Abstract
Background
Kaposi sarcoma (KS) has features of both neoplastic growth and hyperplastic proliferation. It is the most common tumor seen in patients with HIV infection. Whether KS is a real tumor or a benign hyperplastic disease is not known.
Material/Methods
Tissues from KS and cutaneous hemangioma lesion DNA were extracted, and then digested with methylation-sensitive restriction endonuclease HpaII. Human androgen receptor gene (HUMARA) was amplified with PCR method and the product was separated on 10% denaturing polyacrylamide gels and stained with ethylene dibromide (EB) to show the polymorphism of HUMARA. Phosphoglycerate kinase (PGK) was amplified and the product was digested by BStXI, agarose gel and EB stained to show the polymorphism of PGK. Finally, we analyzed the clonality of KS.
Results
In the 14 patients with KS, heterozygosity of the HUMARA gene was observed in 12 (85.7%) cases. Loss of heterozygosity of HUMARA gene on X-chromosome (without HpaII digestion there were 2 bands, after HpaII digestion there were just 1 of the bands), representing monoclonal origin, was present in 11 cases of Kaposi sarcoma. Heterozygosity of the PGK gene was observed in 5 (35.7%) cases, which all represent monoclonal origin. There was no significant difference according to country, stage, or HIV and HHV-8 (P>0.05).
Conclusions
The current findings suggest that Kaposi sarcoma is a clonal neoplasm, not a reactive proliferation.
Keywords: Hyperplasia; Mixed Tumor, Malignant; Sarcoma, Kaposi
Background
Kaposi sarcoma is a type of multicentric tumor with unclear pathogenesis, which was first described by Kaposi in 1872. It is the most common tumor in patients with HIV infection [1,2]. Kaposi sarcoma is a highly vascularized tumor that primarily affects the skin; during disease progression it can also spread to lymph nodes and viscera [3–5]. The occurrence of KS may be associated with factors such as geographical environment, race, and sex. There are 4 types: classic KS, endemic KS, epidemic (AIDS-associated) KS, and iatrogenic (immunosuppressive drug-associated) KS [6].
The classical form of KS is a rare vascular tumor affecting elderly men of Mediterranean and eastern European Jewish ancestry. It is predominantly localized to the lower extremities and has an indolent course. Classic KS has a male-to-female ratio of approximately 10:1 to 15:1. The moist common types in Xinjiang China are classic KS and AIDS-associated KS [7].
Three major cell types are contained by the lesions of the 4 KS forms: spindle cells, endothelial cells, and infiltrating inflammatory cells. Spindle cells derive from lymphatic endothelial cells and are the main cell type present in nodular-stage lesions. The exact cell of origin of KS is not clear. The cell of origin may arise from lymphatic or blood endothelium, vascular smooth muscle cells, mesenchymal cells, or a combination of these cells [8]. However, the currently prevalent opinion is that KS probably originates from lymphatic endothelium [9,10]. In 1994 [11], Chang discovered a new type of virus in KS tissues, which was named KS-associated Herpesvirus (KSHV) HHV-8, or Human herpesvirus 8 (HHV-8). It has subsequently been found in all 4 forms and is the most important causative agent in all 4 forms of KS [12]. HHV-8 infection of blood vascular endothelial cells leads to lymphatic endothelial reprogramming of these cells [9,10], and it is suggested that HHV-8 may preferentially infect endothelial cell precursors and mediate their differentiation towards a lymphatic endothelial cell genotype [10,13].
Although there has been much research on KS, the pathogenesis and biological behavior is still poorly understood [14–20] and it is unclear whether KS is a reactive or neoplastic lesion or a monoclonal neoplasm. Moreover, Duprez et al., in order to determine the clonality of KS cellular, studied the size heterogeneity of the HHV-8 terminal repeat (TR) region. Their results suggested that KS lesions, especially in patients with advanced skin tumors, were reactive proliferations rather than true malignancies with metastatic dissemination, but this conclusion is controversial. Rabkin et al. studied the clonality of KS by investigating the HUMARA gene; their findings suggested KS is a monoclonal neoplasm. To further characterize KS, we evaluated the clonality of 14 samples of KS from female patients by using X-chromosomal inactivation HUMARA and PGK gene loci in female somatic cells.
Material and Methods
Tissues
Formalin-fixed, paraffin-embedded surgical tissues were supplied by the Department of Pathology of the People’s Hospital of Xinjiang Uygur Autonomous Region. We identified 14 female patients with KS and 1 female patient with cutaneous hemangioma (the cutaneous hemangioma specimen was used as control). The clinicopathological features of the selected samples are summarized in Table 1. The mean age of the patients was 48.4 years (range 27–71), and every patient had a single lesion. Of the 14 KS patients, 6 women were HIV seronegative (classic KS) and 8 were HIV seropositive (transmitted by sexual contact with a male (whether bisexual male was not clear).Eleven patients were Uyghur, 3 patients were Kazak; all patients were HHV-8 positive. All cases were diagnosed by 2 experienced pathologists. Moreover, all patients agreed to analyses of the materials.
Table 1.
Clinicopathological features and histopathological phases.
| Case No. | Age | Site | Duration (months) | Stage | HIV | HHV-8 |
|---|---|---|---|---|---|---|
| 1 | 59 | Left wrist | 24 | Plaque | − | + |
| 2 | 68 | Mouth | 48 | Plaque | − | + |
| 3 | 46 | Lower extremity | 4 | Macular | + | + |
| 4 | 62 | Leg | 60 | Plaque | − | + |
| 5 | 34 | Thigh | 4 | Plaque | + | + |
| 6 | 34 | Thigh | 6 | Plaque | + | + |
| 7 | 70 | Pelma | 96 | Plaque | − | + |
| 8 | 33 | Hand | 12 | Plaque | + | + |
| 9 | 27 | Acrotarsium | 12 | Plaque | + | + |
| 10 | 27 | Trunk | 6 | Plaque | + | + |
| 11 | 42 | Underjaw | 3 | Plaque | + | + |
| 12 | 68 | Middle finger | 4 | Nodule | − | + |
| 13 | 36 | Underjaw | 3 | Macular | + | + |
| 14 | 71 | Lower extremity | 10 | Plaque | − | + |
Immunohistochemistry
Immunohistochemical staining was performed on sections which were formalin-fixed paraffin-embedded, using the Envision technique. This study used the primary monoclonal antibody.
2 DNA extraction
DNA was extracted from the tumor and cutaneous hemangioma samples using standard procedures. Briefly, the formalin-fixed, paraffin-embedded tissue blocks were cut into 10-um slices and 5~1010 um slices were placed into the 1.5-ml microcentrifuge tubes. Samples were digested in TES (10mmol/L Tris·Hcl, 1 mmol/L EDTA, 0.5%SDS) and TET (100 mmol/L Tris·Hcl, 1 mmol/L EDTA, 1%TritonX-100). Proteinase K was then added to a final concentration of 20 μg/μl and the samples were kept at 37°C overnight. DNA was extracted by phenol chloroform method.
Enzymatic digestion
To perform clonal analysis, cutaneous hemangioma and KS DNA were divided into 2 fractions and 1 of these was digested with HpaII10U (1 ul) for 3–5 h at 37°C, and followed at 65°C for 20 min to inactivate the Enzyme HpaII, whereas the other fraction was undigested. Both fractions were used for PCR amplification.
PCR amplification HUMARA gene
HUMARA-C sense (5′-CCG CCG TCC AAG ACC TAC -3′) and HUMARA-C antisense (5′-GGT TGC TGT TCC TCA TCC AG-3′). The product length of amplification is about 200–400bp. We amplified 2-ul aliquots of HpaII-digested DNA in a 20-ul polymerase chain reaction volume containing 2 ul 10×PCR buffer, 2 ul of dNTPs, 0.5 ul of HUMARA-C sense-primer, 0.5 ul of HUMARA-C antisense, 2 subunits thiamphenicol 1 ul and 0.4 ul of AmpliTaq Gold. Amplification was performed with an initial denaturation step of 95°C for 5 min followed by 20 cycles of 95°C for 45 s, 65°C for 45 s, and 72°C for 30 s, and a final extension step of 72°C for 7 min, and soaking at 4°C. After migration we used a 10% denaturing polyacrylamide gel and 0.5 mg/L EB staining. Analysis was with Gene Genius Bioimaging System.
PGK (Phosphoglycerate Kinase) gene assay
Nested PCR was used to detect single-nucleotide polymorphism (SNP) sites in the PGK gene. The 5-ul aliquots of digested DNA samples were then subjected to nested PCR. The 50-ul reaction mixture consisted of 4 ul of 10-mM dNTPs, primers PGK1A and PGK1B (0.4 pmol each), 5 ul of 10×buffer, 1.5 lL of 50 mM MgCl2and 2.5 U of Taq DNA polymerase. The amplification was performed using a PT-200 thermal cycler for 35 cycles (denaturation 94°C for 40 s, annealing at 58°C for 50 s, and extension at 72°C for 1 min), with a final extension step of 72°C for 7 min. The first round of PCR products (5-uL) were diluted (1: 20) and used as templates for a second PCR reaction with primers PGK2A and PGK2B using the same amplification protocol, except with an annealing temperature of 56°C.The PCR products were digested with 5 U of BstXI at 48°C for 8–10 h in a 20-ul reaction containing 0.2 ul of BSA (10g/L) and 2 ul of 10×reaction buffer. The digested products were visualized by 2% agarose gel electrophoresis and 0.5 mg/L EB staining. Briefly, 2 pairs of primers were used to amplify the PGK gene: PGK1A, 5′-CTG TTC CTG CCC GCG CGG TGT TCC GCA TTC-3′;PGK1B, 5′-ACG CCT GTT ACG TAA GCT CTG CAG GCC TCC-3′; PGK2A, 5′-AGC TGG ACG TTA AAG GGA AGC GGG TCG TTA-3′; PGK2B, 5′-TAC TCC TGA AGT TAA ATC AAC ATCCTC TTG-3′.
Results
Clinical and pathological data on fourteen KS patients
Table 1 indicates the clinical and pathological characteristics of the 14 patients which were studied. Eight patients had HIV-associated KS and 6 patients had classical KS. HIV-associated KS was present for 3–12 months. Four out of 6 classical KS cases had KS lesions present for 2–8 years, and the other 2 patients had KS present for 4 and 10 months. Eleven cases were Uyghur and the other 3 cases were Kazak. All 14 patients were HHV-8-positive. Eleven cases were examined in the plaque phase, 2 cases were macular, and only 1 case was examined in the nodule stage (Figure 1).
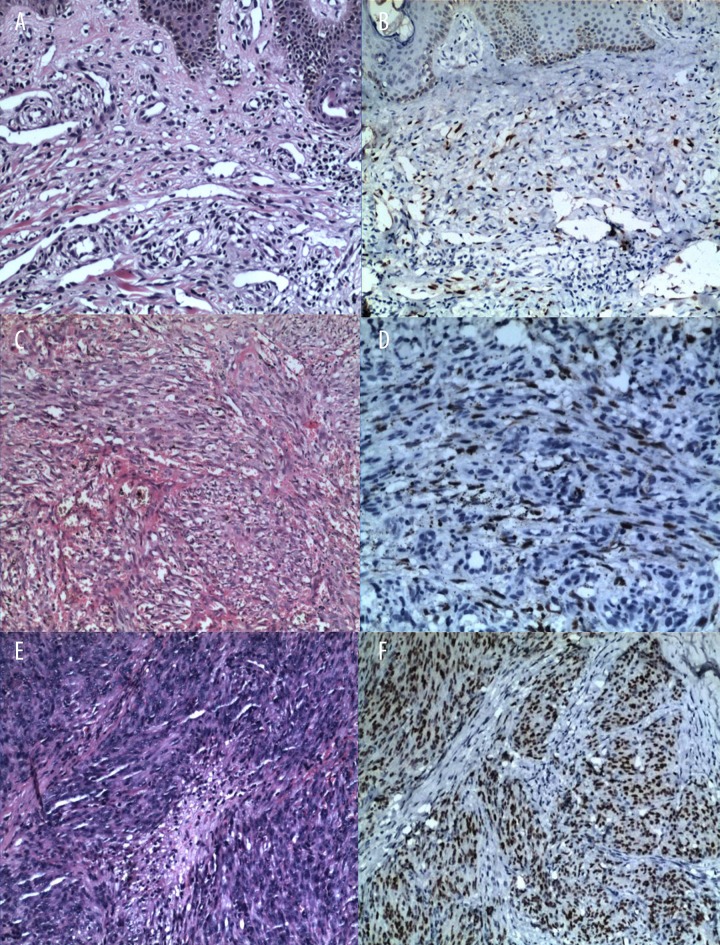
Figure 1.
Representative case of KS in patients of every stage. (A) (Hematoxylin and eosin ×100) showing a macular stage patient. (C) (Hematoxylin and eosin ×200) showing a plaque stage patient. (E) (Hematoxylin and eosin ×100) showing a nodule stage patient. (B–F) The HHV-8 of every stage staining was positive.
Clonality analysis
In 2 of the 14 KS patients (cases 1 and 5) the DNA sample failed to amplify expected PCR products (HUMARA) visible on 2% agarose gel, while in 5 KS patients (1, 3, 4,9, and 12) the DNA sample succeed in amplifying (PGK). Eleven out of 12 KS tissues were analyzed by HUMARA gene and 5 KS tissues were analyzed by PGK gene. Without HpaII digestion there were 2 bands, and after HpaII digestion there was just 1 band (Figure 2). In 1 patient a polyclonal pattern was demonstrated in the KS tissue. These results suggest that KS is a clonal neoplasm rather than a reactive proliferation.
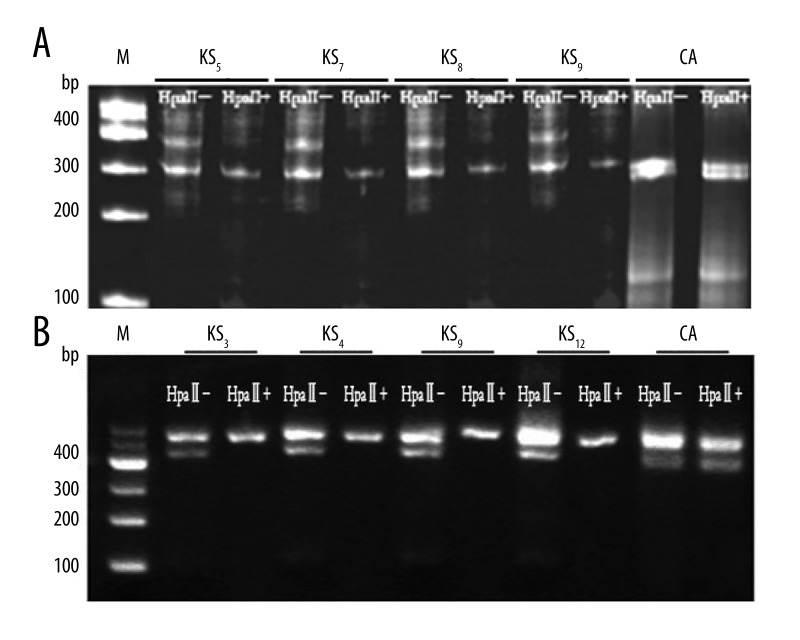
Figure 2.
Clonality analysis in HUMARA (A) and PGK (B) gene of representative KS and cutaneous hemangioma. KS – Kaposi sarcoma; CA – cutaneous hemangioma; M – DNA marker; HpaII+ – with HpaII digestion; HpaII– – without HpaII digestion. KS indicated monoclonal samples. Before HpaII digestion there were 2 bands, and the intensity of the other band was lower. CA indicated polyclonal samples, with and without HpaII digestion; there all were 2 bands, but the latter band was lower intensity than the former band.
As a polyclonal control, and to validate the ability to detect clonal populations from skin with the clonal assay, we have extracted DNA from 1 cutaneous angiosarcoma sample. This analysis showed a typical polyclonal pattern because there were 2 bands with and without HpaII digestion.
The clonality rate of the different nation, stages, HIV infection in KS
Ten Uyghur cases and 3 Kazak cases were monoclonal origin, and there was no significant difference between the 2 groups (P>0.05). Two cases of macular stage lesion, 10 cases of plaque stage lesion, and 1 case of nodule stage lesion were of monoclonal origin; there was no significance among the 3 groups (P>0.05). Seven HIV-positive cases and 6 HIV-negative cases with were of monoclonal origin, and there was no significant difference between the 2 groups (P>0.05).
Discussion
Whether KS is a neoplastic lesion or a reactive proliferation remains unknown. The reasons are as follows: 1) The World Health Organization (WHO) publication about vascular nomenclature [21] proposed KS is a vascular tumor of intermediate malignancy (low-grade malignant). 2) The biological behavior of the 4 types KS are very different. Classic KS is relatively benign; patients can live for another 10–15 years and die from other diseases. Iatrogenic KS is the result of the long-term use of immunosuppressive therapy and it can be reversed if pathogenic factors are removed. The characteristics of classic and iatrogenic forms of KS seems to be more in line with reactive proliferation [22,23], but the epidemic and endemic forms of KS are relatively malignant, and the patients have short survival [24]. 3) The main characteristic of disease lesions are thin-walled neovascular formations and proliferating spindle cells. Many researchers have extensively studied KS phenotype, and some believe KS is a clonal neoplasm [25,26], but some other researchers consider it a reactive proliferation. 4) Many researchers have studied the clonality of KS; they use the same methods but reach different conclusions. Rabkin [19] was the first researcher to propose that KS (at least epidemic KS) is a monoclonal malignant tumor through the analysis of the HUMARA gene. Through the analysis of the HUMARA gene in 7 patients with KS, of which 4 cases were classical and 3 cases were AIDS-related, Delabesse [14] concluded that KS is a polyclonal tumor.
The most important characteristic of neoplastic lesions is clonality, and the reactive hyperplasia is absence of clonality. Among various methods in clonality analysis, X chromosome inactivation analysis is the best for tumor clonality analysis [15]. Analysis of the HUMARA and PGK gene allows us to distinguish between a true neoplastic lesion and a non-neoplastic proliferation lesion. The present study is convincing because we analyzed the corresponding changes based on previous research, and analyzed the polymorphism of PGK gene clonality in different KS tumors from 14 female KS patients. Our findings indicate that KS lesions arise from a single clone of cells and that KS is a monoclonal cancer.
During oosperm implantation, in every female cell 1 of the 2 X-chromosomes is randomly inactivated by DNA methylation; this process can permanently inactivate the oosperm and its daughter cells. Due to the nature of the X-chromosome random inactivation, almost all normal female cells contain inactivated maternal and paternal (2 types) X-chromosomes. However, in asexual propagation, tumors are differentiated by and grown from a single cell, so all tumor cells have either the maternal or the paternal X-chromosome inactivated [15]. The first exon of the human androgen-receptor (HUMARA) gene, which resides on the X-chromosome, contains a highly polymorphic trinucleotide (CAG) repeat. Before the polymorphic CAG repeat region, which contains 5 methylation-sensitive restriction enzyme sites (2 HpaII sites and 3 HhaI sites), in humans the hybrid rate of CAG repeat is up to 90%. In heterozygous females, the paternal and maternal X-chromosome sequences can be distinguished by the CAG repeat polymorphism, and the active and inactive allele can also be distinguished by the methylation status [14]. The X-chromosome chain phosphate glycerol kinase (pGK) gene is located in Xqll-q13, close to the HUMARA gene. The first exon of pGK gene(PGK-l) has a single-nucleotide polymorphism (G/A), which is characterized by the change of the site of a single base in the formation of Bstxl restriction endonuclease restriction sites; it is also called the Bstxl locus polymorphism because of its proximity to the X chromosome inactivation region, combined with characteristics of methylation modification. Random distribution of the PGK-1BstXI polymorphism becomes a detection marker of X-chromosome inactivation. After digestion with methylation-sensitive restriction enzymes HpaII or HhaI, the normal and reactive hyperplastic tissues showed 2 alleles of equal intensity, but the neoplastic tissues show only 1 allele, or 1 of the 2 alleles exhibited reduced intensity.
In our research, the hybrid rate of HUMARA gene is 85.7%, the hybrid rate of PGK gene is 35.7%, which is similar to the results of researchers outside China.
Our samples qualified for DNA analysis of the HUMARA and PGK gene. Among samples from 14 KS patients, 11 samples had 2 bands before HpaII digestion, and 1 of 2 bands disappeared after HpaII digestion in the HUMARA gene. For example, it showed that after HpaII digestion, the upper band disappeared in the representative cases (cases 6–9); 5 samples had 2 bands before HpaII digestion, and 1 of 2 bands disappeared after HpaII digestion in the PGK gene. For example, it showed that after HpaII digestion, the upper band disappeared in the representative cases (case 3, 4, 9, and 12). The result suggested KS (at least classic and epidemic KS) is a clonal neoplasm rather than a reactive proliferation. The circumstantial evidence suggests that KS is a monoclonal malignant tumor, a view shared by Rabkin. The present study found no significant difference in clonality rate by country, stage, or HIV and HHV-8 infection status, perhaps because our sample size was too small. Future research should expand the sample size.
Some researchers reported that use of microdissection to accurately separate the spindle cells can avoid the pollution of adipocyte and vascular endothelial cells, which can improve the sensitivity of experiments. But Rabkin estimated that the tumor DNA content of a sample is greater than 80%, which also can detect unbalanced methylation. In our study, all tissue samples were more than 80%, which was ensured by 2 pathologists.
Analysis of the pattern of X-chromosome inactivation (HUMARA gene and PGK gene) has been used to determine the clonality of KS lesional cells in females. An important drawback of this method is that it can only be performed on females, and since KS affects males with a higher frequency than females, this method cannot be used in most cases of KS. Human herpesvirus 8, also known as KS-associated herpesvirus (HHV-8/KSHV), is present in spindle cells at all KS stages, and is the most important pathogen at all clinical types of KS. Our preliminary study indicated that the positive rate of HHV-8 antibody in KS samples of lesions and serum was more than 80%. Judde et al. [17] studied the clonal nature of HHV-8 terminal repeat sequences in advanced KS lesions and found that such lesions display all patterns of clonality. This finding supports the concept that KS starts as a benign angioproliferative disorder consisting of a polyclonal cell population, and in later stages of HIV-KS disease some, if not all, KS lesional cells will evolve to a clonal cell population that will undergo expansion. Duprez et al. [18], using the same HHV-8 episomal analysis, demonstrated that most of the advanced KS lesions were oligoclonal (82%), each individual lesion having multiple HHV-8-infected clones concurrently. Some (18%) advanced KS lesions were monoclonal, with each lesion composed of a monoclonal expansion of a single HHV-8-infected cell. In addition, individual KS lesions that occur concurrently at different anatomical sites in a subject with multicentric KS were either monoclonal or oligoclonal. The size of the HHV-8 episomes varied even between the multicentric monoclonal KS lesions, clearly indicating that these lesions arose from independent clones and were not metastatic lesions originating from the clonal expansion of a single HHV-8-infected cell.
The main limitation of the present study was the technique that we used to analyze the viral clonality. The method requires a substantial amount of DNA of high molecular weight, which cannot be obtained from all KS biopsy samples. Typically only highly spindle cell-infiltrated lesions are informative. Moreover, fixed biopsies cannot be used in such a technique because they generally provide fragmented nucleic acid [27]. The advantage of the clonality analysis method is that it can be used in male patients, enlarging the sample size. Consequently, clonality origination deserves further research attention.
Conclusions
Our results suggest that KS (at least the classic and epidemic forms) is a neoplastic lesion, not a reactive proliferation. However, additional studies with larger sample sizes are needed to conclusively prove our hypothesis.
Footnotes
Source of support: Departmental sources
Reference
- 1.Stiller CA, Trama A, Brewster DH, et al. Descriptive epidemiology of Kaposi sarcoma in Europe. Report from the RARECARE project. Cancer Epidemiol. 2014;38(6):670–78. doi: 10.1016/j.canep.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Radu O, Pantanowitz L. Kaposi sarcoma. Arch Pathol Lab Med. 2013;137(2):289–94. doi: 10.5858/arpa.2012-0101-RS. [DOI] [PubMed] [Google Scholar]
- 3.Gramolelli S, Schulz TF. The role of Kaposi sarcoma-associated herpesvirus in the pathogenesis of Kaposi sarcoma. J Pathol. 2015;235(2):368–80. doi: 10.1002/path.4441. [DOI] [PubMed] [Google Scholar]
- 4.Ruocco E, Ruocco V, Tornesello ML, et al. Kaposi’s sarcoma: etiology and pathogenesis, inducing factors, causal associations, and treatments: facts and controversies. Clin Dermatol. 2013;31(4):413–22. doi: 10.1016/j.clindermatol.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas JL, Gustin JK, Dezube B, et al. Kaposi’s sarcoma: a model of both malignancy and chronic inflammation. Panminerva Med. 2007;49(3):119–38. [PubMed] [Google Scholar]
- 6.Morand JJ, Lightburn E, Simon F, et al. Update on Kaposi’s sarcoma. Med Trop (Mars) 2007;67(2):123–30. [PubMed] [Google Scholar]
- 7.Wu XJ, Pu XM, Zhao ZF, et al. The expression profiles of microRNAs in Kaposi’s sarcoma. Tumour Biol. 2015;36(1):437–46. doi: 10.1007/s13277-014-2626-1. [DOI] [PubMed] [Google Scholar]
- 8.Gessain A, Duprez R. Spindle cells and their role in Kaposi’s sarcoma. Int J Biochem Cell Biol. 2005;37(12):2457–65. doi: 10.1016/j.biocel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 9.Hong YK, Foreman K, Shin JW, et al. Lymphatic reprogramming of blood vascular endothelium by Kaposi sarcoma-associated herpesvirus. Nat Genet. 2004;36(7):683–85. doi: 10.1038/ng1383. [DOI] [PubMed] [Google Scholar]
- 10.Konstantinopoulos PA, Dezube BJ, Pantanowitz L. Morphologic and immunophenotypic evidence of in-situ Kaposi’s sarcoma. BMC Clin Pathol. 2006;30(6):7. doi: 10.1186/1472-6890-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–69. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 12.Bhutani M, Polizzotto MN, Uldrick TS, et al. Kaposi sarcoma-associated herpesvirus-associated malignancies: epidemiology, pathogenesis, and advances in treatment. Semin Oncol. 2015;42(2):223–46. doi: 10.1053/j.seminoncol.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang HW, Trotter MW, Lagos D, et al. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet. 2004;36(7):687–93. doi: 10.1038/ng1384. [DOI] [PubMed] [Google Scholar]
- 14.Delabesse E, Oksenhendler E, Lebbe C, et al. Molecular analysis of clonality in Kaposi’s sarcoma. J Clin Pathol. 1997;50(8):664–68. doi: 10.1136/jcp.50.8.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabkin CS, Janz S, Lash A, et al. Monoclonal origin of multicentric Kaposi’s sarcoma lesions. N Engl J Med. 1997;336(14):988–93. doi: 10.1056/NEJM199704033361403. [DOI] [PubMed] [Google Scholar]
- 16.Gill PS, Tsai YC, Rao AP, et al. Evidence for multiclonality in multicentric Kaposi’s sarcoma. Proc Natl Acad Sci USA. 1998;95(14):8257–61. doi: 10.1073/pnas.95.14.8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Judde JG, Lacoste V, Briere J, et al. Monoclonality or oligoclonality of human herpesvirus 8 terminal repeat sequences in Kaposi’s sarcoma and other diseases. J Natl Cancer Inst. 2000;92(9):729–36. doi: 10.1093/jnci/92.9.729. [DOI] [PubMed] [Google Scholar]
- 18.Duprez R, Lacoste V, Brirer J, et al. Evidence for a multiclonal origin of multicentric advanced lesions of Kaposi sarcoma. J Natl Cancer Inst. 2007;99(14):1086–94. doi: 10.1093/jnci/djm045. [DOI] [PubMed] [Google Scholar]
- 19.Rabkin CS, Bedi G, Musaba E, et al. AIDS-related Kaposi’s sarcoma is a clonal neoplasm. Clin Cancer Res. 1995;3(1):257–60. [PubMed] [Google Scholar]
- 20.Nihal M, Mikkola D, Qian Z, et al. The clonality of tumor-infiltrating lymphocytes in African Kaposi’s sarcoma. J Cutan Pathol. 2001;28(9):200–5. doi: 10.1034/j.1600-0560.2001.028004200.x. [DOI] [PubMed] [Google Scholar]
- 21.Goh SGN, Calonje E. Cutaneous vascular tumours: an update. Histopathology. 2008;52:661–73. doi: 10.1111/j.1365-2559.2007.02924.x. [DOI] [PubMed] [Google Scholar]
- 22.Jakob L, Metzler G, Chen KM, et al. Non-AIDS associated Kaposi’s sarcoma: clinical features and treatment outcome. PLoS One. 2011;6(4):e18397. doi: 10.1371/journal.pone.0018397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vincenzi B, D’Onofrio L, Frezza AM, et al. Classic Kaposi Sarcoma: to treat or not to treat? BMC Res Notes. 2015;10(8):138. doi: 10.1186/s13104-015-1076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Restrepo CS, Ocazionez D. Kaposi’s sarcoma: imaging overview. Semin Ultrasound CT MR. 2011;32(5):456–69. doi: 10.1053/j.sult.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Simonart T, Hermans P, Schandene L, et al. Phenotypic characteristics of Kaposi’S sarcoma tumour cells derived from patch-, plaque- and nodular-stage lesions: analysis of cell cultures isolated from AIDS and non-AIDS patients and review of the literature. Br J Dermatol. 2000;143(3):557–63. doi: 10.1111/j.1365-2133.2000.03709.x. [DOI] [PubMed] [Google Scholar]
- 26.Thomas S, Sindhu CB, Sreekumar S, et al. AIDS associated Kaposi’s sarcoma. J Assoc Physicians India. 2011;6(59):387–89. [PubMed] [Google Scholar]
- 27.Wood NH, Feller L. The malignant potential of HIV-associated Kaposi sarcoma. Cancer Cell Int. 2008;8:14. doi: 10.1186/1475-2867-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]