based on estimated glomerular filtration rate (concordance 0.779, P < .001).
Conclusions A risk prediction model that takes longitudinal laboratory test results and clinical documentation into consideration can predict CKD progression from stage III to stage IV more accurately than three models that do not take all of these variables into consideration.
Keywords: risk prediction, electronic health records, topic modeling, survival analysis
BACKGROUND AND SIGNIFICANCE
The field of clinical disease risk prediction and progression is well developed, with hundreds of models published across many diseases. Given their history predating electronic health records (EHRs), these models have largely been developed with data easily accessible to clinicians. Likewise, current progression risk models for chronic kidney disease (CKD) largely rely on commonly obtained laboratory or vital sign data.1,2 CKD affects a large portion of the population,3 is associated with significant morbidity and mortality,4 and is a high-risk clinical condition with frequent adverse events.5,6 Despite this, patients with kidney disease frequently go unrecognized, and their care is often suboptimal.7–15 Early identification and more accurate prognostication of these patients using better risk prediction models may improve outcomes by facilitating timelier initiation of appropriate therapies, monitoring, and specialty referral.16
While using readily available data for risk prediction might simplify computation, this might be at the expense of more robust prognostication. EHRs contain much information about patient histories and patient information conveyed both in the discrete elements of the record and in the narratives. With increasing EHR adoption, clinical documentation is a potentially rich, underutilized source of information that can aid in clinical decision support.17 Two aspects of the EHR in particular present an opportunity for automated risk prediction: the presence of longitudinal data, and the rich information conveyed in the clinical narratives. Automated information extraction from narratives using natural language processing (NLP) is an active field of research and has shown promising results in estimating disease risk;18 increasing appropriate cancer screening;19,20 and identifying post-operative complications,21 influenza,22 inflammatory bowel disease,23 pneumonia,24–26 and heart failure.27,28
The goal of the present study was to incorporate longitudinal clinical documentation as a novel feature in disease progression risk calculation. While NLP has been used traditionally to look for the presence of particular pieces of information in the clinical narrative, such as presence of signs of pneumonia,24–26 recent research in NLP and data science have proposed methods that discover patterns from large amounts of data that do not require a specific target. For example, one of the methods used in this study to incorporate information from the narratives into our risk modeling discovers topics discussed in a collection of texts in an unsupervised fashion. Using CKD as a proof of principle, we aimed to develop a risk model that can provide satisfactory prediction of progression from CKD stage III to stage IV using heterogeneous sources of data. This outcome, with a higher incidence and typically far in advance of end-stage renal disease (ESRD), may be more meaningful for general practitioners, providing guidance for therapy and appropriate specialty referral.
In this study, we investigate the following research questions: (i) How can longitudinal data from the patient record be incorporated into risk modeling? (ii) How can EHR data be incorporated into a risk modeling paradigm, focusing on two critical data elements of the EHR, laboratory data and clinical narrative?
The family of models we investigate to represent patient documentation is unsupervised. We cast the problem of risk modeling as a survival analysis task; thus demonstrating that these methods are capable of producing two valuable outcomes: an interpretable set of variables associated with risk of CKD progression at the population level, and an actionable model to estimate risk of progression for individual patients.
METHODS
Study Population
The study cohort was derived from the clinical data warehouse of a single institution, NewYork-Presbyterian (NYP) Hospital. The cohort consisted of patients of an outpatient primary care clinic known as the Associates in Internal Medicine (AIM) clinic. As of January 1, 2013, a total of 21 580 patients were seen at the AIM clinic at least three times with an average record length of 11.71 years (±7.61) and an average of 981.29 laboratory measurements. There are 64% females and 36% males in this population. The earliest recorded observation for this cohort was August 23, 1988.
Patients who developed CKD stage III (defined as estimated glomerular filtration rate (eGFR) consistently <60 ml/min/1.73 m2 for ≥3 months) during the course of their documented clinical record were included in the cohort. The following patients were excluded from the analysis: left censored patients (i.e., whose first documentation in their record shows evidence of CKD stage III), patients who meet definitions for CKD stages III and IV simultaneously, as well as HIV-positive patients and transplant patients. The study cohort was divided randomly into development (90%) and validation (10%) cohorts.
The study was reviewed and approved by the institutional review board at Columbia University Medical Center, and patient informed consent was waived due to the large scale and retrospective nature of the study.
Variables
Independent Variables
Table 1 shows the independent variables included in the five different statistical models that were considered. Demographic variables were age and gender (ethnicity/race variables were omitted because their value in the EHR is left as the default value “Other” for most of the patients). All other variables, as they are time-dependent, were binned by month and the mean value was used if multiple values were observed within a month. To handle missing values, the most recent value was carried forward for each bin prior to stage III onset, and for variables that had no value for a given patient prior to stage III onset, mean values from the development dataset were used instead.
Table 1:
Baseline statistics and independent variables for the five studied models
| Independent Variables | Development cohort (n=2617) | Validation cohort (n=291) | eGFR model | LKF model | TKF model | LTKF model | RLT model |
|---|---|---|---|---|---|---|---|
| Age | 66.95 ± 11.43 | 67.17 ± 11.69 | X | X | X | X | X |
| Gender (M/F) | 912 (35%)/1697 (65%) | 107 (36%)/192 (64%) | X | X | X | X | X |
| eGFR* | 50.34 ± 8.47 | 50.48 ± 7.60 | X | ||||
| Laboratory Test-based factors and biases (24 variables) | X | X | |||||
| Text-based factors and biases (60 variables) | X | X | |||||
| 25OH Vitamin D | 19.18 ± 7.25 | 16.83 ± 5.11 | X | ||||
| Bicarbonate | 25.20 ± 3.06 | 25.23 ± 3.22 | X | ||||
| BUN | 21.45 ± 8.06 | 21.04 ± 7.75 | X | ||||
| Calcium | 9.39 ± 0.42 | 9.37 ± 0.40 | X | ||||
| Chloride | 102.83 ± 3.50 | 102.64 ± 3.25 | X | ||||
| Creatinine | 1.15 ± 0.34 | 1.10 ± 0.36 | X | ||||
| C-reactive protein | 7.70 ± 8.36 | 6.13 ± 9.86 | X | ||||
| Hematocrit | 37.90 ± 4.80 | 37.68 ± 4.67 | X | ||||
| Hemoglobin | 12.35 ± 1.84 | 12.06 ± 1.66 | X | ||||
| (K) Potassium | 4.29 ± 0.45 | 4.27 ± 0.41 | X | ||||
| Magnesium | 1.85 ± 0.25 | 1.86 ± 0.25 | X | ||||
| (Na) Sodium | 138.97 ± 2.81 | 138.81 ± 2.71 | X | ||||
| Phosphate | 3.41 ± 0.67 | 3.39 ± 0.63 | X | ||||
| Protein | 7.29 ± 0.66 | 7.43 ± 0.69 | X | ||||
| Parathyroid Hormone | 140.69 ± 83.64 | 141.98 ± 82.44 | X | ||||
| Triglyceride | 147.85 ± 72.60 | 154.47 ± 78.15 | X | ||||
| Urine Protein/creatinine | 32.51 ± 30.56 | 31.21 ± 29.58 | X | ||||
| Urine protein qualitative | 2.12 ± 1.02 | 2.12 ± 0.93 | X | ||||
| Uric Acid | 6.35 ± 2.02 | 6.14 ± 1.98 | X |
eGFR = estimated glomerular filtration rate; LKF = Laboratory Test Kalman Filter; TKF = Text Kalman Filter; LTKF = Laboratory Test and Text Kalman Filter; RLT = recent laboratory tests.
*eGFR was calculated using the CKD-EPI equation.29
Clinical documents included in the study consisted of discharge summaries, and outpatient primary care and outpatient specialty notes. The free text notes were preprocessed using the probabilistic topic modeling technique called latent Dirichlet allocation (LDA)30,31. LDA is a probabilistic mixed membership model often applied to text analysis. LDA models a corpus as a set of documents, each of which is represented by a probability distribution over a set of K topics. In this analysis, K = 50. Each topic, in turn, is represented by a probability distribution over all the terms in the vocabulary. In the generative model for LDA, each document is generated by drawing K topics according to the weighting associated with the document and drawing N terms, according to the weighting over terms associated with each topic. Given this model of documents and corpora, inference amounts to identifying the weightings over terms associated with each of the K topics, and the weightings over topics associated with each document given the observed documents. A Gibbs sampling approach was employed to identify the appropriate parameters. More detailed discussion of this model and methods for inference can be found in the literature.30,31
Example topics can be seen in Tables 2 and 3, where they are represented by their most highly associated words. As LDA is an unsupervised method, these topics were automatically discovered by the model and did not require any manual guidance. A given patient’s notes within each time bin were thus represented as a distribution over 50 discovered topics.
Table 2:
Topics associated with increased risk of progression. (topic titles shown in parentheses were assigned manually once the topics were generated, and are presented as a way to label the topics)
| Topic 3 (heart failure) | Topic 32 (diabetes) | Topic 29 (dialysis) |
|---|---|---|
| Lasix | Units | q15 |
| Volume | Insulin | Dialysis |
| Edema | Subcutaneous | Fistula |
| Heart | Lantus | Volume |
| Failure | Glucose | Bid |
| Worsening | Diabetes | Lasix |
| Diuresis | Times | Placement |
| Severe | 70/30 | Improved |
| Diastolic | Diabetic | Heparin |
| Overload | Days | Examined |
Laboratory variables were chosen for their relatedness to CKD and associated comorbidities by a board certified nephrologist (see Table 1).
Dependent Variable
The outcome of interest was defined as progression to CKD stage IV (eGFR consistently <30 ml/min/173 m2 for ≥3 months).
Table 3:
Topics associated with decreased risk of progression. (Topic titles shown in parentheses were assigned manually once the topics were generated, and are presented as a way to label the topics)
| Topic 33 (family history) | Topic 35 (health maintenance) | Topic 41 (non-specific) | Topic 43 (gynecological) | Topic 45 (asthma) |
|---|---|---|---|---|
| Died | Died | History | Breast | Albuterol |
| Age | Flu | Pressure | Vaginal | Asthma |
| Years | Visit | Rate | Mammo | Inhaled |
| Mother | Fasting | Count | Cancer | Lung |
| Father | Colonoscopy | Three | hx | Obstructive |
| Brother | Year | Revealed | pap | Wheezing |
| Sister | Shot | Times | nl | Advair |
| Worked | Vaccine | Shortness | Age | Pulm |
| Children | wnl | Discharged | Will | Restrictive |
| Deceased | Check | Creatinine | Endometrial | Puffs |
Statistical Analysis
Survival analysis
The following five multivariate Cox proportional hazards models were fit on the development dataset: eGFR and Recent Laboratory Tests (RLT) models and three time series models (Table 1). All models included demographic variables and all other values, as shown in Table 1, at the time of stage III onset. All variables were standardized to have zero mean and unit variance.
eGFR Model
This model included eGFR value, age, and gender data at stage III onset as independent variables.
RLT Model
This model considered as independent variables the values for the 19 variables included in the study prior to stage III onset.
Time series-based models
In the eGFR and RLT model, a patient’s data immediately prior to CKD stage III onset is considered, but all other previous values are ignored. Also, laboratory test results can be very variable depending on the time of day, recency of meals, or other factors. Furthermore, clinical documentation can vary significantly from note to note depending on the author, specialty, and setting. To address this variability and simultaneously make risk predictions that incorporate longitudinal patient data, we combine time series analysis and survival analysis to construct these risk prediction models.
The time series model used in these experiments is a variant of the well-known Kalman filter (otherwise known as linear dynamical systems), a time series model for noisy temporal observations which is designed to infer a set of smooth latent (unobserved) states from which the noisy observations are based.32,33 In our case, the noisy observations include the laboratory test results and the clinical documentation. The model is employed such that the latent state inferred at the time of CKD three onset provides a representation of a patient at that time, rather than the observed independent variables.
The specification of the model is as follows: Let represents the initial latent state for patient , let be the latent state for patient at time , and let be the observation for patient at time . Given these definitions, the full model specification is:
where is the identity matrix, represents a multivariate Gaussian distribution with mean zero and variance , is a weight matrix representing the linear relationship between the latent state and the observations, represents the observation bias terms for patient , and , , and are variance parameters. After learning the model parameters, the latent state, , at the time of CKD stage III and the per-patient bias terms, , will be used as the final output of these models. Parameter learning in these models was achieved through an approximation technique known as mean field variational inference.34,35
For each of the three time series models considered, a Kalman filter was fit using the development cohort data and applied to the validation data using the previously determined parameters. When applied to the validation data, only observations prior to the onset of stage III CKD are used to estimate the state of the Kalman filters. The first time series model is called the Laboratory Test Kalman Filter (LKF) and included the 19 laboratory test variables as input, the second time series model is called the Text Kalman Filter (TKF) and included the patient notes, as represented by their topic distributions, and the third time series model is called the Laboratory Test and Text Kalman Filter (LTKF) and combines the output of the LKF and TKF models (Table 1). The output of these models are used as independent variables in the subsequent survival analysis and include, for each patient, their inferred state at CKD stage III onset and biases for each of the input variables that represent long term changes.
Prediction model validation and Experimental Setup
The coefficients and the baseline hazard function for each of the Cox proportional hazards models36 were held constant and applied to the validation dataset. Concordance (C statistic), a measure of discriminatory power equivalent to the area under the receiver operating characteristic curve, was used to evaluate model performance on the validation dataset.37 Pairwise comparisons between the models were tested using the corrected resampled t-test38 and P-values were adjusted for multiple comparisons with the Holm-Bonferroni correction.39
All time series models were developed using the Python programming language version 2.6.5 (Python Software Foundation, Delaware, United States) and statistical analyses were performed using R version 3.0.2 (R Foundation for Statistical Computing, Vienna, Austria). P-values < .05 were considered statistically significant.
RESULTS
Cohort Description
The development set included 2617 patients and the validation set included 291 patients. There were 307 stage IV events in the development set and 35 events in the validation set. The average age was 67 and there were more female (65%) than male (35%) patients in both cohorts. (Table 1).
eGFR model
As shown in Figure 1, low eGFR at onset of stage III CKD was associated with high risk of progression (P < .001). Younger age was associated with increased risk of progression (P < .005). Male gender (the variable for gender was encoded as 1 for male and 0 for female gender) was also associated with increased risk of progression (P = .005).
Figure 1:
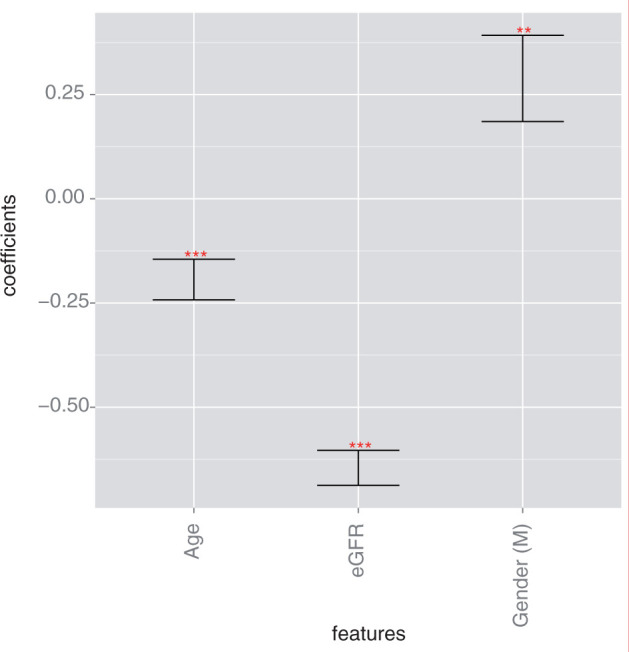
Log hazard ratios for the eGFR Model. Progression of CKD from stage III to stage IV in this model was associated with low eGFR (P < .001), male gender (P = .0051), and younger age (P < .001). (* indicates P < .05, ** indicates P < .01, *** indicates P < .001).
RLT Risk model
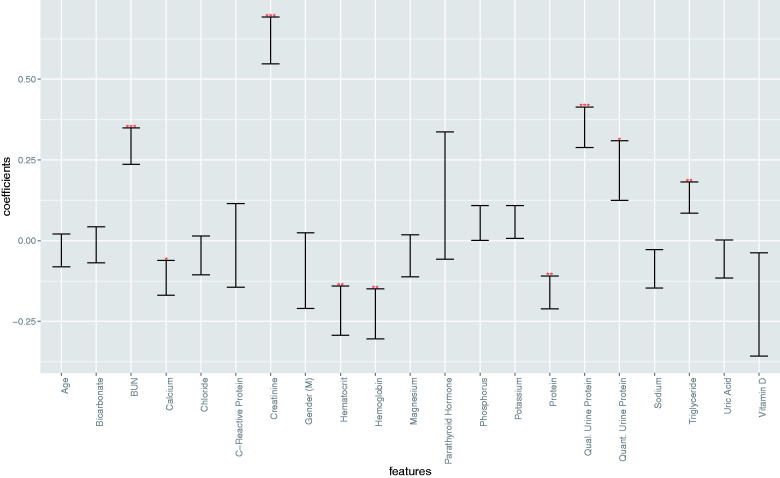
In the RLT model, elevated levels of BUN (P < .001), creatinine (P < .001), triglycerides (P = .0061), and urine protein (quantitative, P = .019; qualitative, P < .001), as well as decreased levels of hematocrit (P = .0043), hemoglobin (P = .0034), calcium (P = .033), and serum protein (P = .0017) were associated with increased risk of CKD progression (Figure 2). When introducing laboratory variables at the time of onset in the survival model, age and gender are not associated with risk of progression anymore.
Figure 2.
Log hazard ratios for the Recent Laboratory Tests Model. Progression of CKD from stage III to stage IV in this model was associated with elevated levels of BUN (P < .001), creatinine (P < .001), triglycerides (P = .0061), and urine protein (quantitative, P = .019; qualitative, P < .001) as well as decreased levels of hematocrit (P = .0043), hemoglobin (P = .0034), calcium (P = .033), and serum protein (P = .0017). (* indicates P < .05, ** indicates P < .01, *** indicates P < .001).
Kalman Filter Risk models
In the TKF Risk model, topics associated with a higher risk of progression are shown in Table 2 and contained terms related to heart failure (P < .001), diabetes (P < .001), and dialysis (P = .028). Mention of these topic words throughout the course of the patient records indicated high risk for progression. Younger age was also found to be associated with increased risk in this model (P < .001). Topics associated with lower risk are shown in Table 3 and included terms having to do with family history (P = .031), health maintenance, (P = .045), gynecological care (P = .038), and asthma (P = .0045). Terms associated with all 50 topics can be found in the online supplement.
In the LKF Model, long-standing lower values for sodium (P = .047) and hematocrit (P = .039) and long-standing elevated values for BUN (P < .001), creatinine (P < .001), and urine protein (quantitative, P < .001; qualitative, P < .001) were associated with higher risk of progression (see supplement; Supplementary Data).
In the LTKF model, which considered both the clinical notes and the laboratory values, lower values of bicarbonate (P = .021), elevated levels of BUN (P = .0041), creatinine (P = .0013), and urine protein (quantitative, P < .001; qualitative, P < .001), and the presence of terms associated with heart failure (P = .0017) and diabetes (P = .0042) were associated with higher risk of progression (eFigure2). As in the TKF Model, the presence of terms associated with asthma (P = .022) was associated with a lower risk of progression. In the LKF model, the learned Kalman-filter states were also predictive of progression (details can be found in the online supplement)
The learned Kalman-filter states also had statistically significant coefficients for predicting progression in all 3 models (details can be found in the online supplement).
Model Performances in the Validation Cohort
Table 4 compares the performances of the 5 models on the validation set. The concordance was highest (i.e., higher predictive ability) for the LTKF model (0.849), followed by the LKF model (0.836), RLT model (0.819), eGFR model (0.779) and TKF model (0.733). The LTKF model performs significantly better than the RLT (P = .031), TKF (P < .001), and eGFR (P < .001) models, the LKF model performs significantly better than the TKF (P < .001) and eGFR (P < .001) models, the RLT model performs significantly better than the TKF (P < .001) and eGFR (P = .007) models.
Table 4.
Concordances and concordance comparisons for the 5 studied models: Laboratory Test and Text Kalman Filter (LTKF), Laboratory Test Kalman Filter (LKF), Text Kalman Filter (TKF), Recent Laboratory Tests (RLT), and estimated Glomerular Filtration Rate (eGFR)
| Δ LTKF | Δ LKF | Δ TKF | Δ RLT | Δ eGFR | Concordance | |
|---|---|---|---|---|---|---|
| LTKF | *** | * | *** | 0.849 | ||
| LKF | *** | ** | 0.836 | |||
| TKF | *** | 0.733 | ||||
| RLT | ** | 0.819 | ||||
| eGFR | 0.779 |
*P < .05, **P < .01, ***P < .001.
DISCUSSION
We have developed and performed an internal validation for five models for CKD progression from stage III to stage IV. Our models leverage different types of variables—demographic, laboratory and/or clinical documentation data that are collected routinely during the course of clinical care as part of the EHR—as well as the longitudinal aspect of the records as encoded through Kalman filters.
In absence of laboratory and documentation information, the simplest model (eGFR model) identifies that low eGFR at time of CKD stage III diagnosis is associated with higher risk of progression. Furthermore, younger patients with impaired kidney function (stage III CKD) progress more rapidly toward stage IV CKD. Consistent with current knowledge of CKD, male gender was found to be associated with more rapid loss of eGFR, and the laboratory test models (RLT) identified laboratory data known to be associated with CKD progression.1,40
We found that text is a valuable predictor for CKD progression and that the use of time series models to characterize patient state can substantially improve predictive accuracy for progression. In particular, the LTKF model which incorporated demographic, laboratory, and clinical documentation data had the highest concordance of the models considered.
With a concordance of 0.733, the TKF model predicts nearly as well as the eGFR model where the difference did not reach statistical significance. Variables of this model with significant coefficients included well-known risk factors and complications of CKD including diabetes, heart failure, and dialysis. Conversely, variables associated with a decreased risk of progression include topics indicative of documentation of health care maintenance and outpatient care. Although the difference did not meet the significance criterion, the LKF model performs better than the RLT model indicating that incorporating previous data in the form of a Kalman filter model can improve prognostication. The LKF model also performs significantly better than the TKF and eGFR models indicating that laboratory tests are a particularly valuable source of prognostic information for CKD progression. Lastly, the LTKF model had the highest concordance at 0.849 and the significant coefficients and their directionality corresponded to variables known to be related to CKD progression.
Risk prediction in CKD has been studied extensively, with dozens of available risk models with acceptable performance (discrimination 0.56–0.94). Most developed classifiers use readily obtainable information, including age, demographics, and laboratory data. Hence, laboratory data, comorbidities, and occasional vital signs are the sole dimensions of contemporary CKD classifiers. Age, sex, and eGFR are included in almost all models, but fewer than half use proteinuria (qualitative assessment or quantitative proteinuria or albuminuria), serum creatinine, serum albumin, or blood pressure. A minority of models incorporate other features, including comorbidities (e.g., diabetes, hypertension, stroke, peripheral vascular disease, or heart failure), common laboratory tests (e.g., serum calcium, bicarbonate, phosphorus, or cholesterol), or other vital signs (e.g., body mass index or weight). Our work here combines a novel feature —clinical documentation—with more established features used for risk prediction to demonstrate the potential of such an approach.
While ESRD or death as hard outcomes have significant value for prediction, complications of CKD begin early and worsen progressively through the various CKD stages.41–44 From stage III to IV CKD, cardiovascular risk increases,4 anemia and bone-mineral disease worsens,42–47 and myriad other considerations—including appropriate dosing of medications6,48 and potential for renal replacement therapy preparation49 —need to be evaluated. It is well known that the reporting of eGFR with routine laboratory results has led to an overall increase in nephrology referrals,11,50,51 with a concern that much of the increase may be inappropriate.52 Patients with non-progressive CKD stage III can often be safely and appropriately managed in the primary care setting, whereas those apt to progress to CKD stage IV may benefit from earlier referral to nephrologists who can assist with the medical management of CKD complications and considerations.16,53
Although the outcome in this study is largely based on laboratory test results (eGFR) and nephrology, as a field, is very laboratory test oriented, it was beneficial to include features drawn the clinical documentation. Many other fields and diseases are not as focused on laboratory test results, and are based much more on clinical documentation. We, therefore, would expect a benefit in extending a similar analysis to other fields and diseases.
Limitations
Because our dataset consists of a non-curated, real-world set of patient characteristics, as recorded through clinical care, there is some potential noise in the collected variables. For instance, given the lack of high quality information about ethnicity, we cannot assess which ethnic groups are well represented in our dataset. This fact may introduce noise in the eGFR calculations.
The models we designed and validated are based on data from a single institution. While there is value in focusing on a single institution at a time (the risk predictions are relevant to the characteristics of the institution’s patient population for instance), the model validity and its generalizability would be better demonstrated over data from several institutions. In particular, because of the potential variations in clinical vocabulary and overall language in the documentation across different institutions, there would likely be a benefit to generalizing the risk model to patient records from other institutions. Our study requires longitudinal documentation (both inpatient and outpatient notes over many years, for a large set of patient records). Since there are no publicly available datasets (even de-identified) with these properties, extending this study to other datasets is outside the scope of this study and an important limitation of the work. Short of training a model for data from different institutions, the models presented in this study are in theory portable to different institutions. In particular, the unsupervised NLP techniques described here (topic modeling) are actually conducive to such an approach, as they identify patterns in the language of any given corpus without any prior knowledge of the topics or vocabulary to expect. To address the potential differences in language from one institution to another, the topic models would have to be learned on documentation from the new institutions.
CONCLUSIONS
A risk prediction model that takes longitudinal laboratory test results and longitudinal clinical documentation into consideration can statistically significantly predict CKD progression from stage III to stage IV more accurately than three models that do not take all of these variables nor their longitudinal aspect into consideration.
COMPETING INTERESTS
None.
FUNDING
The study was funded by the National Science Foundation (IIS-1344668, IIS-0745520, IIS-1247664, IIS-1009542), The Office of Naval Research (N00014-11-1-0651), The Alfred P. Sloan Foundation, and The Defense Advanced Research Projects Agency (FA8750-14-2-0009). The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
CONTRIBUTORs
A.P., R.R., D.B., and N.E. designed the experiments. A.P. and R.R. implemented and conducted the experiments. A.P., J.S.H., and N.E. designed the cohort selection algorithm. A.P., J.S.H., and N.E. wrote the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank Rimma Pivovarov for the helpful discussions.
Supplementary Material
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1. Echouffo-Tcheugui JB, Kengne AP. Risk models to predict chronic kidney disease and its progression: a systematic review. PLoS Med. 2012;9:e1001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tangri N, Kitsios GD, Inker LA, et al. Risk prediction models for patients with chronic kidney disease: a systematic review. Ann Intern Med. 2013;158:596–603. [DOI] [PubMed] [Google Scholar]
- 3. Coresh J, Selvin E, Stevens LA, et al. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298:2038–2047. [DOI] [PubMed] [Google Scholar]
- 4. Go AS, Chertow GM, Fan D, et al. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N Engl J Med. 2004;351:1296–1305. [DOI] [PubMed] [Google Scholar]
- 5. Chapin E, Zhan M, Hsu VD, et al. Adverse safety events in chronic kidney disease: the frequency of “multiple hits.” Clin J Am Soc Nephrol. 2010;5:95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fink JC, Brown J, Hsu VD, et al. CKD as an underrecognized threat to patient safety. Am J Kidney Dis. 2009;53:681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abdel-Kader K, Fischer GS, Johnston JR, et al. Characterizing pre-dialysis care in the era of eGFR reporting: a cohort study. BMC Nephrol. 2011;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Agrawal V, Ghosh AK, Barnes MA, et al. Awareness and knowledge of clinical practice guidelines for CKD among internal medicine residents: a national online survey. Am J Kidney Dis. 2008;52:1061–1069. [DOI] [PubMed] [Google Scholar]
- 9. Boulware LE, Troll MU, Jaar BG, et al. Identification and referral of patients with progressive CKD: a national study. Am J Kidney Dis. 2006;48:192–204. [DOI] [PubMed] [Google Scholar]
- 10. Hemmelgarn BR, Manns BJ, Straus S, et al. Knowledge translation for nephrologists: strategies for improving the identification of patients with proteinuria. J Nephrol. 2012;25:933–943. [DOI] [PubMed] [Google Scholar]
- 11. Kagoma YK, Weir MA, Iansavichus AV, et al. Impact of estimated GFR reporting on patients, clinicians, and health-care systems: a systematic review. Am J Kidney Dis. 2011;57:592–601. [DOI] [PubMed] [Google Scholar]
- 12. Lenz O, Mekala DP, Patel DV, et al. Barriers to successful care for chronic kidney disease. BMC Nephrol 2005;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsai TT, Patel UD, Chang TI, et al. Contemporary incidence, predictors, and outcomes of acute kidney injury in patients undergoing percutaneous coronary interventions: insights from the NCDR Cath-PCI Registry. JACC Cardiovasc Interv. 2014;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patel TG, Pogach LM, Barth RH. CKD screening and management in the Veterans Health Administration: the impact of system organization and an innovative electronic record. Am J Kidney Dis. 2009;53:S78–S85. [DOI] [PubMed] [Google Scholar]
- 15. Rutkowski M, Mann W, Derose S, et al. Implementing KDOQI CKD definition and staging guidelines in Southern California Kaiser Permanente. Am J Kidney Dis. 2009;53:S86–S99. [DOI] [PubMed] [Google Scholar]
- 16. Black C, Sharma P, Scotland G, et al. Early referral strategies for management of people with markers of renal disease: a systematic review of the evidence of clinical effectiveness, cost-effectiveness and economic analysis. Heal Technol Assess. 2010;14:1–184. [DOI] [PubMed] [Google Scholar]
- 17. Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inf. 2009;42:760–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Callaghan FM, Jackson MT, Demner-Fushman D, et al. NLP-derived information improves the estimates of risk of disease compared to estimates based on manually extracted data alone. In: 5th International Symposium on Semantic Mining in Biomedicine. 2012:18–25. [Google Scholar]
- 19. Wagholikar K, Chaudhry R, Boardman L, et al. Clinical decision support for colonoscopy surveillance using natural language processing. In: 2012 IEEE Second International Conference on Healthcare Informatics, Imaging and Systems Biology. 2012:12–21. [Google Scholar]
- 20. Wagholikar KB, MacLaughlin KL, Henry MR, et al. Clinical decision support with automated text processing for cervical cancer screening. J Am Med Inform Assoc. 2012;19:833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA. 2011;306:848–855. [DOI] [PubMed] [Google Scholar]
- 22. Elkin PL, Froehling DA, Wahner-Roedler DL, et al. Comparison of natural language processing biosurveillance methods for identifying influenza from encounter notes. Ann Intern Med. 2012;156:11–18. [DOI] [PubMed] [Google Scholar]
- 23. Ananthakrishnan AN, Cai T, Savova G, et al. Improving case definition of crohn’s disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm Bowel Dis. 2013;19:1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dublin S, Baldwin E, Walker RL, et al. Natural language processing to identify pneumonia from radiology reports. Pharmacoepidemiol Drug Saf. 2013;22:834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu V, Clark M, Mendoza M, et al. Automated identification of pneumonia in chest radiograph reports in critically ill patients. BMC Med Inform Decis Mak. 2013;13:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hripcsak G, Friedman C, Alderson PO, et al. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med. 1995;122:681–688. [DOI] [PubMed] [Google Scholar]
- 27. Steinhubl SR, Williams BA, Sun J, et al. Text and data mining of longitudinal electronic health records (EHRs) in a primary care population can identify heart failure (HF) patients months to years prior to formal diagnosis using the framingham criteria. Circulation. 2011;124:A12035. [Google Scholar]
- 28. Vijayakrishnan R, Steinhubl SR, Ng K, et al. Prevalence of heart failure signs and symptoms in a large primary care population identified through the use of text and data mining of the electronic health record. J Card Fail. 2014;20:459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. J Mach Learn Res. 2003;3:993–1022. [Google Scholar]
- 31. Cohen R, Aviram I, Elhadad M, et al. Redundancy-aware topic modeling for patient record notes. PLoS One. 2014;9:e87555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meinhold RJ, Singpurwalla ND. Understanding the Kalman filter. Am Stat. 1983;37:123–127. [Google Scholar]
- 33. Kalman RE. A new approach to linear filtering and prediction problems. J Fluids Eng. 1960;82:35–45. [Google Scholar]
- 34. Wainwright MJ, Jordan MI. Graphical models, exponential families, and variational inference. Found Trends Mach Learn. 2008;1:1–305. [Google Scholar]
- 35. Hoffman MD, Blei DM, Wang C, et al. Stochastic variational inference. J Mach Learn Res. 2013;14:1303–1347. [Google Scholar]
- 36. Cox DR. Regression models and life-tables. J R Stat Soc Ser B. 1972;34:187–220. [Google Scholar]
- 37. Penciana MJ, D’Agostino RB. Overall C as a measure of discrimination in survival analysis: Model specific population value and confidence interval estimation. Stat Med. 2004;23:2109–2123. [DOI] [PubMed] [Google Scholar]
- 38. Nadeau C, Bengio Y. Inference for the generalization error. Mach Learn. 2003;52:239–281. [Google Scholar]
- 39. Holm S. A Simple sequentially rejective multiple test procedure. Scand J Stat 1979;6:65–70. [Google Scholar]
- 40. Iseki K. Gender differences in chronic kidney disease. Kidney Int. 2008;74:415–417. [DOI] [PubMed] [Google Scholar]
- 41. Babazono T, Hanai K, Suzuki K, et al. Lower haemoglobin level and subsequent decline in kidney function in type 2 diabetic adults without clinical albuminuria. Diabetologia. 2006;49:1387–1393. [DOI] [PubMed] [Google Scholar]
- 42. Levin A, Djurdjev O, Duncan J, et al. Haemoglobin at time of referral prior to dialysis predicts survival: an association of haemoglobin with long-term outcomes. Nephrol Dial Transplant. 2006;21:370–377. [DOI] [PubMed] [Google Scholar]
- 43. Wu I-W, Hsu K-H, Lee C-C, et al. Re-evaluating the predictive roles of metabolic complications and clinical outcome according to eGFR levels – a four-years prospective cohort study in Taiwan. BMC Nephrol. 2013;14:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pottelbergh G V, Vaes B, Jadoul M, et al. The prevalence and detection of chronic kidney disease (CKD)-related metabolic complications as a function of estimated glomerular filtration rate in the oldest old. Arch Gerontol Geriatr. 2012;54:e419–425. [DOI] [PubMed] [Google Scholar]
- 45. Moranne O, Froissart M, Rossert J, et al. Timing of onset of CKD-related metabolic complications. J Am Soc Nephrol. 2009;20:164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Levin A, Bakris GL, Molitch M, et al. Prevalence of abnormal serum vitamin D, PTH, calcium, and phosphorus in patients with chronic kidney disease: results of the study to evaluate early kidney disease. Kidney Int. 2007;71:31–38. [DOI] [PubMed] [Google Scholar]
- 47. Drion I, Joosten H, Dikkeschei LD, et al. eGFR and creatinine clearance in relation to metabolic changes in an unselected patient population. Eur J Intern Med. 2009;20:722–727. [DOI] [PubMed] [Google Scholar]
- 48. Hug BL, Witkowski DJ, Sox CM, et al. Occurrence of adverse, often preventable, events in community hospitals involving nephrotoxic drugs or those excreted by the kidney. Kidney Int. 2009;76:1192–1198. [DOI] [PubMed] [Google Scholar]
- 49. Kinchen KS, Sadler J, Fink N, et al. The timing of specialist evaluation in chronic kidney disease and mortality. Ann Intern Med. 2002;137:479–486. [DOI] [PubMed] [Google Scholar]
- 50. Jain AK, McLeod I, Huo C, et al. When laboratories report estimated glomerular filtration rates in addition to serum creatinines, nephrology consults increase. Kidney Int. 2009;76:318–323. [DOI] [PubMed] [Google Scholar]
- 51. Hemmelgarn BR, Zhang J, Manns BJ, et al. Nephrology visits and health care resource use before and after reporting estimated glomerular filtration rate. JAMA. 2010;303:1151–1158. [DOI] [PubMed] [Google Scholar]
- 52. Greer RC, Powe NR, Jaar BG, et al. Effect of primary care physicians’ use of estimated glomerular filtration rate on the timing of their subspecialty referral decisions. BMC Nephrol. 2011;12:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smart NA, Dieberg G, Ladhani M, et al. Early referral to specialist nephrology services for preventing the progression to end-stage kidney disease. Cochrane Database Syst Rev. 2014;6:CD007333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.