Abstract
Streptococcus suis, more particularly serotype 2, is a major swine pathogen and an emerging zoonotic agent worldwide that mainly causes meningitis, septicemia, endocarditis, and pneumonia. Although several potential virulence factors produced by S. suis have been identified in the last decade, the pathogenesis of S. suis infections is still not fully understood. In the present study, we showed that S. suis produces membrane vesicles (MVs) that range in diameter from 13 to 130 nm and that appear to be coated by capsular material. A proteomic analysis of the MVs revealed that they contain 46 proteins, 9 of which are considered as proven or suspected virulence factors. Biological assays confirmed that S. suis MVs possess active subtilisin-like protease (SspA) and DNase (SsnA). S. suis MVs degraded neutrophil extracellular traps, a property that may contribute to the ability of the bacterium to escape the host defense response. MVs also activated the nuclear factor-kappa B (NF-κB) signaling pathway in both monocytes and macrophages, inducing the secretion of pro-inflammatory cytokines, which may in turn contribute to increase the permeability of the blood brain barrier. The present study brought evidence that S. suis MVs may play a role as a virulence factor in the pathogenesis of S. suis infections, and given their composition be an excellent candidate for vaccine development.
Introduction
Bacterial membrane vesicles were first discovered some five decades ago in Gram-negative bacteria [1]. These globular structures, which range in diameter from 10 to 200 nm, result from outer membrane blebbing and are called outer membrane vesicles (OMV). The biogenesis of OMVs remains unclear due to the difficulty in obtaining mutant strains that do not produce OMVs [2]. Over the years, many roles have been assigned to OMVs, including intercellular communication [3], response to environmental stresses [4], biofilm formation [5], pathogenic processes [6], and horizontal gene transfer [7]. The rising interest in bacterial membrane vesicles lead to the creation of EVpedia, a web-based database that collects published data about vesicles produced by prokaryotes and eukaryotes [8]. Given that OMVs contain large amounts of membrane-associated proteins and virulence factors [9], they are highly immunogenic and may induce immunoprotection against pathogens. This has led to a growing interest in bacterial OMVs as potential vaccine candidates [10]. An OMV-based Neisseria meningitidis vaccine has already proven its effectiveness and is currently being used to prevent meningitis [11].
Despite the fact that vesicles produced by Gram-negative bacteria have been extensively studied, vesicles from Gram-positive bacteria were overlooked for decades since it was thought that the rigidity of the peptidoglycan-rich cell wall would not allow vesicle blebbing [12]. The discovery of cytoplasmic membrane-derived vesicles, or simply membrane vesicles (MVs), in Gram-positive bacteria, suggested that vesicle production is a ubiquitous phenomenon [13]. They were first described in 1990 [14] and studied in more depth in the last decade. MV production by numerous Gram-positive bacteria as well as their putative roles in virulence have been reported for Staphylococcus aureus [12], Bacillus spp. [15, 16], Listeria monocytogenes [17], Clostridium perfringens [18], Streptomyces coelicolor [19] and Streptococcus spp. [20, 21] (for recent review, see [22]).
The Gram-positive bacterium Streptococcus suis is a major swine pathogen worldwide that mainly causes septicemia, meningitis, endocarditis, arthritis, and pneumonia [23]. It is also considered an emerging zoonosis agent since it can infect humans who are in close contact with diseased pigs or their byproducts [24]. Thirty-three serotypes have been described to date based on the composition of their capsular polysaccharides. While the serotype distribution varies depending on the geographical origins of the strains, S. suis serotype 2 is considered the most pathogenic and the most prevalent capsular type recovered from diseased pigs and humans [23, 24]. The massive use of antibiotics in the swine industry has contributed to the emergence of drug resistant strains, making S. suis infections more difficult to treat and causing increasingly severe economic losses [25]. It is thus essential to better understand the pathogenic process of S. suis infections in order to identify new therapeutic strategies.
In this study, we hypothesized that S. suis produces MVs that may play an important role in the pathogenesis of S. suis infections. MVs were isolated using a differential centrifugation and filtration protocol. Their protein composition was analyzed and characterized using a proteomic approach. Given that bacterial vesicles generally carry virulence factors, two virulence-associated activities were assayed. The pro-inflammatory effects of S. suis MVs on monocytes and macrophages were also investigated.
Materials and Methods
Bacterial strain and isolation of membrane vesicles
S. suis P1/7 was grown in Todd-Hewitt Broth (THB, Becton, Dickinson and Company, Sparks, MD, USA) at 37°C. MVs were isolated using centrifugation and filtration protocol as described by Lee et al. [12], with slight modifications. Briefly, a 1-L overnight culture of S. suis (early stationary phase) was centrifuged at 10 000 × g for 20 min at 4°C, and the supernatant was filtered through a 0.45-μm pore size cellulose nitrate membrane and then through a 0.22 μm pore size cellulose nitrate membrane to remove residual bacteria. The filtrate was centrifuged at 150 000 × g for 3 h at 4°C. The pellets were resuspended in 50 mM phosphate-buffered saline (PBS, pH 7.2), further centrifuged similarly, and stored in PBS at –20°C until used. MVs were quantified by dry weight as well as by protein content (Quick Start Bradford Assay; Bio-Rad Laboratories, Mississauga, ON, Canada) according to the manufacturer’s protocol.
Transmission electron microscopy
Vesicles were suspended in 0.1 M sodium cacodylate buffer (pH 7) containing 5% glutaraldehyde and 0.15% ruthenium red. They were then treated with 1 mg/ml of polycationic ferritin and were processed as previously described by Vanrobaeys et al. [26]. Thin sections were prepared and were observed using a JEOL 1230 transmission electron microscope at an accelerating voltage of 80 kV. A 1.27 correction factor was applied to determine MVs diameter, as suggested by Kong et al. [27].
Proteomic analysis
Three independent batches of S. suis MVs with a protein content of 10 μg were washed three times with 50 mM ammonium bicarbonate buffer (pH 8) using Amicon 3-kDa molecular weight cut off centrifugal filters (EMD Millipore Canada; Mississauga, ON, Canada). The samples were vacuum-dried in a Speedvac concentrator (Thermo Fisher Scientific, Waltham, MA, USA) and were stored at –20°C until they were trypsin digested. Proteins were solubilized in 25 μl of 50 mM ammonium bicarbonate containing 1% sodium deoxycholate and were heated at 95°C for 5 min. The samples were reduced by adding 0.2 mM dithiothreitol (37°C for 30 min) and were alkylated by adding 0.9 mM iodoacetamide (37°C for 20 min). Trypsin (1 μg) was then added, and the samples were incubated overnight at 37°C. Proteolytic degradation was stopped by acidification with 3% acetonitrile-1% trifluoroacetic acid (TFA)-0.5% acetic acid. Peptides were purified using C18 spin tips (Thermo Fisher Scientific) and were pooled, vacuum centrifuge-dried, and resuspended in 0.1% formic acid. Peptide samples (800 ng) were fractionated by online reversed-phase nanoscale capillary liquid chromatography (nanoLC). The fractions were analyzed by electrospray mass spectrometry (ES MS/MS; Proteomics platform of the Québec Genomic Center) using a 5600 mass spectrometer (AB Sciex, Framingham, MA, USA) equipped with an Agilent 1200 nano pump, a nanoelectrospray ion source, and a self-pack PicoFrit column (15 cm x 0.075 i.d.; New Objective, Woburn, MA, USA) packed with a Jupiter C18 phase (particle size: 5 μm; pore size: 300 Å; Phenomenex, Torrance, CA). Peptides were eluted with a linear 2–50% linear gradient of acetonitrile-0.1% formic acid for 30 min at 300 nl/min. Mass spectra were acquired using Analyst software version 1.6 with the data dependent acquisition mode (AB Sciex). Each full scan mass spectrum (400 to 1250 m/z) was followed by collision-induced dissociation of the twenty most intense ions. The dynamic exclusion was set at 3 s and the tolerance at 100 ppm.
MS/MS peak list Mascot Generic Format (MGF) files were generated using ProteinPilot version 4.5 (AB Sciex) with the Paragon and Progroup algorithms [28]. The MGF sample files were analyzed using Mascot version 2.4.0 (Matrix Science, London, UK). Mascot and X! Tandem were set up to search the UniRef StreptococcusSuisP17 database (version February 2014, 2039 entries) assuming trypsin digestion. Mascot and X! Tandem were searched with a fragment ion mass tolerance of 0.100 Da and a parent ion tolerance of 0.100 Da. Carbamidomethylated cysteine was specified in Mascot and X! Tandem as a fixed modification. Dehydrated N-terminus, Glu->pyro-Glu of the N-terminus, ammonia-loss of the N-terminus, Gln->pyro-Glu of the N-terminus, deamidated asparagine and glutamine, and oxidation of methionine were specified in X! Tandem as variable modifications.
Proteome Scaffold version 4.3.4 (Proteome Software Inc., Portland, OR USA) was used to validate the MS/MS-based peptide and protein identification. Peptide identification was accepted if it could established there was a 6% or greater probability that the False Discovery Rate (FDR) was less than 1% using the Peptide Prophet algorithm [29] with Scaffold delta-mass correction. Peptide identification was accepted if it could be established that there was a 98% or greater probability of achieving an FDR less than 1% and that the protein contained at least three identified peptides. Proteins were considered present in significant amounts when at least 30 spectra were identified. The relative abundance of proteins was determined by spectral counting. Protein probabilities were assigned by the Protein Prophet algorithm [30]. Proteins that contained similar peptides and could not be differentiated based on an MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins were annotated with Gene Ontology (GO) terms from gene_association.goa_uniprot (downloaded March 18, 2013) [31]. Protein localization was predicted using GO terms as well as the Uniprot database. Proteins containing a LPXTG motif were predicted as cell wall-anchored.
Determination of enzymatic activities
N-succinyl-Ala-Ala-Pro-Phe-pNa, the substrate for the S. suis subtilisin protease SspA, was dissolved at a concentration of 2 mg/ml in 50% dimethylformamide, and 20 μl was added to the wells of a 96-well microplate containing 100 μl of S. suis MVs (protein content of 300 and 80 μg/ml in PBS). Assay mixtures were incubated at 37°C for 4 h. The absorbance at 415 nm (A415) was then recorded using a xMark microplate reader (Bio-Rad Laboratories). DNase activity was assayed as described in a previous study [32]. Briefly, MVs (protein content of 64, 32, and 16 μg/ml in PBS, pH 7.5) were incubated for 4 h at 37°C in the presence of 2 μg of salmon sperm DNA (Sigma-Aldrich Canada Ltd., Oakville, ON, Canada) in 50 mM PBS. One hundred microliters of Quant-iT Pico-Green dsDNA reagent (Life Technologies Inc., Burlington, ON, Canada) prepared following the manufacturer’s protocol was added to the reaction mixtures, which were then incubated for 5 min at room temperature. Fluorescence was recorded using a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT, USA) with the excitation and emission wavelengths set at 485 ± 20 nm and 528 ± 20 nm, respectively.
Neutrophil extracellular traps degradation
The human promyelocytic leukemia cell line HL60 (ATCC CCL-240) was routinely grown in RPMI-1640 medium (Life Technologies Inc.) supplemented with 10% heat-inactivatedfetal bovine serum (FBS) and 100 μg/ml of penicillin-streptomycin at 37°C in a 5% CO2 atmosphere. Cells (106 cells/ml) were treated for 3 days with 1.25% dimethylsulfoxide (DMSO) to induce differentiation into neutrophil-like cells. The efficacy of cell differentiation was evaluated using the nitro-blue tetrazolium (NBT) assay according to Collins et al. [33], with modifications. Briefly, 500 μl of cells (5.106 cells/ml) were incubated for 2 h at 37°C in the presence of 10 μg/ml of Escherichia coli lipopolysaccharide (LPS) and 250 μl of 0.01% NBT (Sigma-Aldrich Canada Ltd.). The reaction was stopped by adding 500 μl of 0.5 M HCl. The mixtures were centrifuged for 4 min at 10 000 × g, and the pellets were resuspended in 200 μl of DMSO. The absorbance at 550 nm (A550) was recorded using a xMark microplate reader (Bio-Rad Laboratories) and was compared to a negative control (non-differentiated cells). Neutrophil-like cells (100 μl, 106 cells/ml) were seeded in a 96-well black wall clear bottom microplate and were treated with 0.5 μM phorbol 12-myristate 13-acetate (PMA) for 3 h at 37°C in a 5% CO2 atmosphere. MVs (protein content of 40, 20, 10, or 1 μg/ml) were then added to the reaction mixtures, and the samples were incubated for an additional 2 h. Extracellular DNA was labeled using 0.5 μM SYTOX Green Nucleic Acid Stain (Life Technologies Inc.) according to the manufacturer’s instructions. After a 10-min incubation at room temperature, fluorescence was recorded using a Synergy 2 microplate reader (BioTek Instruments) with the excitation and emission wavelengths set at 485 ± 20 nm and 528 ± 20 nm, respectively. Unstimulated cells were used as blanks to subtract the background signal.
Activation of the NF-κB signaling pathway in monocytes and macrophages
The human monoblastic leukemia cell line U937-3xκB-LUC (U937 cells stably transfected with a construct containing 3 NF-κB binding sites from the Ig κ light chain promoter coupled with the gene encoding the firefly luciferase (3x-κB-luc) [34]) was kindly provided by Dr. Rune Blomhoff (University of Oslo, Norway). The cells were routinely grown at 37°C in a 5% CO2 atmosphere in RPMI-1640 medium supplemented with 10% FBS, 100 μg/ml of penicillin-streptomycin, and 75 μg/ml of hygromycin B (Sigma-Aldrich Canada Ltd.). The monocytes (2.105 cells/ml) were differentiated into macrophage-like cells by incubating them with 10 ng/ml PMA for 24 h. The cell culture medium was then replaced with fresh medium, and the adherent macrophage-like cells were incubated for an additional 24 h prior to use. U937-3xκB-LUC monocytes and macrophage-like cells were suspended at a concentration of 2.106 cells/ml in RPMI-1640 supplemented with 1% FBS, 100 μg/ml of penicillin-streptomycin, and 75 μg/ml of hygromycin B. The cell suspensions (50 μl) were seeded into wells of black bottom, black wall 96-well microplates. Two-fold serial dilutions of S. suis MVs (protein content ranging from 80 to 0.02 μg/ml) were prepared in cell culture medium, and 50 μl of each dilution was added to the wells. The microplates were incubated for 6 h at 37°C in a 5% CO2 atmosphere. E. coli LPS (10 μg/ml) and S. suis P1/7 whole cells (multiplicity of infection [MOI] of 100) were used as positive controls. NF-κB activation was monitored using the Bright-Glo Luciferase Assay System (Promega, Madison, WI, USA) by adding 100 μl of luciferase substrate to the wells at room temperature. Luminescence was recorded using the luminometer option of a Synergy 2 microplate reader (BioTek Instruments) within 3 min of the addition of the substrate.
Cytokine secretion by macrophages
The human monoblastic leukemia U937 cell line was cultivated in RPMI-1640 supplemented with 10% FBS and 100 μg/ml of penicillin-streptomycin at 37°C in a 5% CO2 atmosphere. The cells were then differentiated into adherent macrophage-like cells as described above. Macrophage-like cells (106 cells/ml) were seeded in 6-well cell culture plates (2 ml/well) and were incubated overnight to allow adherence. The cell culture medium was aspirated and was replaced with S. suis P1/7 cell suspensions at an MOI of 100 (positive control) or S. suis MVs (protein content of 40, 20, 10, or 1 μg/ml) prepared in RPMI-1640 supplemented with 1% FBS and 100 μg/ml of penicillin-streptomycin. Following a 24-h incubation at 37°C in a 5% CO2 atmosphere, the culture supernatants were collected and were used to quantify interleukin (CXCL)-8, tumor necrosis factor (TNF)-α, and interleukin (IL)-1β using enzyme-linked immunosorbent assay (ELISA) kits following the manufacturer’s protocols (eBioscience Inc., San Diego, CA, USA).
Statistical analysis
All assays were performed in triplicate, and the means ± standard deviations were calculated. Differences were analyzed for statistical significance using the Student’s t-test and were considered significant at p < 0.05.
Results
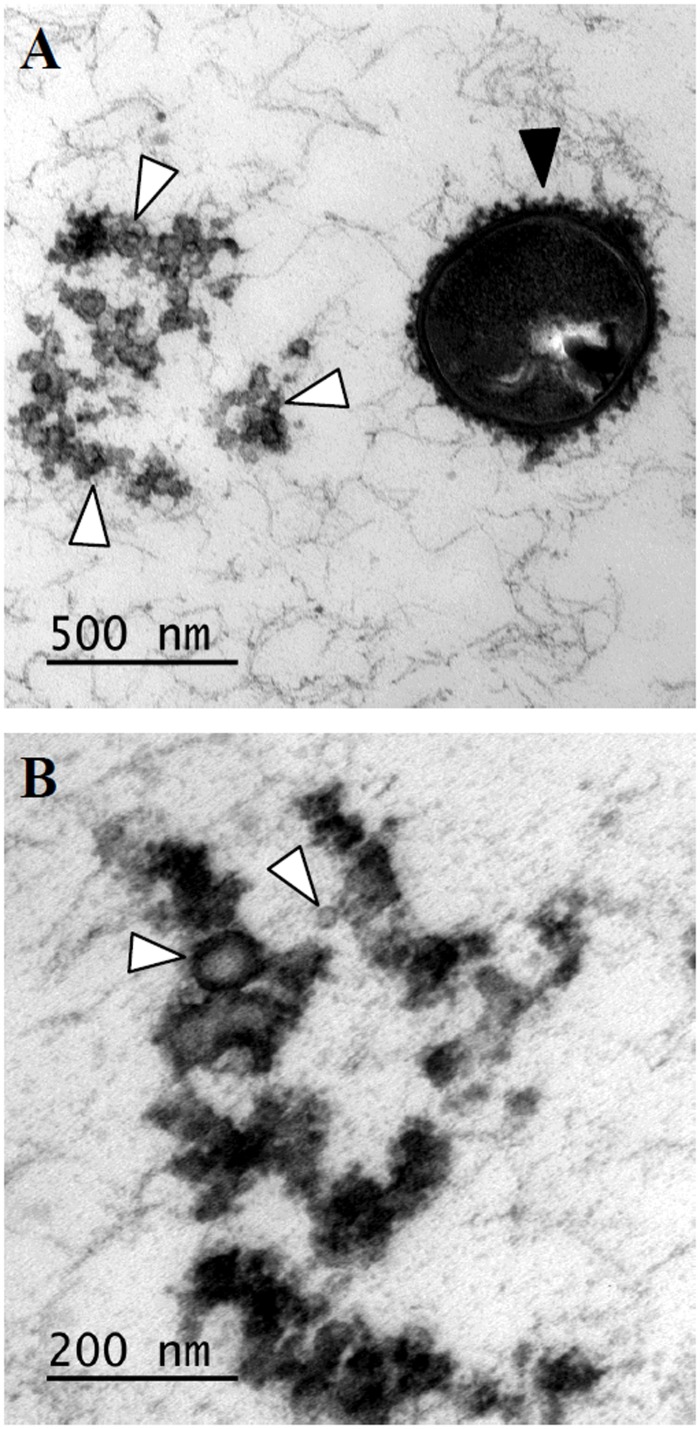
Transmission electron microscopic observations of overnight cultures of S. suis P1/7 revealed the presence of membranous globular structures, suggesting that S. suis P1/7 produces MVs (Fig 1A). The MVs were isolated using a differential centrifugation and filtration protocol. The average recovery rate was 4 mg of MVs (dry weight) per liter of overnight culture, which corresponded to approximately 300 μg of vesicular protein content. As shown in Fig 1B, the MVs appeared to be coated with a dense structure, most likely capsular material, and ranged from 13 to 130 nm in diameter.
Fig 1. Transmission electron micrographs of an overnight culture of S. suis P1/7 (panel A) and the membrane vesicle preparation (panel B).
Black arrow: whole bacterium; white arrows: membrane vesicles.
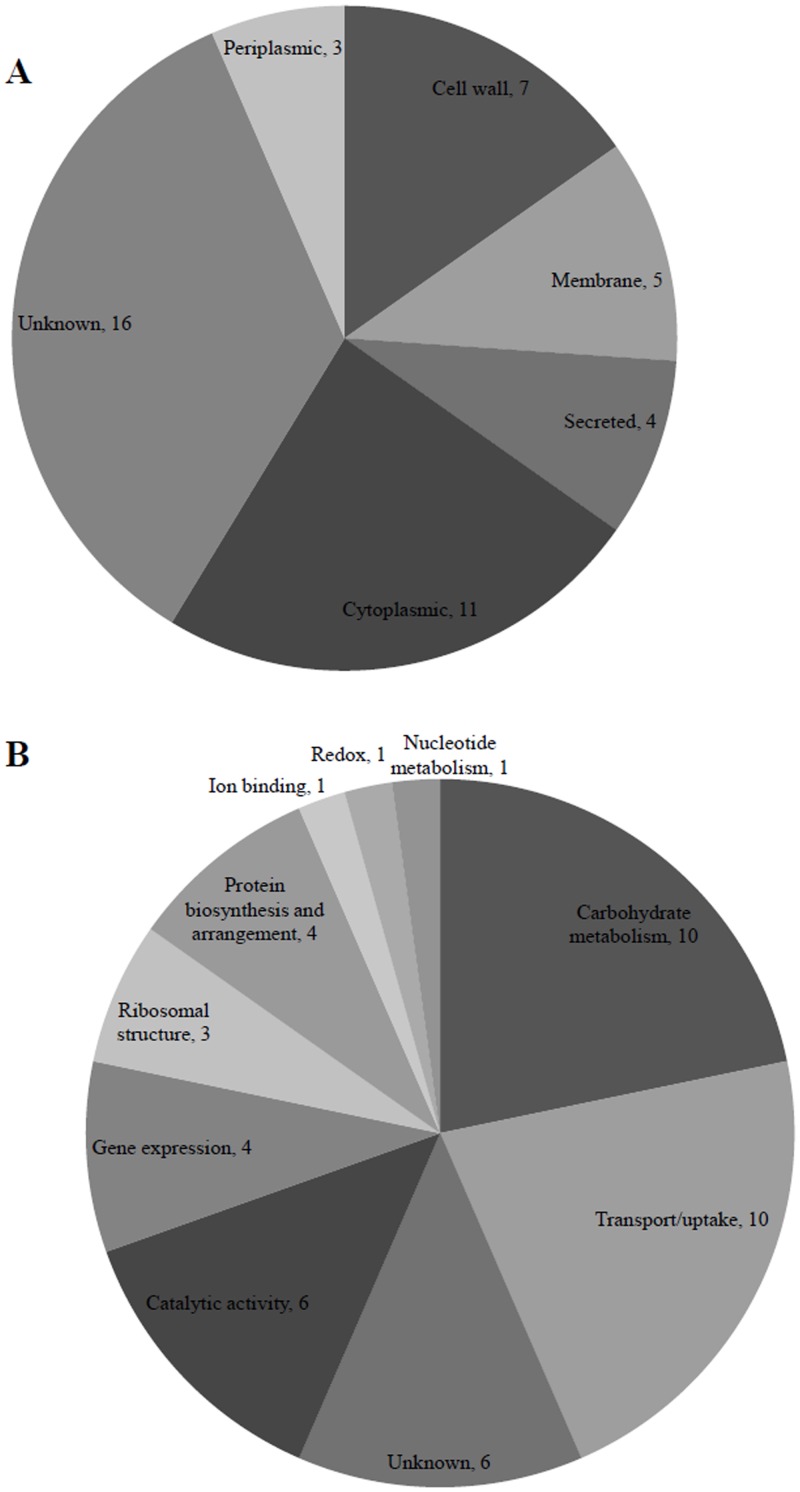
Proteomic sequencing detected 46 proteins in S. suis P1/7 MVs, which are listed in Table 1 by their abundance in each functional group. Only proteins that were detected in at least two batches of MVs and counting 30 or more spectra were considered. Proteins identified using the S. suis P1/7 genome database were either cytoplasmic (n = 11), membrane/cell wall-associated (n = 12), periplasmic (n = 3) or secreted (n = 4). However, the location of 16 proteins could not be assigned (Fig 2A). The biological activities associated with S. suis P1/7 vesicular proteins were mostly involved in the carbohydrate metabolism (n = 10), molecule transport (n = 10), catalytic activities (n = 6), gene expression (n = 4), ribosomal structure (n = 3), protein biosynthesis and arrangement (n = 4), ion binding (n = 1), oxydoreduction (n = 1), nucleotide metabolism (n = 1), and 6 proteins were of unknown function (Fig 2B). In addition, 9 proteins (19.6%) were identified as proven or suspected virulence factors (Table 2), seven of which were located at the cell surface (either cell wall- or membrane-associated) and two were secreted. For the 46 proteins, in terms of relative abundance, the subtilisin-like protease SspA predominated in the MVs, followed by a putative lipoprotein (component of an ABC transporter), the cell wall-anchored DNase SsnA, the IgM protease Ide, putative exported ribonucleases (G and E), a putative N-acetylmuramoyl-L-alanine amidase, the Plr adhesin (glyceraldehyde-3-phosphate dehydrogenase), a putative sugar binding protein, the peptide binding protein OppA, the histidine triad-family protein HtpsC, the muraminidase-released protein Mrp, the elongation factor Tu, a fumarate reductase flavoprotein subunit, an unidentified putative exported protein, and a putative ABC transporter component. Interestingly, of the 15 most abundant proteins identified in the MVs, six are considered virulence factors (SspA, SsnA, Ide, Plr, HtpsC, and Mrp).
Table 1. Protein content of S. suis P1/7 membrane vesicles identified by ES MS/MS in order of abundance in each functional group.
| Protein accession number | Gene accession number | MS/MS identification | Molecular weight | Protein name/function | Predicted localization |
|---|---|---|---|---|---|
| Translational, ribosomal structure, and biogenesis (n = 3) | |||||
| C5VVU1 | SSU0721 | Putative 30S ribosomal protein S1 | 44 kDa | 30S Ribosomal protein S1 | Cytoplasm |
| C5VWE9 | SSU1935 | 30S ribosomal protein S4 | 23 kDa | 30S Ribosomal protein S4 | Cytoplasm |
| C5VVK7 | SSU1771 | 30S ribosomal protein S2 | 29 kDa | 30S Ribosomal protein S2 | Cytoplasm |
| Transport, uptake, and osmoregulation (n = 9) | |||||
| C5VWT7 | SSU0934 | Putative lipoprotein | 36 kDa | Putative ABC transporter | Membrane |
| C5W096 | SSU1664 | Putative oligopeptide-binding protein OppA | 66 kDa | OppA / Peptide binding | Unknown |
| C5W029 | SSU0503 | Putative amino acid ABC transporter | 31 kDa | ABC transporter | Unknown |
| C5VW03 | SSU1853 | Putative amino-acid ABC transporter | 29 kDa | ABC transporter | Periplasm |
| C5VXJ7 | SSU1170 | Extracellular solute-binding protein | 61 kDa | Sugar ABC transporter | Secreted |
| C5VW19 | SSU1869 | Extracellular metal cation-binding protein | 36 kDa | ZnuA / Zinc uptake | Secreted |
| C5VZZ1 | SSU0465 | Putative exported protein | 41 kDa | Predicted membrane fusion protein | Membrane |
| C5VYV7 | SSU0284 | Extracellular solute-binding protein | 35 kDa | ABC transporter | Periplasm |
| C5VYN9 | SSU1364 | Branched-chain amino acid ABC transporter | 41 kDa | ABC transporter | Periplasm |
| Protein biosynthesis (n = 2) | |||||
| C5VXU6 | SSU1196 | Threonine-tRNA ligase | 75 kDa | ThrS / protein biosythesis | Cytoplasm |
| C5VZL9 | SSU0412 | Valine-tRNA ligase | 101 kDa | ValS / protein biosynthesis | Cytoplasm |
| Carbohydrate/sugar metabolism (n = 10) | |||||
| C5VXF6 | SSU1127 | Putative N-acetylmuramoyl-L-alanine amidase | 113 kDa | Peptidoglycan catabolism | Unknown |
| C5VY36 | SSU0153 | Glyceraldehyde-3-phosphate dehydrogenase | 39 kDa | Plr / adhesin | Secreted |
| C5VWC9 | SSU1915 | Putative maltose/maltodextrin-binding protein | 44 kDa | sugar binding | Membrane |
| C5VWE7 | SSU1933 | Putative fumarate reductase flavoprotein subunit | 53 kDa | Fumarate reductase flavoprotein subunit | Unknown |
| C5VXV7 | SSU1207 | Putative lipoprotein | 30 kDa | Predicted glucose-6-phosphate isomerase | Unknown |
| C5VVZ9 | SSU1849 | Putative surface-anchored amylopullulanase | 234 kDa | ApuA / amylopullulanase / Adhesin | Cell wall |
| C5VY37 | SSU0154 | Phosphoglycerate kinase | 42 kDa | Phosphoglycerate kinase | Cytoplasm |
| C5VYJ5 | SSU1320 | Enolase | 47 kDa | Eno / adhesin | Cell wall |
| C5VYP7 | SSU1372 | Multiple sugar-binding protein | 45 kDa | Sugar transporter | Unknown |
| C5VZ34 | SSU1451 | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | 26 kDa | GpmA / glycolytic process | Unknown |
| Nucleotide metabolism (n = 1) | |||||
| C5VX44 | SSU1044 | Ribonucleoside-diphosphate reductase | 82 kDa | DNA synthesis | Cytoplasm |
| Protein folding/arranging (n = 2) | |||||
| C5VX75 | SSU1078 | Foldase protein PrsA | 38 kDa | PrsA / protein folding | Membrane |
| C5VXK4 | SSU1177 | Peptidyl-prolyl cis-trans isomerase | 28 kDa | Protein folding | Unknown |
| Gene expression (n = 4) | |||||
| C5W008 | SSU0482 | Elongation factor Tu | 44 kDa | TufA / elongation factor Tu | Cytoplasm |
| C5VY08 | SSU0123 | DNA-directed RNA polymerase subunit beta | 136 kDa | DNA-dependant RNA polymerase | Cytoplasm |
| C5VY34 | SSU0151 | Elongation factor G | 77 kDa | FusA / elongation factor G | Cytoplasm |
| C5VY07 | SSU0122 | DNA-directed RNA polymerase subunit beta | 133 kDa | RpoB / RNA-polymerase | Cytoplasm |
| Catalytic activity (n = 5) | |||||
| C5VVK9 | SSU1773 | Putative surface-anchored serine protease | 187 kDa | SspA / Subtilisin-like protease | Cell wall |
| C5VVJ6 | SSU1760 | Surface-anchored DNA nuclease | 114 kDa | SsnA / Nuclease | Cell wall |
| C5W022 | SSU0496 | Putative Mac family protein | 124 kDa | IgM protease | Membrane |
| C5W049 | SSU1616 | Putative exported protein | 122 kDa | Predicted ribonuclease G and E | Unknown |
| C5VXH2 | SSU1143 | Putative surface-anchored zinc carboxypeptidase | 118 kDa | Surface anchored zinc carboxypeptidase | Cell wall |
| Defense/virulence mechanisms (n = 4) | |||||
| C5VYR5 | SSU1390 | Streptococcal histidine triad-family protein | 96 kDa | HtpsC | Cell wall |
| C5VVS8 | SSU0706 | Muramidase-released protein | 136 kDa | Mrp | Cell wall |
| C5VZA8 | SSU0370 | Putative penicillin-binding protein 1A | 79 kDa | Pbp1A / Penicillin-binding protein | Unknown |
| C5VXY1 | SSU1231 | Suilysin | 55 kDa | Sly / Hemolysin | Secreted |
| Ion binding (n = 1) | |||||
| C5VY00 | SSU0115 | Zinc-binding protein AdcA | 56 kDa | AdcA/Zinc-binding protein | Unknown |
| Redox (n = 1) | |||||
| C5VYT4 | SSU0261 | Aldehyde-alcohol dehydrogenase | 97 kDa | Oxydoreductase | Unknown |
| Uncharacterized proteins (n = 4) | |||||
| C5VV38 | SSU0595 | Putative exported protein | 13 kDa | Unknown | |
| C5VZS6 | SSU1560 | Putative lipoprotein | 17 kDa | Unknown | |
| C5VV37 | SSU0594 | Putative exported protein | 14 kDa | Unknown | |
| C5VV35 | SSU0592 | Putative exported protein | 12 kDa | Unknown | |
Fig 2. General locations (A) and functions (B) of proteins identified in S. suis P1/7 membrane vesicles by ES MS/MS.
Table 2. Virulence factors identified in S. suis P1/7 membrane vesicles.
| Identified proteins | Factor | Protein accession number | Gene accession number | Molecular weight | Function | References |
|---|---|---|---|---|---|---|
| Surface-anchored serine protease | SspA | C5VVK9 | SSU1773 | 187 kDa | Subtilisin-like protease | [39, 40] |
| Surface-anchored DNA nuclease | SsnA | C5VVJ6 | SSU1760 | 114 kDa | DNase | [41, 42] |
| Mac family protein | Ide | C5W022 | SSU0496 | 124 kDa | IgM protease | [38] |
| Glyceraldehyde-3-phosphate dehydrogenase | Plr | C5VY36 | SSU0153 | 39 kDa | Adhesin | [46] |
| Streptococcal histidine triad-family protein | HtpsC | C5VYR5 | SSU1390 | 96 kDa | Unknown | [37] |
| Muramidase-released protein | Mrp | C5VVS8 | SSU0706 | 136 kDa | Unknown | [43] |
| Surface-anchored amylopullulanase | ApuA | C5VVZ9 | SSU1849 | 234 kDa | Adhesin | [44] |
| Enolase | Eno | C5VYJ5 | SSU1320 | 47 kDa | Adhesin | [45] |
| Suilysin | Sly | C5VXY1 | SSU1231 | 55 kDa | Hemolysin | [47, 48] |
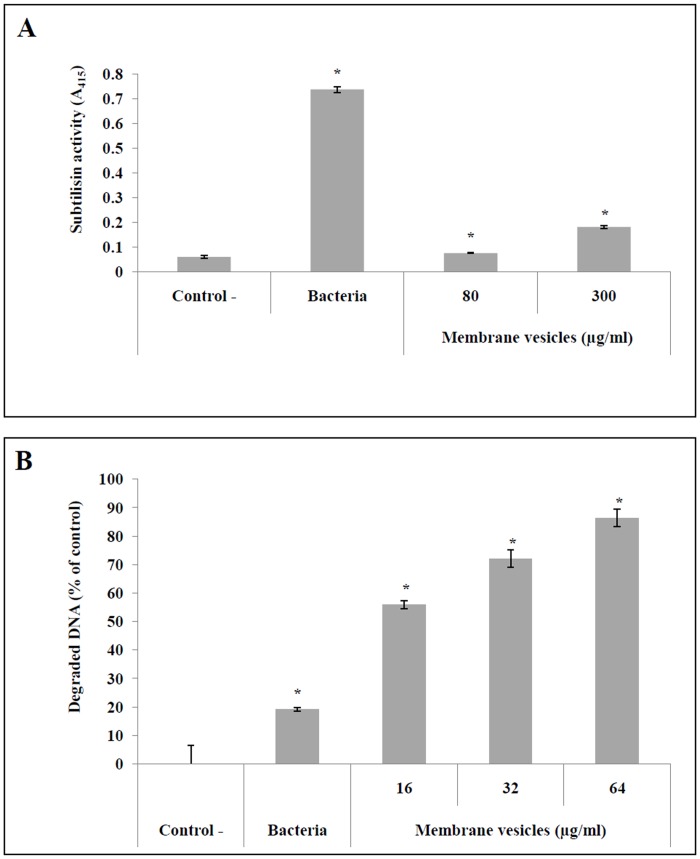
Based on the subtilisin and DNase assays performed to confirm the presence of active virulence factors in S. suis P1/7 MVs, only the highest concentration of MVs (300 μg/ml in protein content) displayed subtilisin activity (Fig 3A). On the other hand, the MVs dose-dependently degraded DNA (Fig 3B). More specifically, at a concentration of 64 μg/ml in protein content, the MVs degraded approximately 90% of the DNA whereas whole bacteria (OD660 = 1) only degraded 20% after a 4 h incubation.
Fig 3. Determination of subtilisin (A) and DNase (B) activities in S. suis membrane vesicles.
*: p < 0.05 compared to negative controls.
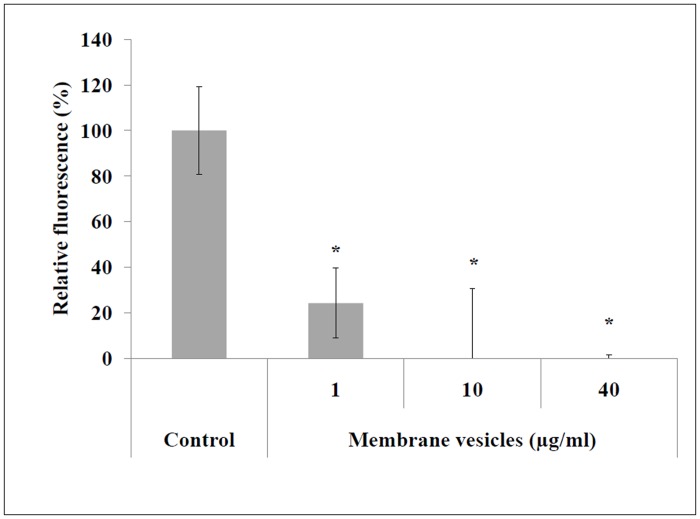
Given that S. suis MVs possessed strong DNase activity, we hypothesized that such structures may induce the degradation of neutrophil extracellular traps (NETs), which consist of DNA as a backbone with embedded histones, antimicrobial peptides, and proteases, and which are part of the first innate immune response at sites of infection [35]. As shown in Fig 4, MVs at a concentration as low as 1 μg/ml in protein content caused the degradation of approximately 70% of extracellular DNA excreted by NETs.
Fig 4. Quantification of NET degradation by S. suis membrane vesicles.
NETs were formed by the PMA-stimulated promyelocytic leukemia cell line HL60. *: p < 0.05 compared to the negative control (no NET degradation).
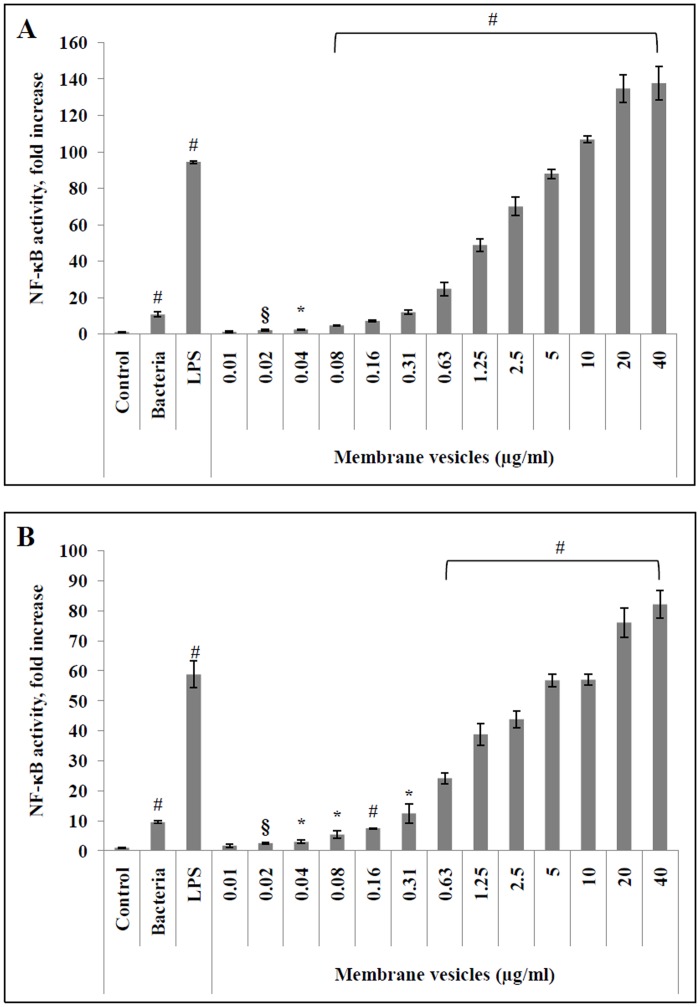
To explore the potential contribution of MVs to the inflammatory process associated with S. suis infections such as meningitis, NF-κB activition was monitored in monocytes and macrophages transfected with a luciferase reporter gene. NF-κB was dose-dependently activated by MVs following a 6-h stimulation of the monocytes and PMA-differentiated macrophages (Fig 5). More specifically, in the presence of 0.02 μg/ml (protein content) of MVs, NF-κB activity increased 2- and 2.5-fold compared to unstimulated monocytes and macrophage-like cells, respectively. In the presence of 40 μg/ml (protein content) MVs, NF-κB activity in monocytes and macrophages increased 137- and 82-fold, respectively, while S. suis P1/7 whole cells at an MOI of 100 increased NF-κB activity by approximately 10-fold in both monocytes and macrophage-like cells. Used as a positive control, E. coli LPS (10 μg/ml), which is a strong pro-inflammatory inducer, caused a 100- and 60-fold increase in NF-κB activity in monocytes and macrophages, respectively.
Fig 5. Quantification of NF-κB activation in U937-3xκB-LUC monocytes (A) and macrophage-like cells (B) by S. suis membrane vesicles (0.01 to 40 μg/ml), E. coli LPS (10 μg/ml), and S. suis P1/7 whole bacteria (MOI = 100).
Results were considered significant at §: p < 0.05, *: p < 0.01, and #: p < 0.001 compared to unstimulated cells.
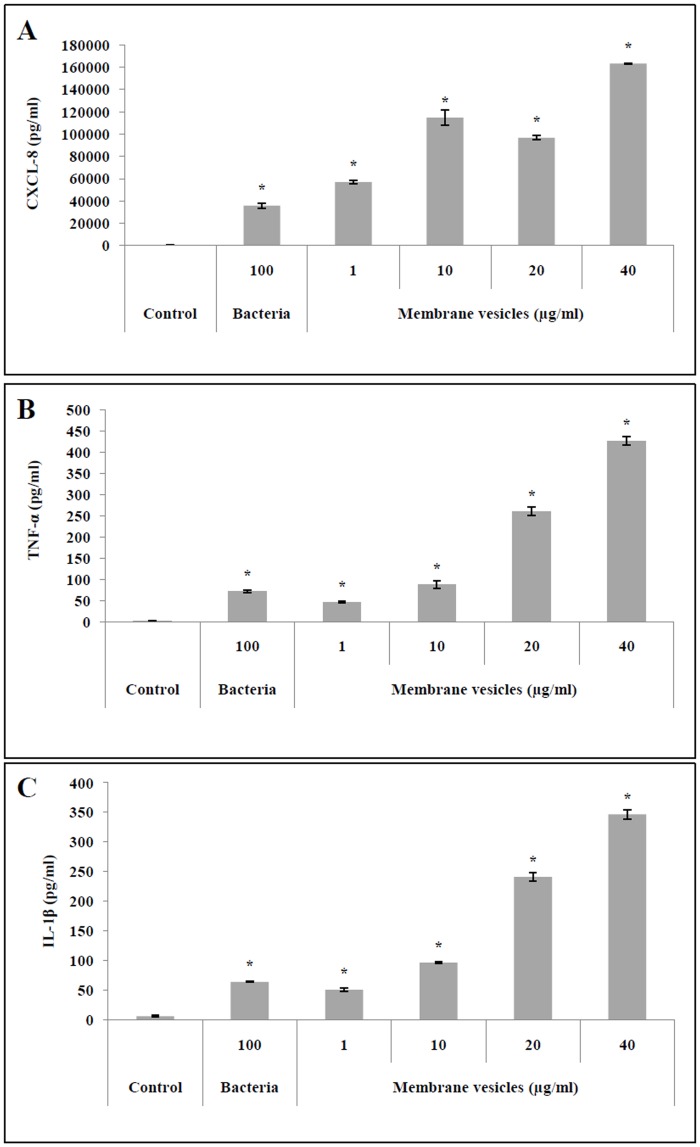
PMA-differentiated U937 cells were stimulated in the presence of S. suis MVs in order to correlate the increased NF-κB activity observed in the U937-3xκB-LUC cell line with the secretion of pro-inflammatory cytokines. As shown in Fig 6, S. suis MVs dose-dependently increased the secretion of CXCL-8, TNF-α, and IL-1β. For example, 40 μg/ml protein content) of MVs increased the secretion of CXCL-8, TNF-α, and IL-1β by macrophage-like cells 4.6-, 5.9-, and 5.4-fold, respectively, compared to the amounts secreted when S. suis P1/7 whole cells (MOI = 100) alone were used to stimulate the macrophage-like cells.
Fig 6. Quantification of CXCL-8 (A), TNF-α (B), and IL-1β (C) secretion by macrophage-like cells stimulated with S. suis membrane vesicles (1 to 40 μg/ml), E. coli LPS (10 μg/ml), and S. suis P1/7 whole bacteria (MOI = 100).
Results were considered significant at *p < 0.05 compared to unstimulated cells.
Discussion
Despite significant advances in our understanding of the pathogenic process of S. suis infections, major unresolved issues remain, and consequently additional studies are required to better characterize the pathogenic strategies of this swine and zoonotic pathogen. In the present study, we hypothesized that S. suis produces MVs as it has been observed with other pathogenic streptococci [20, 21]. Transmission electron microscopic observations of overnight cultures showed that S. suis P1/7 releases extracellular MVs during normal growth. MVs with diameters ranging from 13 to 130 nm were isolated from a culture supernatant of S. suis P1/7 using a simple differential centrifugation and filtration protocol. The MVs appeared to be coated by capsular material that may have bound to the vesicles during the blebbing process. Since MVs were isolated without the use of gradient centrifugation, one should not exclude that this capsular-like material could contain remaining cellular debris, even though isolated MVs were washed once in PBS. Given the relatively low vesicle yield, further studies are required to determine the growth conditions that will increase the production of MVs by S. suis.
A proteomic analysis of the S. suis MVs identified 46 proteins that participate in a broad array of functions, including molecule transport, protein, carbohydrate, and nucleotide metabolism, gene expression and protein folding processes. MVs produced by S. aureus [36] and S. pneumoniae [21] were reported to contain 143 and 211 proteins, respectively. Like S. aureus and S. pneumoniae MVs, S. suis MVs contain mainly cytoplasmic proteins as well as some membrane and cell wall proteins.
Nine proteins that are proven or suspected virulence factors were identified in S. suis MVs, including seven surface proteins (two membrane-bound: HtpsC [37], Ide [38]; and five cell wall-anchored: SspA [39, 40], SsnA [41, 42], Mrp [43], ApuA [44] and Eno [45]), and two extracellular proteins (Plr [46] and Sly [47, 48]). The two most abundant virulence factors in MVs were the subtilisin protease SspA and the cell wall-anchored DNase SsnA, which were also among the most abundant proteins overall in S. suis MVs. Biological assays confirmed the presence of both subtilisin and DNase activities in MVs. SspA can degrade the Aα chain of fibrinogen and has been suggested to play a key role in the pathogenicity of S. suis since an SspA-deficient mutant is avirulent in a standard mouse model [40]. Moreover, recombinant SspA induces the secretion of major pro-inflammatory cytokines and, when present in high concentrations, degrades CCL5 and IL-6 [49]. SsnA is the most important cell surface DNase in S. suis and may also be an important virulence factor since an SsnA-deficient mutant has been shown to be susceptible to predation in an amoebae virulence model [32]. Vesicle-associated DNase activity has been previously identified in OMVs secreted by the periodontopathogenic bacterium Porphyromonas gingivalis [50]. Given that SsnA allows bacteria to escape killing by NETs through the degradation of the DNA backbone [42], we investigated the ability of S. suis MVs to degrade NETs produced by PMA-stimulated neutrophils. S. suis-derived MVs dose-dependently degraded extracellular DNA, suggesting that MVs produced by S. suis may play an important role in the ability of the bacteria to escape NETs.
Given that extracellular vesicles can deliver bacterial effectors to host cells, several studies have shown that these structures can modulate the host immune response [51–53]. The monocytic U937-3xκB-LUC cell line was used to monitor the activation of the NF-κB signaling pathway in order to characterize the immunogenic properties of S. suis MVs. The MVs dose-dependently increased NF-κB activity in both monocytes and PMA-differentiated macrophage-like cells. This is in agreement with a study showing that OMVs produced by Escherichia coli can activate the NF-κB pathway in endothelial cells [51]. Since the NF-κB signaling pathway mediates the secretion of several pro-inflammatory cytokines [54], we assayed cytokine secretion by PMA-differentiated U937 cells stimulated with S. suis MVs. The MVs dose-dependently stimulated CXCL-8, TNF-α, and IL-1β secretion by the U937 cells, which is in agreement with the results reported by Jun et al. (2013), who showed that OMVs secreted by Acinetobacter baumannii, an opportunistic pathogen, elicit a pro-inflammatory response via surface-exposed proteins both in vitro and in vivo [55]. Gram-positive MVs not only carry virulence factors and bacterial effectors [15, 56], but can also act as delivery systems for these molecules towards host cells inducing pro-inflammatory pathways that can lead to cellular death [18, 36, 52]. The activation of host cells by S. suis MVs may result from the interactions of vesicular constituents, which likely contain pathogen-associated molecular patterns (PAMPs) with Toll-like receptors (TLRs) as previously shown for MVs secreted by S. aureus [52]. Since the host inflammatory response is very important in bacterial meningitis [57], the immuno-modulatory activity of S. suis MVs may contribute to the etiopathogenic process of the disease. The accumulation of pro-inflammatory mediators can affect the permeability of the blood brain barrier during meningitis. Increasing the permeability of the blood brain barrier facilitates the migration of bacteria to the central nervous system and may promote disease progression and severity [58].
To the best of our knowledge, while more than twenty-five Gram-negative bacterial species have been shown to produce OMVs [59], the release of MVs by Gram-positive bacteria has been reported for much fewer species, including Bacillus anthracis [15], Bacillus subtilis [16], C. perfringens [18], L. monocytogenes [17], S. aureus [12], S. mutans [20], and S. pneumoniae [21]. The present study reports for the first time that S. suis produces MVs that carry large amounts of proteins and active virulence factors and that may reach areas not accessible to whole bacteria. In addition, MVs may compete for antibodies and impede specific antibacterial immune defenses.
The emergence of drug-resistant strains of S. suis due to the widespread and inappropriate use of antibiotics by the swine industry is a major economic and public health concern [60]. Because of this, it is important to explore the fundamental pathogenic mechanisms of S. suis involved during infections in order to identify new candidates for treating S. suis infections and developing effective vaccines. Since S. suis MVs present a broad array of antigens, they may be a potential candidate for designing vaccines for passive antibody-based therapeutic strategies. Extracellular bacterial vesicles activate immune responses and induce immunological memories [11, 59]. For example, it has recently been shown that MVs released by S. pneumoniae induce a protective immune response and protect mice against infections by a virulent strain [21]. Further characterization of the composition and immunogenicity of S. suis MVs is required to better evaluate the potential of these structures for the development of an effective acellular vaccine to prevent S. suis infections.
Acknowledgments
This study was funded by a Discovery grant to DG from the Natural Sciences and Engineering Research Council of Canada (NSERC). BH is the recipient of a scholarship from the Swine and Poultry Infectious Disease Research Center (CRIPA).
Data Availability
All relevant data are within the paper.
Funding Statement
This study was funded by Discovery grant 194703-2009 to DG from the Natural Sciences and Engineering Research Council of Canada (NSERC). BH is the recipient of a scholarship from the Swine and Poultry Infectious Disease Research Center (CRIPA).
References
- 1. Knox KW, Vesk M, Work E. Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli . J Bacteriol. 1966;92(4):1206–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–84. 10.1146/annurev.micro.091208.073413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437(7057):422–5. [DOI] [PubMed] [Google Scholar]
- 4. McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63(2):545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schooling SR, Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol. 2006;188(16):5945–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. MacDonald IA, Kuehn MJ. Offense and defense: microbial membrane vesicles play both ways. Res Microbiol. 2012;163(9–10):607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dorward DW, Garon CF, Judd RC. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae . J Bacteriol. 1989;171(5):2499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim DK, Kang B, Kim OY, Choi DS, Lee J, Kim SR, et al. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. 2013;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Veith PD, Chen YY, Gorasia DG, Chen D, Glew MD, O'Brien-Simpson NM, et al. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res. 2014;13(5):2420–32. 10.1021/pr401227e [DOI] [PubMed] [Google Scholar]
- 10. Collins BS. Gram-negative outer membrane vesicles in vaccine development. Discov Med. 2011;12(62):7–15. [PubMed] [Google Scholar]
- 11. Acevedo R, Fernandez S, Zayas C, Acosta A, Sarmiento ME, Ferro VA, et al. Bacterial outer membrane vesicles and vaccine applications. Front Immunol. 2014;5:121 10.3389/fimmu.2014.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S, et al. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics. 2009;9(24):5425–36. 10.1002/pmic.200900338 [DOI] [PubMed] [Google Scholar]
- 13. Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun. 2012;80(6):1948–57. 10.1128/IAI.06014-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dorward DW, Garon CF. DNA is packaged within membrane-derived vesicles of Gram-negative but not Gram-positive bacteria. Appl Environ Microbiol. 1990;56(6):1960–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rivera J, Cordero RJ, Nakouzi AS, Frases S, Nicola A, Casadevall A. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci U S A. 2010;107(44):19002–7. 10.1073/pnas.1008843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brown L, Kessler A, Cabezas-Sanchez P, Luque-Garcia JL, Casadevall A. Extracellular vesicles produced by the Gram-positive bacterium Bacillus subtilis are disrupted by the lipopeptide surfactin. Mol Microbiol. 2014;93(1):183–98. 10.1111/mmi.12650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee JH, Choi CW, Lee T, Kim SI, Lee JC, Shin JH. Transcription factor sigmaB plays an important role in the production of extracellular membrane-derived vesicles in Listeria monocytogenes . Plos One. 2013;8(8):e73196 10.1371/journal.pone.0073196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang Y, Kong Q, Roland KL, Curtiss R III. Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int J Med Microbiol. 2014;304(3–4):431–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schrempf H, Koebsch I, Walter S, Engelhardt H, Meschke H. Extracellular Streptomyces vesicles: amphorae for survival and defence. Microb Biotechnol. 2011;4(2):286–99. 10.1111/j.1751-7915.2011.00251.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn SJ, et al. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol. 2014;196(13):2355–66. 10.1128/JB.01493-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olaya-Abril A, Prados-Rosales R, McConnell MJ, Martin-Pena R, Gonzalez-Reyes JA, Jimenez-Munguia I, et al. Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae . J Proteomics. 2014;106:46–60. 10.1016/j.jprot.2014.04.023 [DOI] [PubMed] [Google Scholar]
- 22. Kim JH, Lee J, Park J, Gho YS. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin Cell Dev Biol. 2015;40:97–104. 10.1016/j.semcdb.2015.02.006 [DOI] [PubMed] [Google Scholar]
- 23. Gottschalk M. Streptococcosis In: Karriger L RA, Schwartz KJ, Stevenson G, Zimmerman J, editor. Diseases of swine. NJ, USA: Wiley Publishers; 2012. p. 841–55. [Google Scholar]
- 24. Segura M. Streptococcus suis: an emerging human threat. J Infect Dis. 2009;199(1):4–6. 10.1086/594371 [DOI] [PubMed] [Google Scholar]
- 25. Holden MT, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, et al. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis . Plos One. 2009;4(7):e6072 10.1371/journal.pone.0006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vanrobaeys M, De Herdt P, Charlier G, Ducatelle R, Haesebrouck F. Ultrastructure of surface components of Streptococcus gallolyticus (S. bovis) strains of differing virulence isolated from pigeons. Microbiology. 1999;145 (Pt 2):335–42. [DOI] [PubMed] [Google Scholar]
- 27. Kong M, Bhattacharya RN, James C, Basu A. A statistical approach to estimate the 3D size distribution of spheres from 2D size distributions. Geol Soc Am Bull. 2005;117:244–9. [Google Scholar]
- 28. Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6(9):1638–55. [DOI] [PubMed] [Google Scholar]
- 29. Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–92. [DOI] [PubMed] [Google Scholar]
- 30. Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. [DOI] [PubMed] [Google Scholar]
- 31. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haas B, Bonifait L, Vaillancourt K, Charette SJ, Gottschalk M, Grenier D. Characterization of DNase activity and gene in Streptococcus suis and evidence for a role as virulence factor. BMC research notes. 2014;7:424 10.1186/1756-0500-7-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Collins SJ, Ruscetti FW, Gallagher RE, Gallo RC. Normal functional characteristics of cultured human promyelocytic leukemia cells (HL-60) after induction of differentiation by dimethylsulfoxide. J Exp Med. 1979;149(4):969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carlsen H, Moskaug JO, Fromm SH, Blomhoff R. In vivo imaging of NF-kappa B activity. J Immunol. 2002;168(3):1441–6. [DOI] [PubMed] [Google Scholar]
- 35. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–5. [DOI] [PubMed] [Google Scholar]
- 36. Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, et al. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. Plos One. 2011;6(11):e27958 10.1371/journal.pone.0027958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shao Z, Pan X, Li X, Liu W, Han M, Wang C, et al. HtpS, a novel immunogenic cell surface-exposed protein of Streptococcus suis, confers protection in mice. Fems Microbiol Lett. 2011;314(2):174–82. 10.1111/j.1574-6968.2010.02162.x [DOI] [PubMed] [Google Scholar]
- 38. Seele J, Singpiel A, Spoerry C, von Pawel-Rammingen U, Valentin-Weigand P, Baums CG. Identification of a novel host-specific IgM protease in Streptococcus suis . J Bacteriol. 2013;195(5):930–40. 10.1128/JB.01875-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu QY, Liu P, Yu ZJ, Zhao G, Li J, Teng L, et al. Identification of a cell wall-associated subtilisin-like serine protease involved in the pathogenesis of Streptococcus suis serotype 2. Microb Pathogenesis. 2010;48(3–4):103–9. [DOI] [PubMed] [Google Scholar]
- 40. Bonifait L, Dominguez-Punaro MD, Vaillancourt K, Bart C, Slater J, Frenette M, et al. The cell envelope subtilisin-like proteinase is a virulence determinant for Streptococcus suis . BMC Microbiol. 2010;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fontaine MC, Perez-Casal J, Willson PJ. Investigation of a novel DNase of Streptococcus suis serotype 2. Infect Immun. 2004;72(2):774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Buhr N, Neumann A, Jerjomiceva N, von Kockritz-Blickwede M, Baums CG. Streptococcus suis DNase SsnA contributes to degradation of neutrophil extracellular traps (NETs) and evasion of NET-mediated antimicrobial activity. Microbiology. 2014;160(Pt 2):385–95. 10.1099/mic.0.072199-0 [DOI] [PubMed] [Google Scholar]
- 43. Smith HE, Vecht U, Gielkens AL, Smits MA. Cloning and nucleotide sequence of the gene encoding the 136-kilodalton surface protein (muramidase-released protein) of Streptococcus suis type 2. Infect Immun. 1992;60(6):2361–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ferrando ML, Fuentes S, de Greeff A, Smith H, Wells JM. ApuA, a multifunctional alpha-glucan-degrading enzyme of Streptococcus suis, mediates adhesion to porcine epithelium and mucus. Microbiology. 2010;156(Pt 9):2818–28. 10.1099/mic.0.037960-0 [DOI] [PubMed] [Google Scholar]
- 45. Esgleas M, Li Y, Hancock MA, Harel J, Dubreuil JD, Gottschalk M. Isolation and characterization of alpha-enolase, a novel fibronectin-binding protein from Streptococcus suis . Microbiology. 2008;154(Pt 9):2668–79. 10.1099/mic.0.2008/017145-0 [DOI] [PubMed] [Google Scholar]
- 46. Brassard J, Gottschalk M, Quessy S. Cloning and purification of the Streptococcus suis serotype 2 glyceraldehyde-3-phosphate dehydrogenase and its involvement as an adhesin. Vet Microbiol. 2004;102(1–2):87–94. [DOI] [PubMed] [Google Scholar]
- 47. Lun SC, Perez-Casal J, Connor W, Willson PJ. Role of suilysin in pathogenesis of Streptococcus suis capsular serotype 2. Microb Pathogenesis. 2003;34(1):27–37. [DOI] [PubMed] [Google Scholar]
- 48. Gottschalk MG, Lacouture S, Dubreuil JD. Characterization of Streptococcus suis capsular type-2 hemolysin. Microbiology. 1995;141:189–95. [DOI] [PubMed] [Google Scholar]
- 49. Bonifait L, Grenier D. The SspA subtilisin-like protease of Streptococcus suis triggers a pro-inflammatory response in macrophages through a non-proteolytic mechanism. BMC Microbiol. 2011;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leduc A, Grenier D, Mayrand D. Outer membrane-associated deoxyribonuclease activity of Porphyromonas gingivalis . Anaerobe. 1995;1(2):129–34. [DOI] [PubMed] [Google Scholar]
- 51. Soult MC, Lonergan NE, Shah B, Kim WK, Britt LD, Sullivan CJ. Outer membrane vesicles from pathogenic bacteria initiate an inflammatory response in human endothelial cells. J Surg Res. 2013;184(1):458–66. 10.1016/j.jss.2013.05.035 [DOI] [PubMed] [Google Scholar]
- 52. Kim MR, Hong SW, Choi EB, Lee WH, Kim YS, Jeon SG, et al. Staphylococcus aureus-derived extracellular vesicles induce neutrophilic pulmonary inflammation via both Th1 and Th17 cell responses. Allergy. 2012;67(10):1271–81. 10.1111/all.12001 [DOI] [PubMed] [Google Scholar]
- 53. Winter J, Letley D, Rhead J, Atherton J, Robinson K. Helicobacter pylori membrane vesicles stimulate innate pro- and anti-inflammatory responses and induce apoptosis in Jurkat T cells. Infect Immun. 2014;82(4):1372–81. 10.1128/IAI.01443-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Blackwell TS, Christman JW. The role of nuclear factor-kappa B in cytokine gene regulation. Am J Resp Cell Mol Biol. 1997;17(1):3–9. [DOI] [PubMed] [Google Scholar]
- 55. Jun SH, Lee JH, Kim BR, Kim SI, Park TI, Lee JC, et al. Acinetobacter baumannii outer membrane vesicles elicit a potent innate immune response via membrane proteins. Plos One. 2013;8(8):e71751 10.1371/journal.pone.0071751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee J, Lee EY, Kim SH, Kim DK, Park KS, Kim KP, et al. Staphylococcus aureus extracellular vesicles carry biologically active beta-lactamase. Antimicrob Agents Chemother. 2013;57(6):2589–95. 10.1128/AAC.00522-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koedel U, Scheld WM, Pfister HW. Pathogenesis and pathophysiology of pneumococcal meningitis. Lancet Infect Dis. 2002;2(12):721–36. [DOI] [PubMed] [Google Scholar]
- 58. Gottschalk M, Segura M. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet Microbiol. 2000;76(3):259–72. [DOI] [PubMed] [Google Scholar]
- 59. Kim GH, Choi CW, Park EC, Lee SY, Kim SI. Isolation and proteomic characterization of bacterial extracellular membrane vesicles. Curr Protein Pept Sci. 2014;15(7):719–31. [DOI] [PubMed] [Google Scholar]
- 60. Varela NP, Gadbois P, Thibault C, Gottschalk M, Dick P, Wilson J. Antimicrobial resistance and prudent drug use for Streptococcus suis . Anim Health Res Rev. 2013;14(1):68–77. 10.1017/S1466252313000029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.