Abstract
Inhibitors of RAF inhibit the MAPK pathway that plays an important role in the development and progression of those melanoma carrying the V600E BRAF mutation, but there’s a subset of such patients who do not respond to the therapy. Various mechanisms of drug resistance have been proposed which include the clonal heterogeneity of the tumor. We have studied a population of nodular melanoma to investigate the intratumor and intertumor heterogeneity by Laser Capture Microdissection (LCM) analysis. Our results showed that BRAF and NRAS mutations were detected in 47% and 33% of nodular melanoma, respectively, and that there is a discrepancy in mutational pattern of tumoral sample because in the 36% of patients a different mutation, in at least 1 area of the tumor, was found by LCM analysis, giving evidence of the presence of different clonal cells populations. Moreover, we found that mutations in BRAF and NRAS are not mutually exclusive because they were simultaneously present in the same tumor specimens and we observed that when the 2 different mutations were present one is a high-frequency mutation and the other is a low-frequency mutation. This was more evident in lymphonodal metastasis that resulted from wild type to mutational analysis, but showed different mutations following LCM analysis. Therefore, we believed that, when primary tumoral sample results negative to mutational analysis, if it is possible, metastases should be investigated to verify the presence of mutations. Generally, it should be searched for other mutations, in addition to BRAF V600E, so as to better understand the mechanism of drug resistance.
Key Words: nodular melanoma, BRAF, NRAS
Nodular melanoma is the second commonest subtype after superficial spreading melanoma, and it comprises 20% to 25% of cutaneous melanoma cases.1–3 The V600E mutation (Val600Glu) accounts for over 90% of all BRAF mutations detected in cutaneous melanoma, whereas the most common NRAS mutation in cutaneous melanoma are the Q61R (Glut61Arg) and Q61K (Glut61Lys); these mutations result in activation of the downstream effector of the RAS-RAF-MEK-MAPK pathway.4 This pathway is important in several crucial processes, such as proliferation and differentiation.3 Inhibitors of BRAF (vemurafenib and debrafenib) have been developed and have shown clinical benefits, such as response rate, progression-free survival, and overall survival, compared with chemotherapy treatments in patients with BRAF mutated metastatic melanoma.5 Despite this advances, the great majority of patients treated with these drugs have developed disease progression within 6 to 7 months after the initiation of the treatment.5–7 The mechanisms of resistance are multiples and include the reactivation of alternative signaling pathway as well as MAPK pathway involvement5–7 through different ways like the development of an NRAS mutation.8 Recent results indicate that it is not possible to think of an effective therapeutic strategy or of a more sensitive and specific evaluation of malignant tumors without taking into account more molecular parameters5–7; in fact, the acquisition of multiple genomic alterations can cause the formation of different subclones in the same tumor or in the primitive and metastatic localization resulting in intratumor and intertumor heterogeneity in a single patient; it was hypothesized that this process could be one of the mechanism causing resistance to the drug.9 A better understanding of intratumor and intertumor heterogeneity is essential to advance the effect of molecular diagnostics in determining therapeutic strategies. In this study, we analyzed a population of nodular melanoma tumor specimens to detect mutations in BRAF and NRAS using Laser Capture Microdissection (LCM) and investigating in such way the intratumor heterogeneity to define the presence of different clonal population of tumor cells.
MATERIALS AND METHODS
Population Study
The population of this study included 15 patients with nodular melanoma, 9 men and 6 women, with median age of 71 years (range, 23 to 92 y). We chose this histologic type of melanoma because in this study we needed rich tissue samples to make molecular analysis by LCM. The samples were collected from the files of the Pathology Unit of the Latina Local Health Unit at I.C.O.T hospital. The WHO 2008 classification of cutaneous melanoma was used to classify the lesions. Median tumor size was 2.3 mm (range, 1 to 6 mm); of the 15 patients, 2 (13.5%) were of Clark level III, 11 (73%) were of Clark level IV, and 2 (13.5%) were of Clark level V. The Breslow thickness ranged from 1.6 to 12 mm with a mean of 7.1 mm. The majority of patients (10/15, 66.7%) presented ulceration and the mean value of mitotic index was 7. Twelve patients presented regression area (80%, 12/15), 13 patients (87%) showed tumor-infiltrating lymphocytes, and only one of these was in the brisk category (1/13, 8%). Eleven patients underwent sentinel lymph node biopsy and only 1 resulted positive (1/15; 6%). Four patients did not undergo to sentinel lymph node biopsy because they presented obvious metastasis by image analysis techniques. The tumor stage was IIC in 6/15 (40%), IIB in 3/15 (20%), IIA in 1/15 (6%), IIIB in 1/15 (6%), and IV in 4/15 (27%). Immunoistochemical analysis of S100, HMB45, MART1, and Melan A were performed in each case to confirm the morphologic and clinical diagnoses. The mean value of the proliferation index evaluated by Ki67 was 14.7%. Median follow-up was 31 months (range, 12 to 60 mo) and 7/15 (47%) patients died.
Mutation Analysis
Tumor tissue samples were analyzed for both BRAF (NCBI Gene ID, 673) and NRAS (NCBI Gene ID, 4893) gene mutations by direct sequencing analysis of exons 11 and 15 of the BRAF gene and exons 1 and 2 of the NRAS gene. Representative tumor tissue sections (>80% tumor cells) were cut (10 µm thick) and placed directly into a sterile tube. DNA was extracted using Wizard SV Genomic DNA Purification System (Promega, Madison, CA). DNAs were amplified in a final volume of 50 µL containing 30 ng of DNA, 2 mM dNTP, 250 ng/µL of each primer, 1.5 mM MgCl2, 1× PCR buffer, and 1 U HotStartGoTaq Polymerase (Promega). A total of 40 cycles were performed using the Gene Amp PCR System 9700 (Life Technologies, Foster City, CA) at 95°C for 45 seconds, specific annealing temperature for 45 seconds, 72°C for 1 minute. The PCR products were then purified using Exosap-IT (Affymetrix, Santa Clara, CA) and then sequenced using Big Dye Terminator version 1.1 Cycle Sequencing Kit (Life Technologies). Unincorporated primers and dye terminators were removed using the Montage-SEQ96 Sequencing Reaction Cleanup Kit (Merck Millipore, Billerica, MA). Sequencing was performed on an ABI PRISM 3100 Genetic Analyzer (Life Technologies) with 3100 Genetic Analyzer Data Collection software version 1.1. The sequencing and each reaction were performed in triplicate.
LCM to Analyze Intratumor Heterogeneity
We used LCM to isolate 3 areas of 10,000 tumor cells from each of 15 melanoma tumor specimens. For microdissection by Leica LMD 7000 (Leica Microsystems, MI, Italy), 5-µm-thick sections were performed on specific glasses with thermoplastic membrane activated by a low-energy infrared laser pulse and stained with hematoxylin and eosin. The selected tissue fragments were harvested by simple lifting of the cap, which was then transferred to a microcentrifuge tube containing the buffer solutions required for the isolation of the DNA. Total DNA from the microdissected tissue was extracted by using DNA extraction buffer (Tris-HCl pH 8: 100 mM; EDTA: 1 mM; Tween-20: 1%; Proteinase K: 200 to 300 μg/mL) and incubated at 37°C for 16 hours. The reaction was then inactivated at 95°C for 20 minutes. The DNA was stored at −20°C until it was used. Each dissected tumor sample was evaluated for selective analysis of BRAF and NRAS mutations via sequencing.
RESULTS
Mutations Analysis
We detected mutations in 11/15 (73%) cases; 6 patients (54%, 6/11) showed mutations in exon 15 of BRAF, only 1 patient showed a mutation in exon 11 of BRAF (9%, 1/11); the substitution at codon 600 accounted for 71% (5/7) of BRAF mutations and 3 of these were represented by the valine to glutamic acid substitution (V600E, 60%, 3/5). Five patients (45%, 5/11) revealed mutations in NRAS, only 1 in exon 1 (20%, 1/5) and 4 in exon 2 (80%, 4/5), specifically a glutamine to leucine/arginine/lysine substitution at position 61 (Q61L/R/K). In a same tumor specimen we found mutations in BRAF and NRAS, in particular the substitution at position 467 of serine in leucine in exon 11 of BRAF and the substitution at position 61 of glutamine in arginine in exon 2 of NRAS. Four patients (27%, 4/15) did not have mutation in either BRAF or NRAS and were referred to as wild type (WT).
Mutations Analysis by LCM
We analyzed the 3 microdissected areas of each tumor sample to determine whether they contained a mixture of subclones characterized by a different mutational status. We found that 7/11 (64%) mutated patients showed the same mutation found by mutational analysis without LCM in at least 1 area. Only 4 patients (57%, 4/7) showed the same mutation in all 3 areas, specifically V600E in exon 15 of BRAF in 2 patients and the glycine to arginine substitution at position 13 of NRAS exon 1 (G13R) in the third patient; the patient that showed 2 different mutations in the same tumor specimen revealed this characteristic through the LCM analysis in all 3 areas too (S467L BRAF exon 11 and Q61R NRAS exon 2, Fig. 1A). Two patients (29%, 2/7) showed the same mutation in 2 areas: the lysine to glutamic acid substitution at position 601 of BRAF exon 15 (K601E) and the other showed the substitution of valine in arginine in exon 15 of BRAF (V600R) while 1 area resulting WT. Only 1 patient (14%, 1/7) showed the same mutation in 1 area, the valine to lysine substitution at position 600 of BRAF exon 15 (V600K), whereas the other areas resulted WT. Four patients (36%, 4/11) revealed a different mutational status in at least 1 area compared with mutational analysis without LCM: 1 patient (25%, 1/4) showed the same mutation in 2 areas (V600E in exon 15 of BRAF) and another mutation (V600R) in the same exon in the third area; 2 patients (50%, 2/4) showed the same mutation in all 3 areas (Q61R and Q61K) and also other mutations in BRAF exon 15, in particular the first showed the V600R mutation and the alanine to valine substitution at position 598 (A598V, Fig. 1B) and the last showed the isoleucine to valine substitution at position 592 (I592V); another patient (25%, 1/4) showed the same mutation in 2 areas (Q61L) and also revealed the glycine to arginine substitution at position 455 of BRAF exon 11 (G455R). In the group referred to as WT, only 1 patient (1/4, 25%) showed a V600E mutation in all 3 areas (Fig. 1C). The last 3 patients (3/4, 75%) were also referred to as WT by LCM analysis.
FIGURE 1.
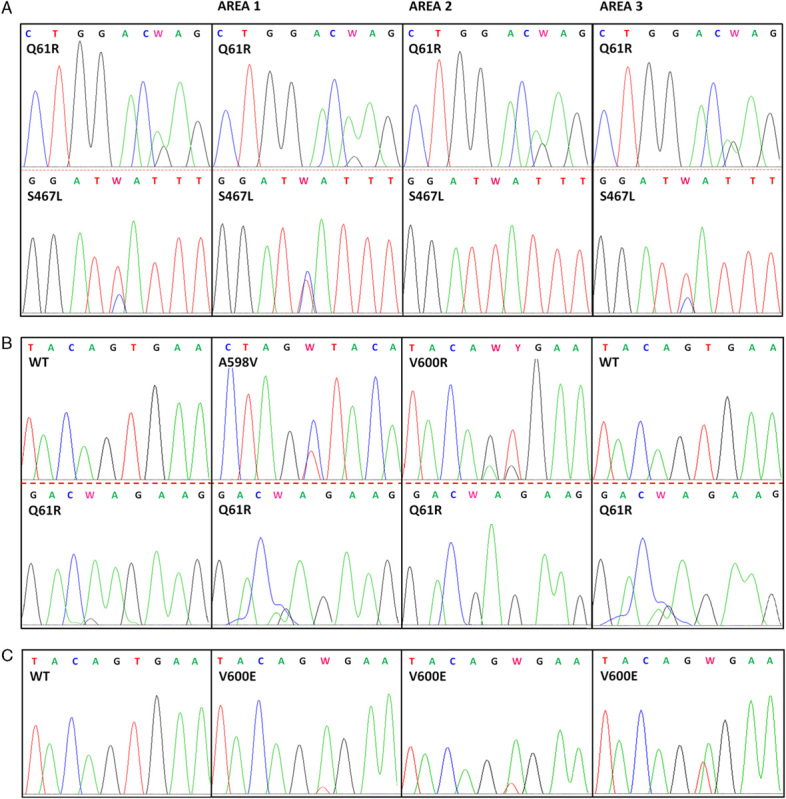
Sequencing electropherograms of the regions of BRAF exons 11 and 15 and NRAS exons 1 and 2. In each line, the first electropherogram corresponds to the tumoral sample, the other 3 electropherograms correspond to the 3 areas of the microdissected tumor. A, Patient that shows the same 2 mutations of tumor sample (S467L of BRAF exon 11, Q61R of NRAS exon 2) in all 3 areas; (B) patient that shows the same mutation found in tumoral sample in all 3 areas (Q61R of NRAS exon 2) and also other 2 mutation in 2 different areas (A598V and V600R on BRAF exon 15, respectively); (C) patient that shows the V600E mutation of BRAF exon 15 in all 3 areas while the tumoral sample is referred to as wild type (WT).
One patient, male, 59 years old, referred as WT in primary tumor specimens and by LCM analysis, showed 6 lymph nodal metastasis (LGH); their mutational analysis did not show mutations, whereas by the LCM analysis we found a different mutational status in 4/6 (67%) LGH:
I LGH: revealed V600E in exon 15 of BRAF in 1 area;
II LGH: showed the valine to isoleucine substitution at position 590 of BRAF exon 15 (V590I) in 1 area;
III LGH: 2 different mutations were revealed in the same area: the serine to proline substitution at position 614 (S614P) and the V600E mutation in exon 15 of BRAF; moreover, this showed the V600E mutation in another area;
IV LGH: mutations in BRAF and NRAS were revealed in 2 different areas, specially the glycine to arginine substitution at position 469 of BRAF exon 11 and the G13R mutation in exon 1 of NRAS;
V LGH: referred as WT;
VI LGH: referred as WT (Fig. 2).
FIGURE 2.
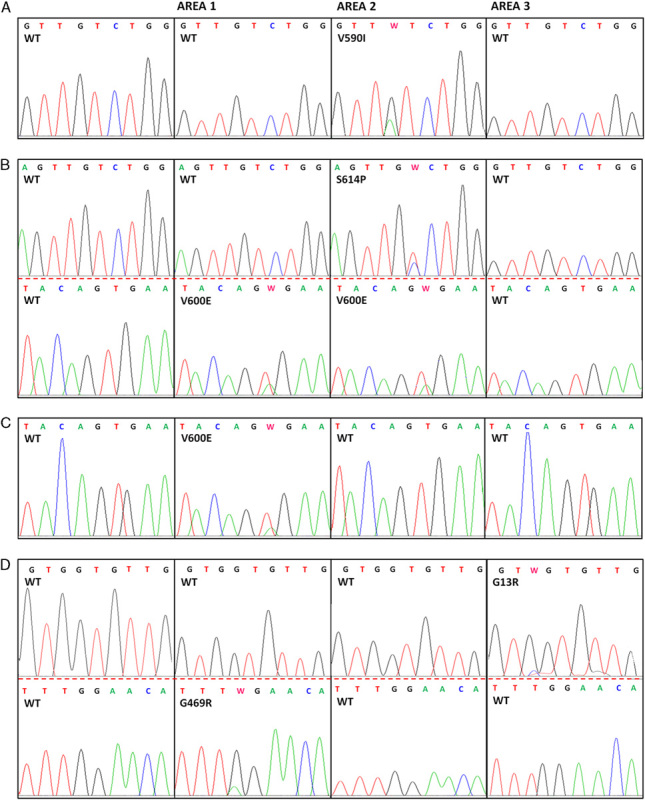
Sequencing electropherograms of the LCM areas that show mutations in BRAF exons 11 and 15 and NRAS exons 1 and 2 of metastatic samples. In each line, the first electropherogram corresponds to the metastatic sample, the other 3 electropherograms correspond to the 3 areas of the microdissected metastasis. A, Metastatic sample that shows the V590I mutation of BRAF exon 15 in 1 area; (B) metastatic sample that shows the V600E mutation of BRAF exon 15 in 2 areas and the S614P mutation of BRAF exon 15 also in area 2; (C) metastatic sample that shows the V600E mutation of BRAF exon 15 only in 1 area; (D) metastatic sample that shows 2 different mutation in 2 different areas, G469R of BRAF exon 11 and G13R of NRAS exon 1.
DISCUSSION
The RAS/RAF/MEK/ERK pathway has been reported to be activated in over 90% of all cutaneous melanomas, and BRAF and NRAS mutations were studied to understand the mechanisms responsible for oncogenesis10,11 and to provide new therapeutic strategies for this deadly disease.12,13
In this study, the frequency of BRAF and NRAS mutations (47%/33%) was consistent with that reported in previous studies in which BRAF mutation ranged from 22% to 72% and NRAS mutation ranged from 0% to 50%4,14,15 and, in agreement with the literature, the majority of BRAF mutations were in codon 600.14,16–19
Surprisingly, 1 patient showed 2 different mutations, 1 in BRAF exon 11 (S467L) and the other in NRAS exon 2 (Q61R) showing that these mutations are not mutually exclusive as previously stated13; moreover, we confirmed this result founding both mutations in all 3 areas of the same tumor, selected with LCM (Fig. 1A). Through LCM analysis, we found a discrepancy in the mutation pattern because the 36% of patients revealed a different mutational status in at least 1 area and even if these mutations have been already reported in literature the frequency appeared to be lower.20–22 Finally, in WT group 1 patient (27%) showed mutations in at least 1 area.
Difference in mutational status was more evident in the analysis of lymph nodal metastasis of a patient; indeed, 4/6 lymph nodal metastasis showed a different mutational pattern from each other and from the primitive sample. Particularly, one of them presented 2 different mutations in the same area, the V600E and the less well-known S614P mutation giving evidence of the intratumor and intertumor heterogeneity as already argued.13,16,17,23
BRAF-targeted therapies showed important results in therapy for melanoma with activating BRAF V600E mutations, but mechanisms of intrinsic and acquired drug resistance have been observed. Our results showed that a second mutation can be present in the same patients and it was postulated that the acquisition of other mutations could be the cause of the MAPK pathway reactivation and tumor progression.8,24 As well known, the investigation of V600E mutation to refer patients to the treatment with inhibitors of BRAF is carried out routinely on the primary tumor; our results suggest that if the primary sample is negative for mutation V600E and the metastatic samples are available, it can be useful also to perform this mutational analysis on the metastatic samples, to give the patient another chance to be eligible for the treatment with inhibitors of BRAF.
We further found that, besides the most frequent mutations BRAF V600E and NRAS Q61R/K,3,4,25–27 there are also other mutations that should be investigated. Often, less frequent mutations are not found because the new methods of mutational analysis are more sensitive but only for the identification of specific known mutations9,28,29 or because the same exons of a gene are not always investigated in different studies.15,30 Our results show that the mutations are not mutually exclusive in nodular melanomas, indeed we found BRAF and NRAS mutations in the same sample; however, we hypothesize that the most frequent mutations could be mutually exclusive, in fact in this study in the case in which 2 different mutations were present, one of these was a high-frequency mutation and the other was a low frequency one.
The mechanism by which the other mutations can participate in the oncogenetic process was not clarified yet.13 Further studies are needed to confirm these preliminary results in larger population and it is important to expand the panel of mutations to be investigated including genetic and clinical information in better understanding the mechanisms of drug resistance in melanoma. For these reasons, it is necessary that mutational analysis is performed with more sensitive methods such as the whole-genome sequencing.
ACKNOWLEDGMENTS
The authors thank the “Serena Talarico Association,” “Fondazione Roma,” and “AISOS” for the precious support to this research.
Footnotes
The authors declare no conflict of interest.
REFERENCES
- 1.Nestle FO, Halpern AC.Bolognia JL, Jorizzo JL, Rapini RP. Melanoma. Dermatology. 2008;22nd edSpain: Mosby;1745–1771. [Google Scholar]
- 2.Menzies SW, Moloney FJ, Byth K, et al. Dermoscopic evaluation of nodular melanoma. JAMA Dermatol. 2013;149:699–709. [DOI] [PubMed] [Google Scholar]
- 3.Saldanha G, Potter L, Daforno P, et al. Cutaneous melanoma subtypes show different BRAF and NRAS mutation frequencies. Clin Cancer Res. 2006;12:4499–4505. [DOI] [PubMed] [Google Scholar]
- 4.Lee JH, Choi JW, Kim YS. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol. 2011;164:776–784. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan RJ, Flaherty KT. Resistance to BRAF-target therapy in melanoma. Eur J Cancer. 2013;49:1297–1304. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stones CJ, Kim JE, Joseph WR, et al. Comparison of responses of human melanoma cell lines to MEK and BRAF inhibitors. Front Genet. 2013;169:287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to BRAF (V600E) inhibition by RTK or NRAS upregulation. Nature. 2010;468:973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yancovitz M, Litterman A, Yoon J, et al. Intra- and Inter-tumor heterogeneity of BRAFV600E mutations in primary and metastatic melanoma. Plos One. 2012;7:e29336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. [DOI] [PubMed] [Google Scholar]
- 11.Broekaert SM, Roy R, Okamoto I, et al. Genetic and morphologic features for melanoma classification. Pigment Cell Melanoma Res. 2010;23:763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhomen N, Marais R. BRAF signaling and targeted therapies in melanoma. Hematol Oncol Clin North Am. 2009;23:529–545. [DOI] [PubMed] [Google Scholar]
- 13.Mandalà M, Voit C. Targeting BRAF in melanoma: biological and clinical challenges. Crit Rev Oncol Hematol. 2013;87:239–255. [DOI] [PubMed] [Google Scholar]
- 14.Wang AX, Qi XY. Targeting RAS/RAF/MEK/ERK signaling in metastatic melanoma. IUBMB Life. 2013;65:748–758. [DOI] [PubMed] [Google Scholar]
- 15.Colombino M, Capone M, Lissia A, et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012;30:2522–2529. [DOI] [PubMed] [Google Scholar]
- 16.Boursault L, Haddad V, Vergier B, et al. Tumor homogeneity between primary and metastatic sites for BRAF status in metastatic melanoma determined by immunohistochemical and molecular testing. PLoS One. 2013;8:e70826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinzerling L, Baiter M, Kühnapfel S, et al. Mutation landscape in melanoma patients clinical implications of heterogeneity of BRAF mutations. Br J Cancer. 2013;109:2833–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greaves WO, Verma S, Patel KP, et al. Frequency and spectrum of BRAF mutations in a retrospective, single-institution study of 1112 cases of melanoma. J Mol Diagn. 2013;15:220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoenewolf NL, Dummer R, Mihic-Probst D, et al. Detecting BRAF mutations in formalin-fixed melanoma: experiences with two state-of-the-art techniques. Case Rep Oncol. 2012;5:280–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas NE, Edmiston SN, Alexander A, et al. Number of nevi and early-life ambient UV exposure are associated with BRAF-mutant melanoma. Cancer Epidemiol Biomarkers Prev. 2007;16:991–997. [DOI] [PubMed] [Google Scholar]
- 21.Tetsu O, Phuchareon J, Chou A, et al. Mutations in the c-Kit gene disrupt mitogen-activated protein kinase signaling during tumor development in adenoid cystic carcinoma of the salivary glands. Neoplasia. 2010;12:708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akslen LA, Puntervoll H, Bachmann IM, et al. Mutation analysis of the EGFR-NRAS-BRAF pathway in melanomas from black Africans and other subgroups of cutaneous melanoma. Melanoma Res. 2008;18:29–35. [DOI] [PubMed] [Google Scholar]
- 23.Lin J, Goto Y, Murata H, et al. Polyclonality of BRAF mutations in primary melanoma and the selection of mutant alleles during progression. Br J Cancer. 2011;104:464–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le K, Blomain ES, Rodeck U, et al. Selective RAF inhibitor impairs ERK1/2 phosphorylation and growth in mutant NRAS, vemurafenib-resistant melanoma cells. Pigment Cell Melanoma Res. 2013;26:509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platz A, Egyhazi S, Ringborg U, et al. Human cutaneous melanoma; a review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site. Mol Oncol. 2008;1:395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goel VK, Lazar AJ, Warneke CL, et al. Examination of mutations in BRAF, NRAS, and PTEN in primary coutaneous melanoma. J Invest Dermatol. 2006;126:154–160. [DOI] [PubMed] [Google Scholar]
- 27.Poynter JN, Elder JT, Fullen DR, et al. BRAF and NRAS mutations in melanoma and melanocytic nevi. Melanoma Res. 2006;16:267–273. [DOI] [PubMed] [Google Scholar]
- 28.Richter A, Grieu F, Carrello A, et al. A multisite blinded study for the detection of BRAF mutations in formalin-fixed, paraffin-embedded malignant melanoma. Sci Rep. 2013;3:1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machnicki MM, Glodkowska-Mrowka E, Lewandowski T, et al. ARMS-PCR for detection of BRAF V600E hotspot mutation in comparison with real-time PCR-based techniques. Acta Biochim Pol. 2013;60:57–64. [PubMed] [Google Scholar]
- 30.Bucheit AD, Syklawer E, Jakob JA, et al. Clinical characteristics and outcomes with specific BRAF and NRAS mutations in patients with metastatic melanoma. Cancer. 2013;119:3821–3829. [DOI] [PubMed] [Google Scholar]


