Introduction
The paradigm of individualized drug therapy based on genetics is an ideal now potentially possible. However, translation of pharmacogenomics into practice has been limited by barriers of availability, cost, and time delays of genetic testing, disagreement about result interpretation, and even lack of understanding about pharmacogenomics in general. We describe our institutional pharmacogenomics implementation project, “The 1200 Patients Project”, a model designed to overcome these barriers and facilitate availability of pharmacogenomic information for personalized prescribing.
Project Context: Overcoming Implementation Barriers
To date, pharmacogenomic information has mostly been cataloged or included in FDA drug labels. Commercial entities have begun to offer fee-for-service pharmacogenomic testing to the public (for some variants/drugs). Yet on a medical system scale, pharmacogenomics offers the potential to better inform the hundreds of thousands of prescribing decisions faced every day during office visits.
We hypothesized that patients and providers are eager to incorporate genetic information into prescribing decisions, but that despite this motivation, current tools for translation of pharmacogenomics in most medical systems make this nearly impossible. We envisioned that development of a new ‘medical system model’ for personalized care is necessary, in which patient genetic information is woven into the clinic visit encounter and advanced information technology catalyzes patient-specific and drug-specific pharmacogenomic consultations to occur.
Our objective was to establish a model system which eliminated practical barriers to pharmacogenomic implementation (Figure 1) and, within a research context, conduct a feasibility study of the incorporation of prospective pharmacogenomic testing. Our model has two hallmarks:
preemptive comprehensive pharmacogenomic genotyping of all enrolled patient participants; and
an interactive informatics portal serving as both a repository for patient-specific pharmacogenomic results and an instantaneous delivery-consultation system to interpret such results for possible clinical translation.
Utilization of these features removes the most common barriers to implementation: physicians no longer have to be aware of (or remember) the existence of a pharmacogenomic relationship; and, because testing is performed preemptively—up-front—there is no time delay for receiving results.
Figure 1. “The 1200 Patients Project” Model for Realizing Pharmacogenomic Implementation.
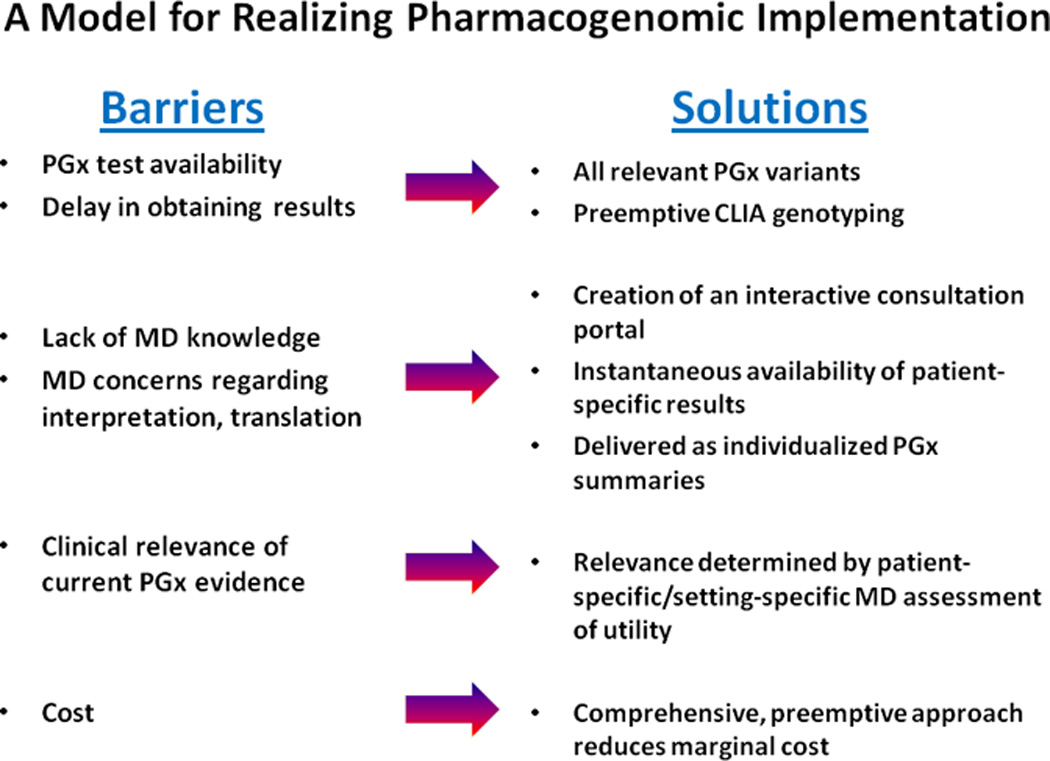
Our study attempts to address and overcome the common barriers to more routine use of pharmacogenomic information in clinical practice. PGx = pharmacogenomic.
Project Overview: Clinical Research Study of Patient-Provider Pairs
The project is developed around an IRB-approved clinical study open at The University of Chicago (clinicaltrials.gov #NCT01280825). The study, entitled “The 1200 Patients Project”, is prospectively recruiting 1200 adults receiving outpatient medical care. Participants must be taking 1–6 prescription medications and must be under the care of one of 12 recruited “early-adopter” physicians (2 cardiologists/1 pulmonologist/1 hepatologist/1 gastroenterologist/3 oncologists/4 general internists). Recruitment of the 12 physicians up-front was essential, since studying use of pharmacogenomic results requires, in our view, study of “patient-provider pairs”, not just patients.
Patients consent to preemptive genotyping across a panel of variants selected based upon clinically relevant evidence of their pharmacogenomic role. The panel provides comprehensive genotype results for a large number of germline polymorphisms that have been identified as impacting response or toxicity for various commonly-used drugs. All patients are genotyped across the full panel, allowing results to be obtained for each patient both for medications the patient is currently taking, and for medications he may be prescribed in the future. The included drugs have published clinical evidence supporting a pharmacogenomic interaction (medications for which there is not yet such information are excluded; but regular updates to the panel are possible and planned). Patient-specific results are then made available (exclusively) to their enrolling provider through a research web-portal, or genomic prescribing system (GPS)1. At each patient visit, providers are monitored for whether they access the GPS to query pharmacogenomic information during treatment decision-making. This phase of the project is centered around the return of pharmacogenomic information and examination of its use. Whether physicians consider pharmacogenomic information during clinic visits and whether this information results in altered patterns of prescribing in patients genetically at high risk of adverse drug outcomes/non-response are the primary and secondary endpoints. Although we acknowledge that a randomized study will be ultimately necessary to demonstrate that genotype-based prescribing changes clinical outcomes, we hypothesize that patients for whom their physicians have available pharmacogenomic results predicting a higher risk of adverse drug events or non-response will be less likely to be started on such medications, and more likely to be instructed to discontinue such medications, compared to concurrently-enrolled matched control patients of the same physicians for whom pharmacogenomic results are not available.
This implementation approach is founded on the premise that before pharmacogenomic diagnostics can be formally tested for outcomes utility, feasibility of delivery-translation must first be demonstrated2. It must first be shown whether busy clinicians will even choose to consider patient-specific pharmacogenomics when prescribing, just as they would consider their patient’s creatinine or hemoglobin level.
Preemptive Genotyping
One of the first challenges to realization of our model was identifying a high-throughput method to preemptively measure, in a Clinical Laboratory Improvement Amendments (CLIA) setting, large numbers of pharmacogenomic genotypes. We sought a panel of significant breadth so as to comprehensively include both variants in well-known drug-metabolizing genes plus other variants having published clinical pharmacogenomic relationships. Some had replicated, high-levels of evidence; the remaining were second-tier, carefully selected variants from comprehensive review of the literature.
Many of the desired variants were available for bundled testing using a commercial ‘ADME pharmacogenomics panel’ from Sequenom3. The remaining were designed into a custom Sequenom panel. A partner laboratory at Oregon Health Sciences University (C. Corless, Director) is performing the CLIA genotyping using the Sequenom technology. CLIA-laboratory results were necessary so that study providers could potentially utilize the study’s research-setting results in clinical decision-making.
It is perhaps obvious that a key advantage of a comprehensive preemptive genotyping method is that all potentially desired pharmacogenomic information is obtained for the patient’s entire drug prescription lifetime with one single test. Although newly-discovered pharmacogenomic variants not currently included in our panel would require future additions to the panel and subsequent repeat genotyping of samples, our decision to be broadly inclusive at this stage was based on our assessment that some variants—if not ready for clinical implementation now—may likely be ready in the near future, as new evidence emerges.
Our broad preemptive approach aims to reduce or eliminate marginal costs associated with alternative genotyping approaches wherein a patient is genotyped each time a relevant prescription is being considered. The scalability of our approach is illustrated by the fact that we are obtaining hundreds of genotype calls generated in a CLIA setting for <$500/patient, comparable to the current cost of most individual CLIA genotype tests.
We have carefully considered the alternative option of whole genome sequencing (WGS). Although WGS has the advantage of detecting rare variants that may have extreme function, it has the disadvantage of requiring substantially greater costs in quality control/bioinformatics analysis, and it remains unclear what level of coverage will ultimately be needed to detect with the required accuracy even well-characterized variants in drug-metabolizing genes, as many of these genes are associated with copy number variation and variable numbers of pseudo-genes, all of which tends to reduce the quality of next-generation sequence data. Moreover, WGS data leads to challenges related to incidental findings that may predict for future illness. This information could be used to discriminate against patients wishing to apply for life, disability, and/or long-term care insurance or wishing to enter the military (the 2008 Genomics Information Nondiscrimination Act applies only to employment and health insurance).
Development of the GPS
Although a recent survey suggests that 98% of physicians believe “patient genetic profiles may influence drug therapy”, only 13% had ordered a pharmacogenomic test, and only 10% felt adequately informed about pharmacogenomics4. We believe these data reflect an unintentional gap that has formed between pharmacogenomic discovery and its translation into practice. Our model relies on a point-of-care informatics support system that bridges three important elements of pharmacogenomic implementation:
Information Dissemination (Provider Education)
Instantaneous Availability of Results
Clinical Interpretation and Guidance (Rx Advice).
Our study’s protected-access web-based portal (the GPS) is tailored to provide rapid, patient-specific, clinically usable pharmacogenomic data to study providers (Figure 2). The GPS provides not raw genotypes but rather a patient-specific interpretation of the genomic data for that drug, distilled into a summary providers can read in <30 seconds. In addition to providing information for drugs the patient is already taking, a dynamic feature is that the GPS allows physicians to query their patient’s results for any other drugs they might be considering prescribing. Because all genotyping for the patient is done up-front, any included drug-variant pairs are immediately searchable at any time. Providers can therefore use the system in real-time, with instantaneous availability of pharmacogenomic results as new treatments are weighed. The GPS fulfills at once a test result function, an interpretive function, and an educational function. Providers can repeatedly access the GPS over the lifetime of a patient, each time treatment decisions arise during care.
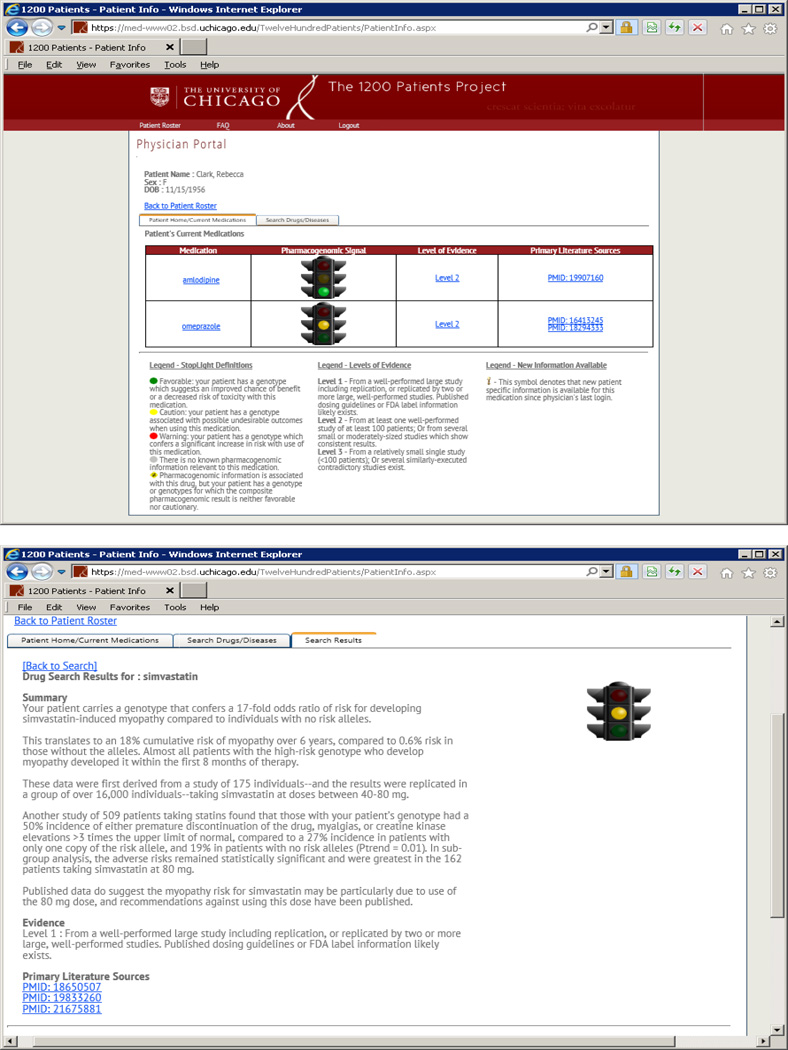
Figure 2. The genomic prescribing system of “The 1200 Patients Project” used for instantaneous delivery of pharmacogenomic results and virtual consultation when considering clinical implementation.
All of a patient’s genetic results impacting current medications are summarized and made available to the provider each time the provider logs-in (top panel). The GPS provides not raw genotypes but rather a patient-specific interpretation of the complex genomic data for that drug, distilled into a summary the provider can read in 30 seconds or less (bottom panel). Each of these clinical pharmacogenomic drug summaries characterizes the nature of the drug-variant association(s) for that patient, the clinical impact of the variant on drug disposition, response, or toxicity, and a brief description of the studies which led to these associations. Providers have the opportunity to view the source publications supporting each drug-variant pair through direct link-outs to PubMed. In this way, the provider can choose (based on interest, familiarity with the subject, and time constraints) the level of detail they desire about each drug-variant summary. A dynamic feature of the GPS is that it allows the physician to query their patient’s preemptively-tested results to determine if any results impact other drugs that they might be considering prescribing. For example, if a provider wants to search whether their patient has a genotype impacting potential use of ‘simvastatin’, the drug’s name can be entered, and the patient-specific “30 second clinical summary” results for simvastatin are produced (bottom panel). Diseases can also be searched if a provider wants to compare pharmacogenomic information about multiple drugs that may treat a given disease. Providers can therefore use the GPS to consider pharmacogenomic information while they are considering prescribing any drug or as they are considering treating any condition. Additionally, each time providers log-in they are alerted if there is new pharmacogenomic information relevant to their patients since the last log-in. Note: the displayed patient name in the top panel has been fictionalized.
Implementation Science
Since our project can be viewed as a “phase I” study of preemptive pharmacogenomic implementation, it will be important to examine the process itself of delivering pharmacogenomic information. This will inform a collective understanding of how to best disseminate pharmacogenomic information. We posit that utility may differ based upon the clinical context, patient, disease being treated, and drug(s). By studying early-adopter provider-patient pairs incorporating a broad range of pharmacogenomic information, we will gain important insights into implementation processes. For many drugs and situations, “there may be little downside to providing additional [pharmacogenomic] information for the prescriber to consider”5, while for others a higher bar for changing decision-making must be met. This project will begin to inform the distinction between the two. It is certainly possible that the early-adopter physicians and their attitudes and behaviors about pharmacogenomics will not be representative of general clinicians (e.g., in community practices), however, studying these early-adopters will still be highly valuable as it is these types of providers that are likely to lead the advance of clinical implementation of pharmacogenomics. We imagine this project and the developed infrastructure as a foundation for the next phase of implementation examination—a larger, randomized trial (within diverse practice settings) to determine whether availability of pharmacogenomic results changes patient health outcomes.
Conclusions
Our project reflects a new, individualized health care model using broad, preemptive pharmacogenomic testing aimed to demonstrate that pharmacogenomics will be utilized if results are timely and if robust informatics support is available to guide interpretation. We believe this model represents an exciting and paradigm-shifting first step toward the way health care should be approached in the 21st century.
Acknowledgments
Supported by NIH K12 CA139160 (PHO), The Conquer Cancer Foundation of the American Society for Clinical Oncology (MJR), and The William F. O’Connor Foundation (MJR).
References
- 1.Ratain MJ. Personalized medicine: Building the gps to take us there. Clin Pharmacol Ther. 2007;81:321–322. doi: 10.1038/sj.clpt.6100092. [DOI] [PubMed] [Google Scholar]
- 2.Greco PJ, Eisenberg JM. Changing physicians' practices. N Engl J Med. 1993;329:1271–1273. doi: 10.1056/NEJM199310213291714. [DOI] [PubMed] [Google Scholar]
- 3.Cantor CR. Company profile: Sequenom, Inc. Pharmacogenomics. 2012;13:529–531. doi: 10.2217/pgs.12.8. [DOI] [PubMed] [Google Scholar]
- 4.Stanek E, et al. Who are adopters of pharmacogenomics among U.S. Physicians?; Presented at the 59th Annual Meeting of The American Society of Human Genetics; 2009. [abstract]. [Google Scholar]
- 5.Altman RB. Pharmacogenomics: "Noninferiority" is sufficient for initial implementation. Clin Pharmacol Ther. 2011;89:348–350. doi: 10.1038/clpt.2010.310. [DOI] [PubMed] [Google Scholar]