Abstract
Exploration of protein function and interaction is critical for discovering links among genomics, proteomics, and disease state; yet, the immense complexity of proteomics found in biological systems currently limits our investigational capacity. Although affinity and autofluorescent tags are widely employed for protein analysis, these methods have been met with limited success because they lack specificity and require multiple fusion tags and genetic constructs. As an alternative approach, the innovative HaloTag protein fusion platform allows protein function and interaction to be comprehensively analyzed using a single genetic construct with multiple capabilities. This is accomplished using a simplified process, in which a variable HaloTag ligand binds rapidly to the HaloTag protein (usually linked to the protein of interest) with high affinity and specificity. In this review, we examine all current applications of the HaloTag technology platform for biomedical applications, such as the study of protein isolation and purification, protein function, protein–protein and protein–DNA interactions, biological assays, in vitro cellular imaging, and in vivo molecular imaging. In addition, novel uses of the HaloTag platform are briefly discussed along with potential future applications.
Introduction
Proper functioning of complex biological systems is dependent upon an array of proteins responsible for maintaining cellular homeostasis.1−3 The complexity of protein–protein interactions in living cells has hindered research into finding new diagnostic and treatment options for many diseases.4,5 In addition, inefficient methods of protein labeling for in vitro and in vivo applications have limited proteomic analysis. While purification is often tedious and requires resources beyond the scope of many laboratories, the process is essential for evaluating protein function. For this reason, newer methodologies are currently being explored for protein analysis.6,7
Traditional protein tagging systems are often limited by low yield or relatively high impurity levels.8 In addition, larger molecular weight protein tagging systems can alter the conformation and functionality of targeted proteins.9 The polyhistidine tag (His-tag) is commonly used for protein analysis because it rarely affects protein function due to its small size. While His-tag is effective for isolation of proteins, this method suffers from high impurity levels due to nonspecific binding of other proteins.10 In addition, His-tag is limited to the isolation and purification of proteins, and an additional tagging system must be employed for cellular imaging or other applications. The HaloTag system was developed to overcome the current limitations of traditional protein tagging platforms by allowing researchers to perform comprehensive protein analysis using a single genetic construct (Figure 1A).
Figure 1.
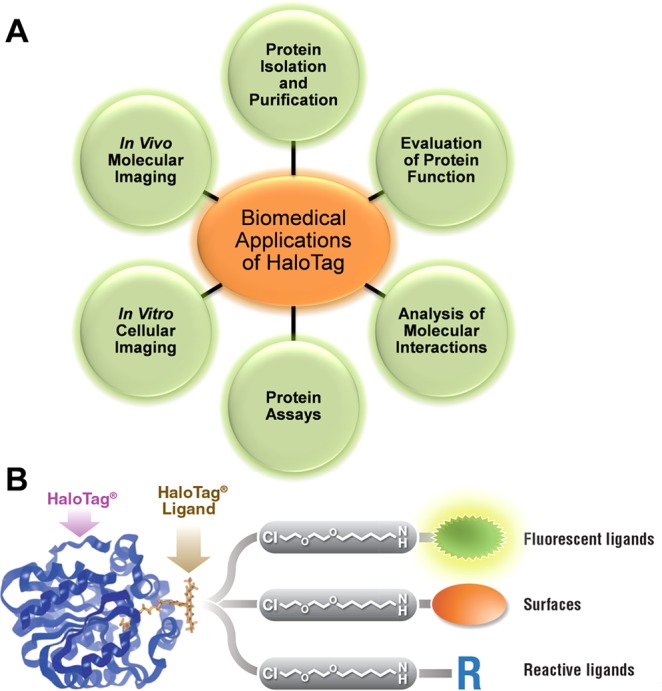
Applications of the versatile HaloTag platform. (A) The HaloTag protein tagging system is utilized for several applications, including protein isolation and purification, evaluation of protein function, analysis of molecular interactions, protein assays, in vitro cellular imaging, and in vivo molecular imaging. (B) Representation of the HaloTag system, in which the HaloTag protein forms a covalent bond with a specific HaloTag ligand. Each HaloTag ligand contains a binding group and functional moiety, such as fluorescent molecules for intracellular and extracellular purposes, surface ligands for protein immobilization with resins or slides, and reactive ligands for imaging purposes. Reprinted with permission from ref (11). Copyright 2012 Urh and Rosenberg.
This is accomplished using a two-step approach, which includes the development of a 33 kDa HaloTag genetically fused to the protein of interest and an application-specific HaloTag ligand (Figure 1B).11,12 A covalent bond is formed between the HaloTag protein and HaloTag ligand when these two moieties come in contact, resulting in rapid and irreversible binding.13 The molecular mechanism of the HaloTag system is based on a mutant bacterial haloalkane dehalogenase enzyme from Rhodococcus rhodochrous, in which Phe272 is substituted by His272.14 During the interaction of the enzyme and ligand, an alkyl-enzyme intermediate is formed during the nucleophilic displacement of a terminal chloride with Asp106. Normally, His272 would function as a general base in wild-type dehalogenase to catalyze the hydrolysis, thus releasing the enzyme. This reaction is altered in the mutant dehalogenase, as the substituted Phe272 does not catalyze the hydrolysis, thus resulting in a covalent adduct with high stability.14
Currently, several HaloTag ligands are offered for different applications ranging from protein isolation to molecular imaging.15 The covalent linkage between the HaloTag ligand and HaloTag protein enables rapid isolation and purification.11 A few examples of HaloTag ligands include HaloTag Alexa Fluor 488 for cell-surface labeling, HaloTag TMR ligand for labeling of cytosolic proteins, and HaloTag resins for immobilization of proteins.14
The HaloTag system for protein tagging has several advantages over traditional protein tagging systems.16 Because different HaloTag ligands can be utilized for studying different aspects of the protein of interest, only a single genetic construct is required. Second, binding of the HaloTag protein with a HaloTag ligand is rapid and irreversible, allowing for sequential labeling experiments to analyze protein synthesis and degradation. Lastly, this technology can be utilized for cellular imaging of both live and fixed cells, as studies have shown that the HaloTag complex remains relatively stable under harsh conditions (e.g., acidic microenvironment).14 The versatility of the HaloTag platform makes it optimal for several protein analysis applications both in vitro and in vivo (Figure 1). This review examines all current application of HaloTag technology for protein isolation and purification, analysis of protein function, studying protein–protein and protein–DNA interactions, performing biological assays, in vitro cellular imaging, and in vivo molecular imaging.
Protein Isolation and Purification
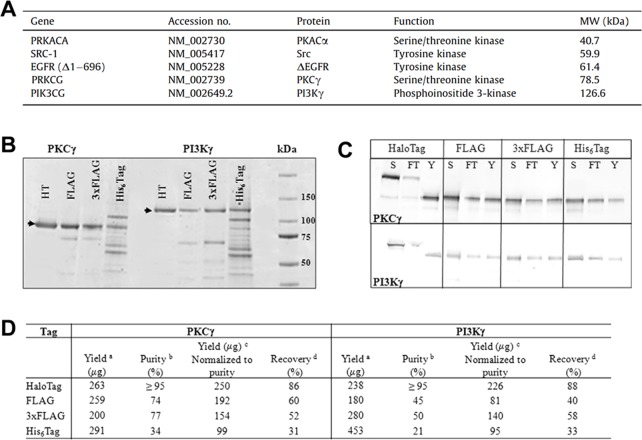
Improvements in protein isolation and purification using the HaloTag platform makes it possible to isolate and purify proteins at levels unachievable by traditional protein isolation methods (e.g., His-tag).17 This is attributed to the highly specific covalent interaction between HaloTag proteins and HaloTag ligands,18 making it feasible to isolate proteins expressed at low levels in mammalian cells. Functionality remains critical for analyzing proteins, yet many tagging systems result in altered activity or inactive proteins. For example, Locatelli-Hoops et al. demonstrated that a stable form of human cannabinoid receptor CB2 could be isolated and purified using the HaloTag system.19 They found that the functionality of the protein was dependent on the terminus of the protein at which the HaloTag was located. While genetically fusing the HaloTag to the N-terminus resulted in an inactive protein, protein activity was maintained when the HaloTag was positioned at the C-terminus. Furthermore, they utilized HaloTag resin with detergents to capture the protein, along with specific tobacco etch virus (TEV) for eluting the receptor after purification. Similarly, five functional human kinases were isolated and purified from mammalian cells using the HaloTag system (Figure 2A).20 In this study, Ohana et al.20 compared the quantity of isolated protein using different protein tagging systems, including FLAG, 3× FLAG, His-tag, and HaloTag (Figure 2B–D). It was found that HaloTag was superior to other protein tagging systems, providing higher quantity yields and superior purification of the protein of interest (Figure 2D).
Figure 2.
Functional human kinases isolated and purified from HEK-293 cells using the HaloTag platform. (A) Five human kinases were selected for isolation and purification. (B) To compare the efficiency of HaloTag to that of other protein tagging systems, PKCγ and PI3Kγ were transiently expressed in HEK-293 cells using four protein labeling protocols, including HaloTag, FLAG, 3× FLAG, and His-Tag. Purified proteins were analyzed by SDS-PAGE. Each protein tagging method resulted in purified protein, yet only HaloTag displayed a single band. The arrow denotes the expected molecular weight of the protein. (C) Protein recovery was determined using normalized volumes of soluble lysate (S), unbound fractions (FT), and purified protein (Y) with the addition of a protease using SDS-PAGE and western blot analysis. (D) The HaloTag platform provided the highest purity of protein for both kinases, as compared to that with the other systems. The percent recovery was also shown to be much higher for the HaloTag system. Reprinted with permission from ref (20). Copyright 2011 Elsevier.
There is a shortage of methods suitable for isolating and purifying full-length proteins, as larger proteins are often more strenuous to capture in their functional state.21 The HaloTag system was shown to effectively isolate the full-length mouse coactivator-associated arginine methyl transferase 1 (CARM1) from mammalian cells using HaloLink resin.22 In addition, highly efficient isolation and purification of the protein was achieved through stringent washes. While traditional protein tagging systems result in loss of protein during washing steps, the covalent interaction between HaloTag resin and HaloTag protein limits the amount of unintentional protein loss. Because the HaloTag platform is rapid and effective, Saul et al. attempted to express 31 full-length proteins using HaloTag in three distinct systems, Escherichia coli and two commercial cell-free systems.23 Ultimately, they were able to purify 42% of the test collection of proteins with purity levels greater than 90%.
In addition to isolating proteins for purification, HaloTag-modified proteins can be immobilized onto various surfaces for other purposes. For example, Nath et al. showed that cell-free protein expression systems can be utilized for capturing select proteins on hydrogel-coated slides containing HaloTag resins.24 Similarly, HaloTag polyproteins were immobilized onto a mica substrate for analysis with atomic force microscopy (AFM)-based single molecule force spectroscopy.25 As stated earlier, proteins isolated using the HaloTag platform are often utilized for several applications, including the study of protein–protein and protein–DNA interactions.26 For example, Saito et al. isolated recombinant ameloblastin from COS-7 cells to study the inhibitory effects of ameloblastin on epithelial cell proliferation.27 In this study, HaloTag was utilized for both the isolation and cellular tracking of ameloblastin, allowing researchers to show that ameloblastin induced cell cycle arrest in epithelial cells that led to periodontitis.
Enzyme activity is critical for cell survival, as enzymes are involved in nearly all intracellular chemical reactions, yet isolation of functional enzymes can be challenging.28,29 A study by Motejadded et al. described a methodology for immobilizing enzymes using HaloTag resins.30 In this study, 13 μg of functional protein per milligram of HaloLink magnetic beads was rapidly isolated for investigating enzyme activity. Several additional studies have employed the HaloTag platform to isolate proteins for functionality studies.31,32 Antibodies are often employed for active targeting and treatment of various diseases, yet isolation and purification of monoclonal or polyclonal antibodies are costly and challenging.33 To demonstrate the feasibility of utilizing HaloTag for antibody isolation and purification, Hata and Nakayama successfully purified small volumes of polyclonal antibodies from E. coli using HaloLink resin.34 The authors noted that the quantity of purified antibody was low, yet the procedure could be scaled up in the future using higher quantities of HaloLink resin.
The HaloTag system has been used extensively in studying the cellular processes and movement of ribosomes in live cells.35,36 Recently, the HaloTag system was used to examine the general process of translation by anchoring HaloTag ribosomes to a glass surface.36 A similar study used the same technique to further explore the process of trans-translation.35 The movement of HaloTag-modified ribosomes was analyzed in living cells using time-lapse microscopy by Gallo et al.37 This study revealed that each HaloTag construct must be characterized before experimentation, as some constructs may result in overexpression or irregular degradation of the protein of interest.
In addition to isolation and purification of single proteins, the HaloTag system can be implemented for isolating protein complexes and cross-linked protein–DNA complexes directly from cell lysates.38 In addition, purified protein–DNA complexes are useful for determining protein binding sites in the genome using microarray analysis and other molecular techniques. In one instance, the multiprotein complex consisting of human eukaryotic RNA polymerases (RNAP) I, II, and III was captured using the HaloTag system from mammalian cells.39 Similarly, a hybrid DNA–protein device based on the activity of cytochrome P450 BM3 was created by Erkelenz et al. using HaloTag technology, exploiting the potential use of HaloTag for genetic engineering in the future.40
Evaluation of Protein Function
Understanding protein function is critical for the design of new therapeutic agents.41 Evaluation of protein function using the HaloTag system can be executed after the protein of interest is efficiently captured and purified. For example, Ai et al. investigated the role of proprotein convertase subtilisin-like kexin type 9 (PCSK9) in both extracellular and cytosolic locations within individual cells using the HaloTag system.42 This was accomplished using a stable cell line expressing HaloTag–PCSK9 in combination with two HaloTag ligands. Cells were incubated with either a cell-permeable ligand (HaloTag TMR) or cell-impermeable ligand (HaloTag Alexa Fluor 488), allowing interactions between PCSK9 and low density lipoprotein receptor (LDLR) to be examined both in the extracellular and intracellular spaces, respectively. Similarly, Mossuto et al. used HaloTag to examine the activity of proteins within early secretory compartments, essentially following the secretion and degradation of endoplasmic reticulum (ER) proteins.43 In this study, both individual proteins and protein aggregates were examined to determine possible protein–protein interactions.
The HaloTag system can be tailored by researchers for studying different disease models, including models in bacteriology and virology. For example, Liu et al. studied the membrane topology of glycoprotein-41 (gp41) of human immunodeficiency virus (HIV) in mammalian cells, striving to discover the number of membrane-spanning domains (MSD) of gp41 using HaloTag.44 Utilizing two distinct HaloTag ligands, this study revealed that gp41 possesses a single MSD. A similar study utilizing HaloTag for studying HIV proteins compared the relative intensity of fluorescence from the HaloTag system with that of green fluorescent protein (GFP). HaloTag was shown to be superior in fluorescence intensity, and the authors noted that HaloTag functioned better than GFP under acidic conditions.45 In addition, HaloTag was employed for examining the mechanism of HIV-1 glycoprotein membrane fusion, allowing scientists to determine critical residues necessary for inhibiting membrane fusion.46
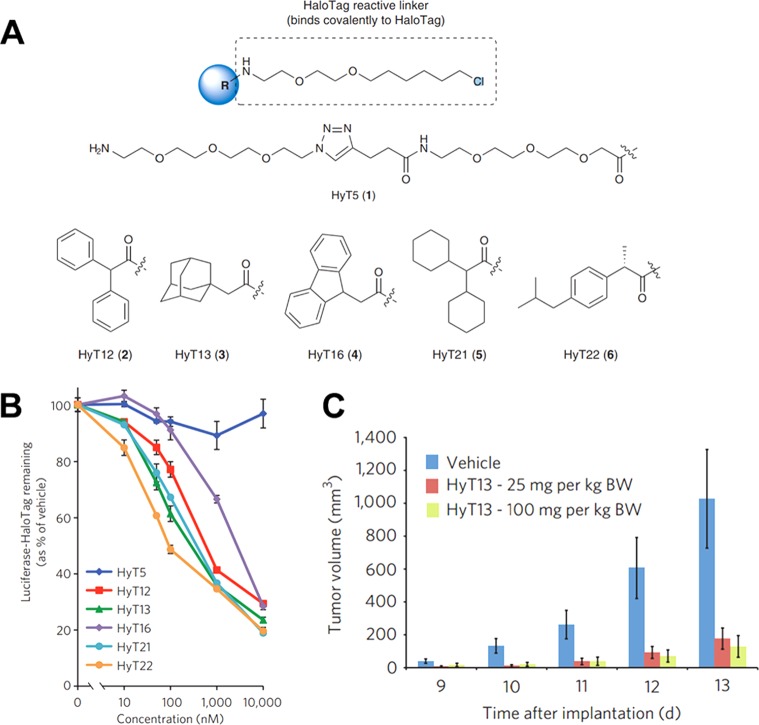
Examination of protein degradation provides clues into the dynamics of protein function and mechanisms of cell apoptosis. Inducing protein degradation in certain disease models could be a potential treatment option in the future.47,48 For example, Neklesa et al. revealed that attaching small hydrophobic molecules to the surface of a specific protein could result in protein degradation via cellular processes (e.g., proteasomes and lysosomes).49 To demonstrate this concept, small hydrophobic ligands were designed to specifically bind a HaloTag protein (Figure 3A). Most of the small hydrophobic ligands could induce protein degradation at concentrations above 10 nM, resulting in cell death (Figure 3B). In addition, one of the novel hydrophobic ligands (HyT13) was shown to inhibit tumor growth of NIH-3T3 flank tumors that expressed HaloTag protein (Figure 3C). Results from this study affirmed that hydrophobic tagging of HaloTag proteins could result in the degradation of both cytosolic and membrane-bound proteins. Furthermore, an additional study from this group examined additional hydrophobic tags capable of inducing protein degradation.50 Interestingly, it was found that only certain cases of protein degradation were directly linked to cellular processes. Other cases of degradation were caused by the direct binding of the hydrophobic molecule to the protein, which resulted in conformational changes.
Figure 3.
Hydrophobic molecules induce degradation of HaloTag proteins. (A) Chemical structure of six hydrophobic HaloTag ligands. (B) Human embryonic kidney cell line, HEK 293T, stably expressing luciferase-modified HaloTag protein was used to measure the biological activity of hydrophobic HaloTag ligands. (C) NIH-3T3 xenografts expressing HaloTag protein were implanted into mice. Tumor growth was monitored in the presence of a hydrophobic HaloTag ligand (HyT13). Reprinted with permission from ref (49). Copyright 2011 Macmillan Publishers Limited.
The HaloTag platform has also been employed to study membrane-bound proteins. For example, HaloTag was utilized to examine the role of surface-bound glycosaminoglycan in causing accelerated embryonic stem cell differentiation into neurons.51 Similarly, membrane electrical potentials were studied in conjunction with the HaloTag system using Förster resonance energy transfer (FRET).52 In addition to the study of membrane-bound proteins, researchers have chemically synthesized mega-molecules with well-defined structures by combining recombinant proteins with linkers in cooperation with the HaloTag platform.53
Analyzing Protein–Protein and Protein–DNA Interactions
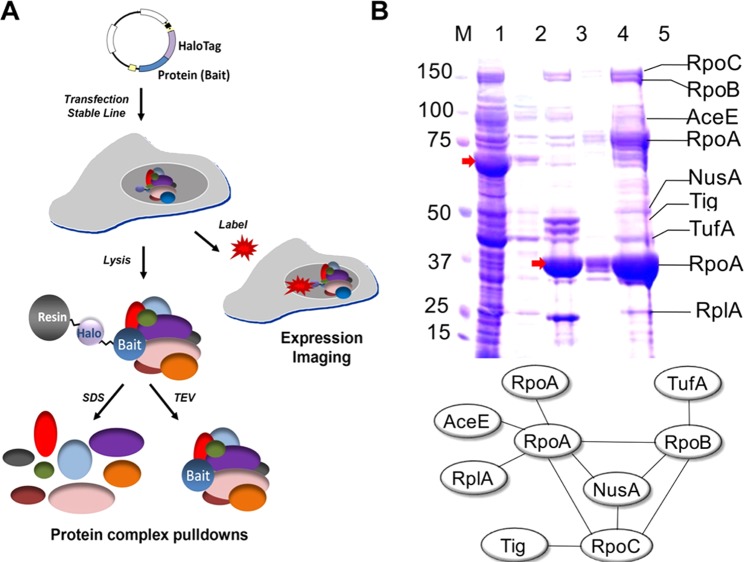
Most cellular processes are dependent on the formation of protein complexes.54 A common protein–protein interaction occurs during cellular signaling when an endogenous ligand binds to a membrane-bound surface receptor, resulting in an intracellular effect.55 Monitoring protein–protein and protein–DNA interactions requires complex techniques, such as pull-down assays.56 For example, interactions of bromodomain protein (BRD4) and histone deacetylase (HDAC1) with additional proteins were examined with the assistance of HaloTag.57 In this pull-down assay, the bait protein linked to HaloTag formed standard protein–protein interactions within the cell. Next, cells were lysed to release the protein complexes and analyzed with liquid chromatography–mass spectrometry (LC–MS) to determine interacting proteins (Figure 4A).57 Pull-down assays are relatively common for protein analysis, as they provide details regarding complex protein interactions and possibly novel interactions. Additional studies evaluating the use of HaloTag for the extraction and purification of protein complexes using pull-down assays have been performed in both bacteria and mammalian cells.58−60
Figure 4.
Pull-down assays for the discovery of protein complexes. (A) Schematic illustration of HaloTag pull-down assays, in which a single HaloTag construct encoding a bait protein is stably transfected into a cell line. The bait protein interacts with additional proteins, at which time cells are lysed and captured using HaloLink resin. Pure proteins can be eluted using a detergent (e.g., SDS), or protein complexes attached the bait protein can be eluted using TEV cleavage. Reprinted with permission from ref (57). Copyright 2014 JoVE. (B) A pull-down assay was performed using HaloTag-modified RpoA to determine the efficiency of HaloTag to extract multiprotein complexes. M, molecular weight marker; 1, unbound proteins; 2, washed proteins; 3, eluted proteins after TEV cleavage; 4, eluted proteins after removal of TEV; and 5, concentrated protein sample. Arrows indicate recombinant HaloTag in lane 1 and cleaved RpoA in lane 3. In addition, a protein interaction map was constructed from data using MALDI-MS/MS. Reprinted with permission from ref (61). Copyright 2012 Peterson and Kwon.
The capability of covalently linking fluorescent tags to HaloTag proteins makes it possible to monitor protein movement in vitro. While this will be discussed further in the In Vivo Molecular Imaging section, many studies examining functionality of proteins utilize fluorescence tagging. For example, HaloTag and SNAP-tag (HaXS) were used in combination as heterodimers to analyze protein targeting to the cytoskeleton, cytoplasm, and lysosomes.62 An additional study using a similar technique developed a novel photocleavable chemical inducer of dimerization (CID) using HaloTag and SNAP-tag systems. This system was utilized to study the translocation of several cellular organelles while monitoring protein movement and relocation with heightened spatiotemporal precision.63 Furthermore, the delivery of electrophiles to cellular target proteins upon photoactivation was demonstrated by Fang et al. using the HaloTag platform.64
To study possible interactions of Yersinia pestis type 3 secretion system (T3SS) with other proteins, Peterson and Kwon developed a novel microarray system in union with HaloTag technology.61 This study evaluated the use of HaloTag for capturing a large protein complex, RNA polymerase, using RpoA as the bait protein. Using SDS-PAGE in combination with MALDI-MS/MS, protein interactions were identified, and a protein interaction map was constructed (Figure 4B).61 Similarly, Camarda et al. employed GFP and HaloTag protein fusions to discover novel protein interactions during gametocyte development in Plasmodium falciparum.65
Fluorescence resonance energy transfer (FRET) is beneficial for analyzing protein–protein interactions in living cells.66,67 Padilla-Parra et al. utilized FRET and demonstrated that HaloTag protein linked with enhanced green fluorescent protein (GFP) displayed superior fluorescence stability in comparison to that of other bioconjugated dyes.68 In addition, FRET was also employed for the detection of nucleic acid using the HaloTag system.69
Examination of protein–protein interactions occurring in the plasma membrane can be challenging due to the poor permeability properties of various targeting ligands.70 The HaloTag platform has surmounted these limitations in several instances, allowing researchers to visualize interactions occurring among extracellular, intracellular, and transmembrane proteins through micropatterning techniques.71 For instance, HaloTag was adapted to investigate the epidermal growth factor receptor (EGFR)–Ras–extracellular signal-regulated kinase (ERK) mitogen-activated protein (MAP) kinase pathway in living cells to measure the dissociation constants of several protein complexes.72 Values obtained from this study were adapted for computation simulations to assist in analyzing possible competitive effects in signal transduction pathways. In addition, HaloTag has been employed for the investigation of protein–DNA interactions using high-throughput methods for functional analysis of human genes73 and conjugation of DNA oligonucleotides to Fab fragments as a potential cancer diagnostic tool.74
Proteomic Analysis Using Protein Assays
Dismal survival rates of specific diseases can often be linked to insufficient tools for their early detection.75 To overcome this pitfall, researchers are investigating novel diagnostic assays as a route for detecting disease at earlier stages, when treatment is optimal.76,77 This is particularly important for individuals who may be more susceptible to disease, such as cigarette smokers who have an elevated risk of developing lung cancer as compared to that of nonsmokers.78,79 For this reason, HaloTag was utilized in the development of a diagnostic assay for detecting lung cancer in patient samples. The assay was constructed to examine 14 tumor-associated markers using the high-throughput Luminex and HaloTag platforms.43 In combination, the assay accurately distinguished >80% of lung cancer cases from the health control group. Similarly, an additional HaloTag-modified assay to measure CREB binding was shown to function equivalently to the commonly utilized high-throughput promoter–luciferase reporter assay.80
Development of novel drug molecules is dependent on high-throughput testing to investigate the efficacy of potential drug compounds in cell lines. This process eliminates potential drug molecules having infinitesimal activity versus highly efficacious drug candidates.81,82 HaloTag was utilized by Wagner et al. in the development of a novel assay to screen potential small molecule binding inhibitors.83 Similarly, Wang et al. developed a protein microarray using HaloTag-modified proteins conjugated to the matrix of the assay plate.84 This assay was functional for both denatured and nondenatured proteins, adding to the versatility of the technique. In addition, Gu et al. adapted the HaloTag platform to create DNA-barcoded proteins for the rapid quantification of protein interactions in cells.85
Small molecule microarrays (SMM) are convenient for screening protein interactions with small molecules.86 Using the HaloTag system, Noblin et al. was able to construct SMM with enhanced sensitivity for the multiplex screening of 20 000 compounds.87 An additional application of the HaloTag technology is cell sorting by labeling cells using cell-surface HaloTag proteins alone or in combination with cell viability markers. In conjunction with flow and laser scanning cytometry, statistical measurements of protein expression in individual or groups of cells can be analyzed.88 Similar techniques were utilized in the development of dual reporter genes for evaluating the process of mRNA splicing using the HaloTag platform.89
In Vitro Cellular Imaging
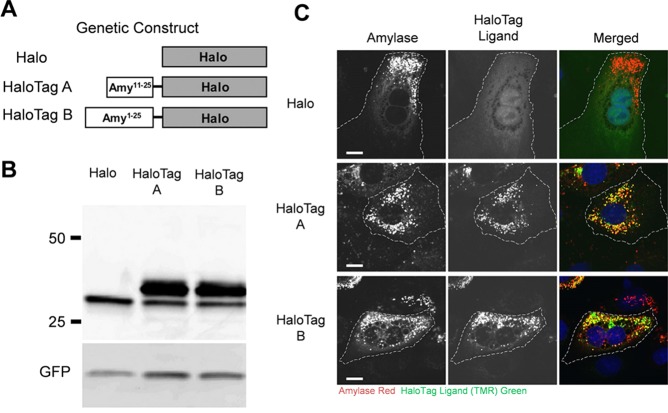
Imaging protein activity in cells is critical for understanding the complex dynamics of cellular signaling.90 Individual proteins can be tagged using the HaloTag system to monitor them efficiently with microscopic techniques. For example, the HaloTag system was employed to investigate the primary cellular localization of matrix metalloproteinase-2 (MMP-2) in cardiomyocytes.91 Through immunofluorescence analysis, MMP-2 was shown to localize primarily in the endoplasmic reticulum (ER), with minimal localization in the mitochondria. Using similar methods, the mechanism of accumulation into secretory granules of exocrine cells by amylase was visualized using a partial sequence of salivary amylase protein fused with HaloTag (Figure 5).92 In this study, two amylase-modified HaloTag proteins were investigated after ensuring that the HaloTag conjugation was successful (Figure 5A,B). Through characterization using fluorescent microscopy, they showed that the HaloTag–amylase protein was similar to endogenous amylase, as it colocalized in similar cellular regions (Figure 5C).
Figure 5.
Intracellular imaging of HaloTag-modified amylase using immunofluorescence microscopy. (A) Two HaloTag–amylase proteins (HaloTag A and HaloTag B) were constructed, as the exact translational start site for salivary amylase has not been identified. (B) Expression of nonconjugated HaloTag (Halo) and both HaloTag A and B proteins was examined. The top band represents the HaloTag complex, whereas the smaller band is indicative of pure HaloTag without amylase attached. (C) Halo, HaloTag A, and HaloTag B were labeled with a HaloTag ligand (TMR-Green), and secretory granules were labeled with an anti-amylase antibody (shown in red). Both HaloTag A and B show colocalization with endogenous amylase, indicating that both were in secretory granules. Scale bar = 10 μm. Reprinted with permission from ref (92). Copyright 2013 the American Physiological Society.
The HaloTag system can be adapted for investigating protein synthesis and degradation in vitro, as HaloTag proteins emit light only when bound to fluorescent HaloTag ligands.93 For example, HaloTag was utilized to fluorescently label peroxisomes in mammalian cells to examine peroxisome protein synthesis at various times points.94 Similarly, Takemoto et al. described a process in which a photosensitizing agent (eosin) modified with HaloTag could be employed for chromophore reassisted light inactivation.95
To assist in developing new imaging agents for use with the HaloTag platform, Singh et al. developed a novel class of oligodeoxyfluorosides (ODFs) that could be used for fluorescence imaging.96 The novel ODFs were composed of short oligomers containing fluorescent chromophores in place of natural nucleic acid bases. The ODFs could be used for either cell-surface or cytoplasmic labeling with a broad range of emission colors and contained a single excitation wavelength. It was shown that many ODFs undergo color changes or experience elevated intensities of their fluorescence when the HaloTag protein interacted with other cellular proteins.96 In a similar study, Liu et al. developed quantum dots targeted with lipoic acid ligase using the HaloTag platform.97 Two quantum dots possessing distinct emission spectra were utilized for imaging single molecules of neurexin in live cells. There have been additional studies investigating the potential uses of quantum dots in combination with the HaloTag system for cellular imaging.98,99 A study by Chen et al. analyzed tissue factor assembly on DNA target sites using both in vitro and in vivo single molecule imaging with the HaloTag system.100
While most protein tagging systems require additional genetic constructs for comprehensive protein analysis, the HaloTag system requires only a separate ligand for each application. For example, HaloTag was employed for investigating the expression and spatial trafficking of integrin in individual cells using a fluorescent HaloTag ligand, and an additional affinity tag was utilized to capture and sort the cells.101 Several additional studies have utilized the HaloTag platform for imaging cellular events, including visualization of clathrin-coated pits,102 monitoring chaperone-mediated autophagy,103 and investigating peroxisome growth and degradation.104,105
Recently, Li et al. designed a Zn2+ fluorescent indicator using HaloTag technology to study the dynamics of secretory granules in living cells.106 Using similar techniques, an examination of retrograde protein transport in rat sympathetic neurons was accomplished using the HaloTag system.107 It is also important to note that the binding of HaloTag protein to fluorescent HaloTag ligands can often overcome the photoswitching artifacts commonly seen with other fluorescent markers, such as GFP.108
The HaloTag system has also been studied in both prokaryotic and plant cells. Using the HaloTag system, Stagge et al. developed an electroporation-based labeling technique for monitoring individual proteins in yeast with super-resolution microscopy.109 Furthermore, HaloTag was also shown to effectively cross the cell wall, allowing for real-time imaging of plant cells in vitro.110 Additional studies have described methods for super-resolution imaging of live bacteria and parasites using the HaloTag system.111,112
In Vivo Molecular Imaging
Tracking of proteins in vivo remains problematic due to the complexity of biological systems. Molecular imaging remains the most promising noninvasive method for monitoring disease progression while providing insight into molecular pathways occurring in vivo.113 Molecular imaging with the HaloTag system can be accomplished by transplanting cells expressing HaloTag proteins into the animal model and subsequently injecting a dose of the HaloTag ligand.114 Currently, HaloTag has been successfully utilized for optical/fluorescent, positron emission tomography (PET), and magnetic resonance (MR) imaging.
Several studies have adapted the HaloTag technology for optical imaging techniques. For example, Tseng et al. employed a HCT116 xenograft model expressing HaloTag protein for whole animal fluorescence imaging.115 The HaloTag ligand displayed enhanced uptake in the HaloTag-expressing tumor, with minimal uptake in the control tumor. Similarly, tumor nodules from cancer cells highly expressing HaloTag receptors were conjugated with four different fluorophore ligands to evaluate cellular growth at specific time points.116 In addition, monitoring hypoxia in vivo was accomplished by successfully targeting hypoxic regions of the tumor using HaloTag ligands.117 Similar studies have utilized the HaloTag platform to investigate autophagy and cellular hypoxia using automated fluorescence microscopy and flow cytometry.118,119
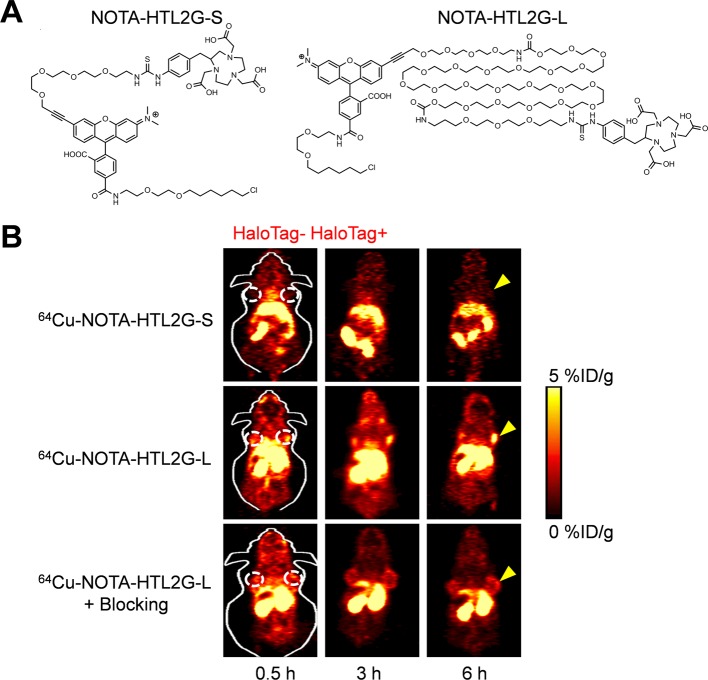
The HaloTag platform has also been evaluated for use in PET and MR imaging as potential imaging agents. Hong et al. employed a breast cancer model (4T1) expressing HaloTag to investigate the uptake of novel radiolabeled HaloTag ligands for PET imaging.120,121 Two novel radiolabeled ligands were synthesized and termed NOTA-HTL2G-S and NOTA-HTL2G-L. The ligands were identical except for the length of poly(ethylene glycol) (PEG) chains, with ligands having short (S) or long (L) PEG chains (Figure 6A).120 Uptake of NOTA-HTL2G-L was significantly higher than uptake of NOTA-HTL2G-S in the HaloTag-expressing tumor (Figure 6B), successfully validating the potential use of HaloTag for PET imaging.120 In addition, blocking studies confirmed that NOTA-HTL2G-L binds specifically to the HaloTag-expressing cell line, providing additional evidence that accumulation was not attributed to passive targeting effects.
Figure 6.
PET imaging of 4T1 tumor-bearing mice with novel HaloTag ligands. (A) Chemical structure of NOTA-HTL2G-S and NOTA-HTL2G-L, with different lengths of PEG. (B) PET imaging of mice with two 4T1 xenografts; the left tumor does not express HaloTag protein, and the right tumor expresses HaloTag protein. Mice were injected with the short (64Cu-NOTA-HTL2G-S) or long (64Cu-NOTA-HTL2G-L) form of the HaloTag ligand. In addition, a blocking agent was used with 64Cu-NOTA-HTL2G-L. Accumulation of 64Cu-NOTA-HTL2G-L ligand can be seen in HaloTag tumors from 3 to 24 h postinjection. Reprinted with permission from ref (120). Copyright 2013 AJTR.
In comparison to PET imaging, MRI suffers from low sensitivity and elevated background noise. Contrast agents are provided to improve sensitivity in some instances. Recently, a novel HaloTag ligand was investigated as a potential MR imaging agent by Strauch et al., who developed a HaloTag–Gd(III) complex known as 2CHTGd.122 The novel contrast agent, acting as a HaloTag ligand, resulted in a 6-fold increase in relativity when the ligand reached the target tissue. In addition, the ligand possessed a prolonged circulation time, allowing for more imaging opportunities.
Conclusions
HaloTag is a versatile protein labeling system that can be utilized for several biomedical applications, as summarized in Table 1. While traditional protein tagging systems are limited to protein isolation and purification, the HaloTag system has overcome this limitation, as a single genetic construct allows proteins to be comprehensively analyzed. This review has presented the current applications of HaloTag for protein analysis, demonstrating that HaloTag can be employed for protein isolation and purification, evaluating protein function, investigating protein–protein and protein–DNA interactions, detecting disease through assays, monitoring protein movement and localization in vitro, and imaging in vivo.
Table 1. Select Proteins Successfully Isolated and Purified Using the HaloTag Platform.
| protein category | examples | refs |
|---|---|---|
| cell membrane | CB2, gp41 | (19, 44, 46) |
| intracellular | SRC-1, PKCγ, PI3Kγ, CARM-1, PCSK9, BRD4 | (20−22, 42, 57) |
| extracellular | Ameloblastin, MMP-2, glycosaminoglycans, Amylase | (27, 51, 91, 92) |
| enzymes | RNA Polymerase, RpoA, Cytochrome P450 | (39, 40, 61) |
| antibodies | Polyclonal, Fab fragments | (33, 34, 74) |
| ribosomes | (35−37) |
While this review has mentioned several advantages of the HaloTag platform for protein labeling, there are some limitations that should be addressed in future studies. One limitation of the HaloTag system is the requirement of various ligands for different applications; thus, its applications may be limited by the availability of ligands. Researchers may design their own HaloTag ligands if a ligand does not currently exist for the application of interest. An additional limitation of the HaloTag system occurs when the addition of a protein tag results in an inactive protein. In most cases, functionality of the protein can be restored by attaching the HaloTag protein to the opposite terminus of the protein. For example, Locatelli-Hoops et al. had to attach the HaloTag at the C-terminus of the protein, as attachment at the N-terminal location resulted in an inactive protein.19 Additionally, the HaloTag protein can demonstrate alterations in protein function, as the tag may result in conformational changes.
While this review has focused on HaloTag technology, there are several other commercially available protein tagging systems, including His-tag,123 FLAG-tag,124 SNAP-tag,125 LigandLink,126 and AviTag,127,128 among others. His-tag is the most widely employed system, as the small size of the tag results in minimal alterations in protein conformation or function.129 While it is effective for isolation of most proteins, some disadvantages of the His-tag system include low purity due to the co-elution of other histidine-rich proteins, potential degradation of the tag, and dimer or trimer formation.129 FLAG-tag is another widely employed protein tagging system, as it produces highly purified protein for cellular imaging and other applications, yet this system is often limited by expensive, low-capacity resins.9 A newer system called LigandLink utilizes the binding ability of a modified bacterial dihydrofolate reductase for trimethoprim, through construction of trimethoprim-coupled fluorescent ligands.126 Additionally, there are several other protein tagging systems currently available or in development, yet each system is often application-specific.
Because each protein tagging system has advantages and disadvantages, researchers should choose a system that will best align with their experimental goals. In general, all fusion tag-based systems are designed to specifically label a protein of interest for various applications. The main differences between systems are the type of tag utilized (peptide, chemical) and the molecular mechanism of the tagging system.126 In general, antibodies remain an optimal method for protein isolation and analysis, yet they are expensive and not available for all targets. This review focused on the versatility of the HaloTag system, as other systems are often limited to one or two applications. In addition, there are several potential uses of the HaloTag platform. Some potential applications of the HaloTag system include the formation of novel assays for detecting genetic diseases, photoacoustic imaging of orthotopic tumors expressing HaloTag in vivo, and further evaluation of highly complex protein interactions. Since this technology is relatively new, there is still limited research into all of the potential biomedical applications of HaloTag.
Acknowledgments
This work was supported, in part, by the University of Wisconsin–Madison, the Department of Defense (W81XWH-11-1-0644), the National Institutes of Health (NIBIB/NCI R01CA169365, P30CA014520, T32CA009206), and the American Cancer Society (125246-RSG-13-099-01-CCE).
The authors declare no competing financial interest.
References
- Toyama B. H.; Hetzer M. W. (2013) Protein homeostasis: live long, won’t prosper. Nat. Rev. Mol. Cell Biol. 14, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C. V.; Sali A.; Baumeister W. (2007) The molecular sociology of the cell. Nature 450, 973–982. [DOI] [PubMed] [Google Scholar]
- Hanash S. (2003) Disease proteomics. Nature 422, 226–232. [DOI] [PubMed] [Google Scholar]
- Hanash S. M.; Pitteri S. J.; Faca V. M. (2008) Mining the plasma proteome for cancer biomarkers. Nature 452, 571–579. [DOI] [PubMed] [Google Scholar]
- Kolch W.; Mischak H.; Pitt A. R. (2005) The molecular make-up of a tumour: proteomics in cancer research. Clin. Sci. 108, 369–383. [DOI] [PubMed] [Google Scholar]
- McHugh C. A.; Russell P.; Guttman M. (2014) Methods for comprehensive experimental identification of RNA–protein interactions. Genome Biol. 15, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngounou Wetie A. G.; Sokolowska I.; Woods A. G.; Roy U.; Deinhardt K.; Darie C. C. (2014) Protein–protein interactions: switch from classical methods to proteomics and bioinformatics-based approaches. Cell. Mol. Life Sci. 71, 205–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornhorst J. A.; Falke J. J. (2000) Purification of proteins using polyhistidine affinity tags. Methods Enzymol. 326, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichty J. J.; Malecki J. L.; Agnew H. D.; Michelson-Horowitz D. J.; Tan S. (2005) Comparison of affinity tags for protein purification. Protein Expression Purif. 41, 98–105. [DOI] [PubMed] [Google Scholar]
- Li Y. F. (2010) Commonly used tag combinations for tandem affinity purification. Biotechnol. Appl. Biochem. 55, 73–83. [DOI] [PubMed] [Google Scholar]
- Urh M.; Rosenberg M. (2012) HaloTag, a platform technology for protein analysis. Curr. Chem. Genomics 6, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encell L. P.; Friedman Ohana R.; Zimmerman K.; Otto P.; Vidugiris G.; Wood M. G.; Los G. V.; McDougall M. G.; Zimprich C.; Karassina N.; et al. (2012) Development of a dehalogenase-based protein fusion tag capable of rapid, selective and covalent attachment to customizable ligands. Curr. Chem. Genomics 6, 55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa I.; Berkovich R.; Alegre-Cebollada J.; Badilla C. L.; Rivas-Pardo J. A.; Taniguchi Y.; Kawakami M.; Fernandez J. M. (2013) Nanomechanics of HaloTag tethers. J. Am. Chem. Soc. 135, 12762–12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los G. V.; Encell L. P.; McDougall M. G.; Hartzell D. D.; Karassina N.; Zimprich C.; Wood M. G.; Learish R.; Ohana R. F.; Urh M.; et al. (2008) HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382. [DOI] [PubMed] [Google Scholar]
- Benink H. A.; Urh M. (2015) HaloTag technology for specific and covalent labeling of fusion proteins. Methods Mol. Biol. 1266, 119–128. [DOI] [PubMed] [Google Scholar]
- Los G. V.; Wood K. (2007) The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol. Biol. 356, 195–208. [DOI] [PubMed] [Google Scholar]
- Pina A. S.; Lowe C. R.; Roque A. C. (2014) Challenges and opportunities in the purification of recombinant tagged proteins. Biotechnol. Adv. 32, 366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey M.; Emoto K.; Takahashi H.; Castner D. G.; Grainger D. W. (2009) Affinity-based protein surface pattern formation by ligand self-selection from mixed protein solutions. Adv. Funct. Mater. 19, 3046–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli-Hoops S.; Sheen F. C.; Zoubak L.; Gawrisch K.; Yeliseev A. A. (2013) Application of HaloTag technology to expression and purification of cannabinoid receptor CB2. Protein Expression Purif. 89, 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohana R. F.; Hurst R.; Vidugiriene J.; Slater M. R.; Wood K. V.; Urh M. (2011) HaloTag-based purification of functional human kinases from mammalian cells. Protein Expression Purif. 76, 154–164. [DOI] [PubMed] [Google Scholar]
- Seddon A. M.; Curnow P.; Booth P. J. (2004) Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta, Biomembr. 1666, 105–117. [DOI] [PubMed] [Google Scholar]
- Chumanov R. S.; Kuhn P. A.; Xu W.; Burgess R. R. (2011) Expression and purification of full-length mouse CARM1 from transiently transfected HEK293T cells using HaloTag technology. Protein Expression Purif. 76, 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul J.; Petritis B.; Sau S.; Rauf F.; Gaskin M.; Ober-Reynolds B.; Mineyev I.; Magee M.; Chaput J.; Qiu J.; et al. (2014) Development of a full-length human protein production pipeline. Protein Sci. 23, 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath N.; Hurst R.; Hook B.; Meisenheimer P.; Zhao K. Q.; Nassif N.; Bulleit R. F.; Storts D. R. (2008) Improving protein array performance: focus on washing and storage conditions. J. Proteome Res. 7, 4475–4482. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y.; Kawakami M. (2010) Application of HaloTag protein to covalent immobilization of recombinant proteins for single molecule force spectroscopy. Langmuir 26, 10433–10446. [DOI] [PubMed] [Google Scholar]
- Urh M.; Hartzell D.; Mendez J.; Klaubert D. H.; Wood K. (2008) Methods for detection of protein-protein and protein-DNA interactions using HaloTag. Methods Mol. Biol. 421, 191–209. [DOI] [PubMed] [Google Scholar]
- Saito N.; Ariyoshi W.; Okinaga T.; Kamegawa M.; Matsukizono M.; Akebiyama Y.; Kitamura C.; Nishihara T. (2014) Inhibitory effects of ameloblastin on epithelial cell proliferation. Arch Oral Biol. 59, 835–840. [DOI] [PubMed] [Google Scholar]
- Olsen M. J.; Stephens D.; Griffiths D.; Daugherty P.; Georgiou G.; Iverson B. L. (2000) Function-based isolation of novel enzymes from a large library. Nat. Biotechnol. 18, 1071–1074. [DOI] [PubMed] [Google Scholar]
- Milner S. E.; Maguire A. R. (2012) Recent trends in whole cell and isolated enzymes in enantioselective synthesis. Arkivoc 321–382. [Google Scholar]
- Motejadded H.; Kranz B.; Berensmeier S.; Franzreb M.; Altenbuchner J. (2010) Expression, one-step purification, and immobilization of HaloTag (TM) fusion proteins on chloroalkane-functionalized magnetic beads. Appl. Biochem. Biotechnol. 162, 2098–2110. [DOI] [PubMed] [Google Scholar]
- Leippe D. M.; Zhao K. Q.; Hsiao K.; Slater M. R. (2010) Cell-free expression of protein kinase a for rapid activity assays. Anal Chem. Insights 5, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigham J. L.; Perera B. G.; Maly D. J. (2013) A hexylchloride-based catch-and-release system for chemical proteomic applications. ACS Chem. Biol. 8, 691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag R.; Hilbrig F. (2012) Isolation and purification of recombinant proteins, antibodies and plasmid DNA with hydroxyapatite chromatography. Biotechnol. J. 7, 90–102. [DOI] [PubMed] [Google Scholar]
- Hata T.; Nakayama M. (2007) Rapid single-tube method for small-scale affinity purification of polyclonal antibodies using HaloTag (TM) Technology. J. Biochem. Biophys. Methods 70, 679–682. [DOI] [PubMed] [Google Scholar]
- Imataka H. (2012) Single-molecule imaging with a tagged ribosome to explore trans-translation. J. Biochem. 152, 293–295. [DOI] [PubMed] [Google Scholar]
- Zhou Z. P.; Shimizu Y.; Tadakuma H.; Taguchi H.; Ito K.; Ueda T. (2011) Single molecule imaging of the trans-translation entry process via anchoring of the tagged ribosome. J. Biochem. 149, 609–618. [DOI] [PubMed] [Google Scholar]
- Gallo S.; Beugnet A.; Biffo S. (2011) Tagging of functional ribosomes in living cells by HaloTagA (R) technology. In Vitro Cell. Dev. Biol.: Anim. 47, 132–138. [DOI] [PubMed] [Google Scholar]
- Daniels D.; Urh M. (2013) Isolation of intracellular protein–DNA complexes using HaloCHIP, an antibody-free alternative to chromatin immunoprecipitation. Methods Mol. Biol. 977, 111–124. [DOI] [PubMed] [Google Scholar]
- Daniels D. L.; Mendez J.; Mosley A. L.; Ramisetty S. R.; Murphy N.; Benink H.; Wood K. V.; Urh M.; Washburn M. P. (2012) Examining the complexity of human RNA polymerase complexes using HaloTag technology coupled to label free quantitative proteomics. J. Proteome Res. 11, 564–575. [DOI] [PubMed] [Google Scholar]
- Erkelenz M.; Kuo C. H.; Niemeyer C. M. (2011) DNA-mediated assembly of cytochrome P450 BM3 subdomains. J. Am. Chem. Soc. 133, 16111–16118. [DOI] [PubMed] [Google Scholar]
- Takeda-Shitaka M.; Takaya D.; Chiba C.; Tanaka H.; Umeyama H. (2004) Protein structure prediction in structure based drug design. Curr. Med. Chem. 11, 551–558. [DOI] [PubMed] [Google Scholar]
- Ai X.; Fischer P.; Palyha O. C.; Wisniewski D.; Hubbard B.; Akinsanya K.; Strack A. M.; Ehrhardt A. G. (2012) Utilizing HaloTag technology to track the fate of PCSK9 from intracellular vs. extracellular sources. Curr. Chem. Genomics 6, 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossuto M. F.; Sannino S.; Mazza D.; Fagioli C.; Vitale M.; Yoboue E. D.; Sitia R.; Anelli T. (2014) A dynamic study of protein secretion and aggregation in the secretory pathway. PLoS One 9, e108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Kondo N.; Long Y.; Xiao D.; Iwamoto A.; Matsuda Z. (2010) Membrane topology analysis of HIV-1 envelope glycoprotein gp41. Retrovirology 7, 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H.; Meng F. X.; Kondo N.; Iwamoto A.; Matsuda Z. (2012) Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP. Protein Eng., Des. Sel. 25, 813–820. [DOI] [PubMed] [Google Scholar]
- Wang H.; Li X.; Nakane S.; Liu S.; Ishikawa H.; Iwamoto A.; Matsuda Z. (2014) Co-expression of foreign proteins tethered to HIV-1 envelope glycoprotein on the cell surface by introducing an intervening second membrane-spanning domain. PLoS One 9, e96790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson L. I.; Davies F. E. (2012) DangER: protein ovERload. Targeting protein degradation to treat myeloma. Haematologica 97, 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K. (2010) Protein degradation: time for trimming. Nat. Rev. Mol. Cell Biol. 11, 754–755. [DOI] [PubMed] [Google Scholar]
- Neklesa T. K.; Tae H. S.; Schneekloth A. R.; Stulberg M. J.; Corson T. W.; Sundberg T. B.; Raina K.; Holley S. A.; Crews C. M. (2011) Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat. Chem. Biol. 7, 538–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tae H. S.; Sundberg T. B.; Neklesa T. K.; Noblin D. J.; Gustafson J. L.; Roth A. G.; Raina K.; Crews C. M. (2012) Identification of hydrophobic tags for the degradation of stabilized proteins. ChemBioChem 13, 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsipher A.; Griffin M. E.; Stone S. E.; Hsieh-Wilson L. C. (2014) Long-lived engineering of glycans to direct stem cell fate. Angew. Chem., Int. Ed. 1466–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H.; Jinno Y.; Tomita A.; Okamura Y. (2013) Optically detected structural change in the N-terminal region of the voltage-sensor domain. Biophys. J. 105, 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modica J. A.; Skarpathiotis S.; Mrksich M. (2012) Modular assembly of protein building blocks to create precisely defined megamolecules. ChemBioChem 13, 2331–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.; Thornton J. M. (1995) Protein–protein interactions: a review of protein dimer structures. Prog. Biophys. Mol. Biol. 63, 31–65. [DOI] [PubMed] [Google Scholar]
- Kvernmo T.; Hartter S.; Burger E. (2006) A review of the receptor-binding and pharmacokinetic properties of dopamine agonists. Clin. Ther. 28, 1065–1078. [DOI] [PubMed] [Google Scholar]
- Pollard T. D. (2010) A guide to simple and informative binding assays. Mol. Biol. Cell 21, 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. L.; Méndez J.; Benink H.; Niles A.; Murphy N.; Ford M.; Jones R.; Amunugama R.; Allen D.; et al. (2014) Discovering protein interactions and characterizing protein function using HaloTag technology. J. Visualized Exp. e51553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E.; van Kessel S. P.; van Bezouwen L. S.; Kumar N.; Boekema E. J.; Scheffers D. J. (2012) Bacillus subtilis SepF binds to the C-terminus of FtsZ. PLoS One 7, e43293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst R.; Hook B.; Slater M. R.; Hartnett J.; Storts D. R.; Nath N. (2009) Protein–protein interaction studies on protein arrays: effect of detection strategies on signal-to-background ratios. Anal. Biochem. 392, 45–53. [DOI] [PubMed] [Google Scholar]
- Ose R.; Oharaa O.; Nagase T. (2012) Galectin-1 and galectin-3 mediate protocadherin-24-dependent membrane localization of beta-catenin in colon cancer cell line HCT116. Curr. Chem. Genomics 6, 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.; Kwon K. (2012) The HaloTag: improving soluble expression and applications in protein functional analysis. Curr. Chem. Genomics 6, 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhart D.; Zimmermann M.; Jacques O.; Wittwer M. B.; Ernst B.; Constable E.; Zvelebil M.; Beaufils F.; Wymann M. P. (2013) Chemical development of intracellular protein heterodimerizers. Chem. Biol. 20, 549–557. [DOI] [PubMed] [Google Scholar]
- Zimmermann M.; Cal R.; Janett E.; Hoffmann V.; Bochet C. G.; Constable E.; Beaufils F.; Wymann M. P. (2014) Cell-permeant and photocleavable chemical inducer of dimerization. Angew. Chem., Int. Ed. 53, 4717–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X.; Fu Y.; Long M. J.; Haegele J. A.; Ge E. J.; Parvez S.; Aye Y. (2013) Temporally controlled targeting of 4-hydroxynonenal to specific proteins in living cells. J. Am. Chem. Soc. 135, 14496–14489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarda G.; Bertuccini L.; Singh S. K.; Salzano A. M.; Lanfrancotti A.; Olivieri A.; Scaloni A.; Sharma A.; Alano P. (2010) Regulated oligomerisation and molecular interactions of the early gametocyte protein Pfg27 in Plasmodium falciparum sexual differentiation. Int. J. Parasitol. 40, 663–673. [DOI] [PubMed] [Google Scholar]
- Jares-Erijman E. A.; Jovin T. M. (2003) FRET imaging. Nat. Biotechnol. 21, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Taraska J. W.; Puljung M. C.; Olivier N. B.; Flynn G. E.; Zagotta W. N. (2009) Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat. Methods 6, 532–U594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Parra S.; Auduge N.; Lalucque H.; Mevel J. C.; Coppey-Moisan M.; Tramier M. (2009) Quantitative comparison of different fluorescent protein couples for fast FRET-FLIM acquisition. Biophys. J. 97, 2368–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstock D.; Chen W. (2014) Halo-tag mediated self-labeling of fluorescent proteins to molecular beacons for nucleic acid detection. Chem. Commun. 50, 13735–13738. [DOI] [PubMed] [Google Scholar]
- Lin S. H.; Guidotti G. (2009) Purification of membrane proteins. Methods Enzymol. 463, 619–629. [DOI] [PubMed] [Google Scholar]
- Lochte S.; Waichman S.; Beutel O.; You C. J.; Piehler J. (2014) Live cell micropatterning reveals the dynamics of signaling complexes at the plasma membrane. J. Cell Biol. 207, 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie W.; Harada Y.; Matsuda M.; Aoki K. (2014) Quantitative in vivo fluorescence cross-correlation analyses highlight the importance of competitive effects in the regulation of protein-protein interactions. Mol. Cell. Biol. 34, 3272–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase T.; Yamakawa H.; Tadokoro S.; Nakajima D.; Inoue S.; Yamaguchi K.; Itokawa Y.; Kikuno R. F.; Koga H.; Ohara O. (2008) Exploration of human ORFeome: high-throughput preparation of ORF clones and efficient characterization of their protein products. DNA Res. 15, 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakkila H.; Peltomaa R.; Lamminmaki U.; Soukka T. (2014) Precise construction of oligonucleotide–Fab fragment conjugate for homogeneous immunoassay using HaloTag technology. Anal. Biochem. 37–44. [DOI] [PubMed] [Google Scholar]
- Etzioni R.; Urban N.; Ramsey S.; McIntosh M.; Schwartz S.; Reid B.; Radich J.; Anderson G.; Hartwell L. (2003) The case for early detection. Nat. Rev. Cancer 3, 243–252. [DOI] [PubMed] [Google Scholar]
- Cote G. A.; Gore A. J.; McElyea S. D.; Heathers L. E.; Xu H.; Sherman S.; Korc M. (2014) A pilot study to develop a diagnostic test for pancreatic ductal adenocarcinoma based on differential expression of select miRNA in plasma and bile. Am. J. Gastroenterol. 109, 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianidou E. S.; Strati A.; Markou A. (2014) Circulating tumor cells as promising novel biomarkers in solid cancers. Crit. Rev. Clin. Lab Sci. 51, 160–171. [DOI] [PubMed] [Google Scholar]
- Reck M.; Heigener D. F.; Mok T.; Soria J. C.; Rabe K. F. (2013) Management of non-small-cell lung cancer: recent developments. Lancet 382, 709–719. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Fillmore C. M.; Hammerman P. S.; Kim C. F.; Wong K. K. (2014) Non-small-cell lung cancers: a heterogeneous set of diseases. Nat. Rev. Cancer 14, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell D. D.; Trinklein N. D.; Mendez J.; Murphy N.; Aldred S. F.; Wood K.; Urh M. (2009) A functional analysis of the CREB signaling pathway using HaloCHIP-chip and high throughput reporter assays. BMC Genomics 10, e2774331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.; Nim S.; Teyra J.; Datti A.; Wrana J. L.; Sidhu S. S.; Moffat J.; Kim P. M. (2014) A systematic approach to identify novel cancer drug targets using machine learning, inhibitor design and high-throughput screening. Genome Med. 6, e13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols A. C.; Black M.; Yoo J.; Pinto N.; Fernandes A.; Haibe-Kains B.; Boutros P. C.; Barrett J. W. (2014) Exploiting high-throughput cell line drug screening studies to identify candidate therapeutic agents in head and neck cancer. BMC Pharmacol. Toxicol. 15, e4258049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. K.; Nath N.; Flemming R.; Feltenberger J. B.; Denu J. M. (2012) Identification and characterization of small molecule inhibitors of a plant homeodomain finger. Biochemistry 51, 8293–8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Barker K.; Steel J.; Park J.; Saul J.; Festa F.; Wallstrom G.; Yu X.; Bian X.; Anderson K. S.; et al. (2013) A versatile protein microarray platform enabling antibody profiling against denatured proteins. Proteomics: Clin. Appl. 7, 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L.; Li C.; Aach J.; Hill D. E.; Vidal M.; Church G. M. (2014) Multiplex single-molecule interaction profiling of DNA-barcoded proteins. Nature 515, 554–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegas A. J.; Fuller J. H.; Koehler A. N. (2008) Small-molecule microarrays as tools in ligand discovery. Chem. Soc. Rev. 37, 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noblin D. J.; Page C. M.; Tae H. S.; Gareiss P. C.; Schneekloth J. S.; Crews C. M. (2012) A HaloTag-based small molecule microarray screening methodology with increased sensitivity and multiplex capabilities. ACS Chem. Biol. 7, 2055–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko E.; Ranjbar S.; Jasenosky L.; Goldfeld A.; Vorobjev I.; Barteneva N. (2011) The use of HaloTag-based technology in flow and laser scanning cytometry analysis of live and fixed cells. BMC Res. Notes 4, 340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K.; Nagase T.; Imai K.; Nonoyama S.; Obara M.; Mizukami T.; Nunoi H.; Kanegane H.; Kuribayashi F.; Amemiya S.; et al. (2012) A dual reporter splicing assay using HaloTag-containing proteins. Curr. Chem. Genomics 6, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M.; Karagiannis P.; Taniguchi Y. (2014) Protein expression analyses at the single cell level. Molecules 19, 13932–13947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes B. G.; Fan X. H.; Cho W. J.; Schulz R. (2014) MMP-2 is localized to the mitochondria-associated membrane of the heart. Am. J. Physiol.: Heart Circ. Physiol. 306, H764–H770. [DOI] [PubMed] [Google Scholar]
- Fujita-Yoshigaki J.; Matsuki-Fukushima M.; Yokoyama M.; Katsumata-Kato O. (2013) Sorting of a HaloTag protein that has only a signal peptide sequence into exocrine secretory granules without protein aggregation. Am. J. Physiol.: Gastrointest. Liver Physiol. 305, G685–G696. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K.; Inoue S.; Ohara O.; Nagase T. (2009) Pulse–chase experiment for the analysis of protein stability in cultured mammalian cells by covalent fluorescent labeling of fusion proteins. Methods Mol. Biol. 577, 121–131. [DOI] [PubMed] [Google Scholar]
- Fransen M. (2014) HaloTag as a tool to investigate peroxisome dynamics in cultured mammalian cells. Methods Mol. Biol. 1174, 157–170. [DOI] [PubMed] [Google Scholar]
- Takemoto K.; Matsuda T.; McDougall M.; Klaubert D. H.; Hasegawa A.; Los G. V.; Wood K. V.; Miyawaki A.; Nagai T. (2011) Chromophore-assisted light inactivation of HaloTag fusion proteins labeled with eosin in living cells. ACS Chem. Biol. 6, 401–406. [DOI] [PubMed] [Google Scholar]
- Singh V.; Wang S. L.; Kool E. T. (2013) Genetically encoded multispectral labeling of proteins with polyfluorophores on a DNA backbone. J. Am. Chem. Soc. 135, 6184–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. S.; Phipps W. S.; Loh K. H.; Howarth M.; Ting A. Y. (2012) Quantum dot targeting with lipoic acid ligase and HaloTag for single-molecule imaging on living cells. ACS Nano 6, 11080–11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So M. K.; Yao H.; Rao J. (2008) HaloTag protein-mediated specific labeling of living cells with quantum dots. Biochem. Biophys. Res. Commun. 374, 419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; So M. K.; Loening A. M.; Yao H. Q.; Gambhir S. S.; Rao J. H. (2006) HaloTag protein-mediated site-specific conjugation of bioluminescent proteins to quantum dots. Angew. Chem., Int. Ed. 45, 4936–4940. [DOI] [PubMed] [Google Scholar]
- Chen J. J.; Zhang Z. J.; Li L.; Chen B. C.; Revyakin A.; Hajj B.; Legant W.; Dahan M.; Lionnet T.; Betzig E.; et al. (2014) Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen S.; Zimprich C.; McDougall M. G.; Klaubert D. H.; Los G. V. (2008) Spatial separation and bidirectional trafficking of proteins using a multi-functional reporter. BMC Cell Biol. 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes S.; Staufenbiel M.; Lisse D.; Richter C. P.; Beutel O.; Busch K. B.; Hess S. T.; Piehler J. (2012) Triple-color super-resolution imaging of live cells: resolving submicroscopic receptor organization in the plasma membrane. Angew. Chem., Int. Ed. 51, 4868–4871. [DOI] [PubMed] [Google Scholar]
- Seki T.; Yoshino K. I.; Tanaka S.; Dohi E.; Onji T.; Yamamoto K.; Hide I.; Paulson H. L.; Saito N.; Sakai N. (2012) Establishment of a novel fluorescence-based method to evaluate chaperone-mediated autophagy in a single neuron. PLoS One 7, e31232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delille H. K.; Agricola B.; Guimaraes S. C.; Borta H.; Luers G. H.; Fransen M.; Schrader M. (2010) Pex11pbeta-mediated growth and division of mammalian peroxisomes follows a maturation pathway. J. Cell Sci. 123, 2750–2762. [DOI] [PubMed] [Google Scholar]
- Huybrechts S. J.; Van Veldhoven P. P.; Brees C.; Mannaerts G. P.; Los G. V.; Fransen M. (2009) Peroxisome dynamics in cultured mammalian cells. Traffic 10, 1722–1733. [DOI] [PubMed] [Google Scholar]
- Li D.; Liu L.; Li W. H. (2015) Genetic targeting of a small fluorescent zinc indicator to cell surface for monitoring zinc secretion. ACS Chem. Biol. 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S. A.; Lund K.; LaPointe P.; Campenot R. B. (2013) A HaloTag (R) method for assessing the retrograde axonal transport of the p75 neurotrophin receptor and other proteins in compartmented cultures of rat sympathetic neurons. J. Neurosci. Methods 214, 91–104. [DOI] [PubMed] [Google Scholar]
- Morisaki T.; McNally J. G. (2014) Photoswitching-free FRAP analysis with a genetically encoded fluorescent tag. PLoS One 9, e0107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagge F.; Mitronova G. Y.; Belov V. N.; Wurm C. A.; Jakobs S. (2013) Snap-, CLIP- and Halo-Tag labelling of budding yeast cells. PLoS One 8, e0078745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C.; Schulze J.; Mendel R. R.; Hansch R. (2006) HaloTag (TM): a new versatile reporter gene system in plant cells. J. Exp. Bot. 57, 2985–2992. [DOI] [PubMed] [Google Scholar]
- Lee H. L. D.; Lord S. J.; Iwanaga S.; Zhan K.; Xie H. X.; Williams J. C.; Wang H.; Bowman G. R.; Goley E. D.; Shapiro L.; et al. (2010) Superresolution imaging of targeted proteins in fixed and living cells using photoactivatable organic fluorophores. J. Am. Chem. Soc. 132, 15099–15101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincova E.; Voleman L.; Najdrova V.; De Napoli M.; Eshar S.; Gualdron M.; Hopp C. S.; Sanin D. E.; Tembo D. L.; Van Tyne D.; et al. (2012) Live imaging of mitosomes and hydrogenosomes by HaloTag technology. PLoS One 7, e36314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson P.; McGhee E. J.; Anderson K. I. (2011) Imaging molecular dynamics in vivo—from cell biology to animal models. J. Cell Sci. 124, 2877–2890. [DOI] [PubMed] [Google Scholar]
- Honma N. (2007) HaloTag: a novel technology for in vivo bioimaging and protein functional analysis. Tanpakushitsu Kakusan Koso 52, 1575–1580. [PubMed] [Google Scholar]
- Tseng J. C.; Benink H. A.; McDougall M. G.; Chico-Calero I.; Kung A. L. (2012) In vivo fluorescent labeling of tumor cells with the HaloTag(R) technology. Curr. Chem. Genomics 6, 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N.; Ogawa M.; Choyke P. L.; Karassina N.; Corona C.; McDougall M.; Lynch D. T.; Hoyt C. C.; Levenson R. M.; Los G. V.; et al. (2009) In vivo stable tumor-specific painting in various colors using dehalogenase-based protein-tag fluorescent ligands. Bioconjugate Chem. 20, 1367–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchimaru T.; Kadonosono T.; Tanaka S.; Ushiki T.; Hiraoka M.; Kizaka-Kondoh S. (2010) In vivo imaging of HIF-active tumors by an oxygen-dependent degradation protein probe with an interchangeable labeling system. PLoS One 5, e0015736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BenYounès A.; Tajeddine N.; Tailler M.; Malik S. A.; Shen S.; Métivier D.; Kepp O.; Vitale I.; Maiuri M. C.; Kroemer G. (2011) A fluorescence-microscopic and cytofluorometric system for monitoring the turnover of the autophagic substrate p62/SQSTM1. Autophagy 7, 883–891. [DOI] [PubMed] [Google Scholar]
- Dohi E.; Tanaka S.; Seki T.; Miyagi T.; Hide I.; Takahashi T.; Matsumoto M.; Sakai N. (2012) Hypoxic stress activates chaperone-mediated autophagy and modulates neuronal cell survival. Neurochem. Int. 60, 431–442. [DOI] [PubMed] [Google Scholar]
- Hong H.; Benink H. A.; Uyeda H. T.; Valdovinos H. F.; Zhang Y.; Meisenheimer P.; Barnhart T. E.; Fan F.; Cai W. B. (2013) HaloTag as a reporter gene: positron emission tomography imaging with Cu-64-labeled second generation HaloTag ligands. Am. J. Transl. Res. 5, 291–302. [PMC free article] [PubMed] [Google Scholar]
- Hong H.; Benink H. A.; Zhang Y.; Yang Y. N.; Uyeda H. T.; Engle J. W.; Severin G. W.; McDougall M. G.; Barnhart T. E.; Klaubert D. H.; et al. (2011) HaloTag: a novel reporter gene for positron emission tomography. Am. J. Transl. Res. 3, 392–403. [PMC free article] [PubMed] [Google Scholar]
- Strauch R. C.; Mastarone D. J.; Sukerkar P. A.; Song Y.; Ipsaro J. J.; Meade T. J. (2011) Reporter protein-targeted probes for magnetic resonance imaging. J. Am. Chem. Soc. 133, 16346–16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornhorst J. A., and Falke J. J. (2000) Purification of proteins using polyhistidine affinity tags, in Applications of Chimeric Genes and Hybrid Proteins. Part A, Gene Expression and Protein Purification (Thorner J., Emr S. D., and Abelson J. N., Eds.) Vol. 326, pp 245–254, Academic Press, San Diego, CA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P. M.; Sparrow L. G.; Attwood R. M.; Xiao X. W.; Adams T. E.; McKimm-Breschkin J. L. (2012) Taking down the FLAG! How insect cell expression challenges an established tag-system. PLoS One 7, e37779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein B.; Willig K. I.; Wurm C. A.; Westphal V.; Jakobs S.; Hell S. W. (2010) Stimulated emission depletion nanoscopy of living cells using SNAP-tag fusion proteins. Biophys. J. 98, 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing C. R.; Cornish V. W. (2011) Chemical tags for labeling proteins inside living cells. Acc. Chem. Res. 44, 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaegent M.; Christopoulos T. K. (2002) Recombinant Gaussia luciferase. Overexpression, purification, and analytical application of a bioluminescent reporter for DNA hybridization. Anal. Chem. 74, 4378–4385. [DOI] [PubMed] [Google Scholar]
- Beckett D.; Kovaleva E.; Schatz P. J. (1999) A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpe K. (2003) Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 60, 523–533. [DOI] [PubMed] [Google Scholar]