Abstract
Melanoma is a devastating form of skin cancer with limited therapeutic options. Fifteen to twenty percent of melanoma patients have an activating mutation in the GTPase, NRAS. The major downstream effectors of RAS are RAFs (ARAF, BRAF, CRAF), phosphatidylinositol 3-kinase (PI3K), and the Ral guanine exchange factors (RalGEFs). TANK-binding kinase 1 (TBK1) is an atypical IκB kinase family member that acts downstream of RalGEFs. While many studies have analyzed RAF and PI3K signaling in mutant NRAS melanoma, the role of RalGEF/Ral is understudied and TBK1 has not been examined. To address this, TBK1 was modulated with knockdown approaches and targeted therapies to determine TBK1's role in motility, apoptosis and signaling. In melanoma, NRAS overexpression increased TBK1 phosphorylation. TBK1 depletion inhibited migration and invasion, while its constitutive overexpression led to an increase in invasion. In three dimensional (3D) systems that mimic the dermal microenvironment, TBK1 depletion or inhibition cooperated with MEK inhibitors to promote apoptosis, particularly in the context of MEK-insensitive mutant NRAS. This effect was absent in melanoma cells that are wild-type for NRAS. These results suggest the utility of TBK1 inhibitors as part of a treatment regimen for mutant NRAS melanoma patients, for whom there are no current effective therapies.
Keywords: NRAS, TBK1, MEK, Melanoma, Apoptosis, Invasion
INTRODUCTION
Melanoma is the deadliest form of skin cancer and its incidence is increasing at a greater rate than other cancers (1). It is notorious for its propensity to migrate and invade as well as its resistance to apoptosis. If melanoma is confined to expansion within the epidermis, it is easily cured with surgical intervention (2, 3). However, without early diagnosis, the cells can invade through the basement membrane and expand through deeper dermal layers. The mechanisms underlying melanoma invasion from the epidermis into the dermis remain poorly characterized. In addition to its aggressive nature, melanoma is also notorious for its evasion of apoptosis, a property that has made treatment extremely difficult. Conventional chemotherapies, such as alkylating agents, antibiotics, taxanes, and platinum drugs, do not elicit a significant therapeutic benefit (4).
Fifteen to twenty percent of melanoma patients have mutations in NRAS, most frequently Q61R/K/L that trap the protein in a GTP-bound state leading to its constitutive activation (5, 6). Additionally, mutant BRAF melanoma patients who have been treated with the RAF inhibitor vemurafenib often gain resistance through acquisition of a NRAS mutation or selection for cells with a co-existing NRAS mutation, which permits maintenance of MEK-ERK1/2 pathway activation in the presence of RAF inhibitors (7). While there are no targeted therapies that are FDA-approved for melanoma patients with a primary mutation in NRAS or those that develop it secondarily, one form of targeted therapy that has been explored are MEK inhibitors. In contrast to findings in mutant BRAF V600E/K melanomas (8), Phase I to III studies of MEK inhibitors in mutant NRAS melanomas have not shown consistent clinical efficacy (9, 10). The underlying basis for the varied response is not known. NRAS activates multiple effector pathways and some mutant NRAS melanoma cells may be less reliant on MEK-ERK1/2 signaling. The major downstream effectors of RAS are RAFs (ARAF, BRAF, CRAF), phosphatidylinositol 3-kinase (PI3K), and the Ral guanine exchange factors (RalGEFs) (11). In mutant NRAS melanoma cells that are resistant to MEK inhibitor treatment, PI3K and AKT activity may be important compensatory pathways. Recent evidence has suggested that RalA/B-mediated pathways may also play a key role in mutant NRAS melanomas (12, 13).
RalB signals, in part, by interacting with Sec5, a component of the exocyst complex involved in the maintenance of cell polarity, cell motility and cytokinesis (14). Sec5 directly recruits and activates TANK-binding kinase 1 (TBK1), an atypical IκB kinase family member (15). Thus, TBK1 is activated downstream of RalB and has been implicated in the phosphorylation of AKT at both Thr308 and Ser473 (16-18). Furthermore, TBK1 has been implicated in oncogenesis since its depletion reduces the tumorigenic potential of KRAS-transformed fibroblasts (14), mutant KRAS non-small cell lung carcinoma cell lines (16), and mutant KRAS pancreatic cancer cells (17). Given its role in KRAS-mediated oncogenesis, we examined the role of TBK1 in the malignant properties of mutant NRAS melanoma.
Here, we show that TBK1 is regulated by activated NRAS expression in melanoma cells. In mutant NRAS cells, TBK1 depletion leads to a decrease in tumor cell migration and invasion. In contrast, the expression of a constitutively active form of TBK1 led to an increase in tumor cell invasion. In 3D collagen cultures, the combination of TBK1 and MEK inhibition cooperated to enhance apoptosis in mutant NRAS melanoma cells as well as vemurafenib-resistant mutant BRAF melanoma cells harboring secondary mutation in NRAS. This effect was absent in melanoma cell lines that are wild type for NRAS. These results suggest that TBK1 promotes melanoma cell motility and is a potential target as part of a combination regimen for a subset of melanoma patients with no current effective therapy options.
MATERIALS AND METHODS
Cell culture
SKMel30 and SKMel173 were kindly donated by Dr. David Solit (Memorial Sloan-Kettering Cancer Center, New York, NY). WM852 and WM3670 were purchased from Coriell Institute (Camden, NJ). Other WM melanoma lines were kindly donated by Dr. Meenhard Herlyn (Wistar Institute, Philadelphia, PA). Additional SKMel cell lines were purchased from ATCC (Manassas, VA). WM cell lines,1205Lu, SKMel28, and SBcl2 were cultured in MCDB 153 medium containing 20% Leibovitz L-15 medium, 2% fetal bovine serum (FBS) and 0.2% sodium bicarbonate (WM medium). WM793-Res#5 and WM793-Res #12 were cultured in WM medium in the presence of 5 μM PLX4720 (19). SKMel2 cells were cultured in MEM with 10% FBS. SKMel30, SKMel32, SKMel173, and SKMel207 were cultured in RPMI-1640 with 10% FBS. SKMel3 cells were cultured in McCoy's 5A medium containing 15% FBS. SKMel5 cells were cultured in MEM with 10% FBS, 10% non-essential amino acids, 1 mM sodium pyruvate, and 0.15% sodium bicarbonate.
Western blot analysis
Cells lysed in sample buffer were separated by SDS-PAGE, and proteins were transferred electrophoretically onto Immobilon P membranes (Millipore Corp., Bedford, MA). Membranes were blocked with phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 0.1% Tween 20 for 1 hour and subsequently incubated with primary antibody overnight at 4°C. Antibodies for TBK1/NAK (3504), phospho-TBK1 Ser172 (5483), AKT (9272), phospho-AKT Ser473 (4060), phospho-ERK (9101), IRF3 (4302), and cleaved PARP (9541) were obtained from Cell Signaling Tech. (Danvers, MA); ERK1 (K-23) and NRAS (C-20) from Santa Cruz Biotechnology (Santa Cruz, CA); actin (A2066) from Sigma-Aldrich (St. Louis, MO); and phospho-IRF3 Ser386 (9076443) from Abcam (Cambridge, UK). Membranes were washed in PBS/Tween and incubated with anti-mouse or anti-rabbit IgG peroxidase conjugates (Calbiochem, San Diego, CA) for 1 hour at room temperature. Western blots were developed using SuperSignal chemiluminescent substrate (Pierce, Rockford, IL). Immunoreactivity was detected and quantified using a Fluor-S Multi-Imager and Quantity-One software (Bio-Rad, Hercules, CA).
Cloning and stable cell line generation
A mutant NRASQ61K construct was generated, as previously described (19). Human TBK1 with a tag for myristoylation was cloned from cDNA using the following primers: forward 5’-CACCATGGGAAGCAGCAAGAGCAAGCCCAAGGACCCCAGCCAGCGCGCCCAGAGCACTT CTAATCATCTGG-3’ and reverse 5’-AAGACAGTCAACGTTGCGAAG-3’. All DNA constructs were sequence verified. Lentiviral particles and tetracycline repressor–expressing (TR-expressing) sublines WM1361A TR and WM1366 TR expressing Dox-inducible TBK1-myr were generated, as previously described (20). Transgene expression was induced with 0.1 μg/ml doxycycline in the cell culture medium.
Immunofluorescence
Melanoma cells growing on glass coverslips were washed with PBS and fixed with 3.7% formaldehyde for 20 min. Cells were then permeabilized with 0.2% Triton X-100 for 5 min and non-specific staining blocked with 1% BSA/PBS for 2 hours at room temperature. Coverslips were then incubated with primary antibodies diluted in 1% BSA/PBS overnight at 4°C. After primary antibody, coverslips were washed three times with PBS before incubation with appropriate Alexa Fluor-conjugated secondary antibodies (Invitrogen, Carlsbad, CA) for 1 hour at room temperature. In some instances, the coverslips were incubated with DAPI to visualize nuclei. Coverslips were mounted and visualized on an Eclipse Ti inverted microscope with NIS-Elements AR 3.00 software (Nikon, Melville, NY).
RNA interference
Mutant NRAS cells were transfected for 4 hours with chemically synthesized short-interfering RNA (siRNA) at a final concentration of 25 nM using OligofectAMINE (Invitrogen). Cells were harvested after a further 68 hours. The TBK1 siRNAs (#1: GAACGUAGAUUAGCUUAUAUU; #2: UGACAGAGAUUUACUAUCA) were purchased from Dharmacon Inc. (Lafayette, CO).
Migration and invasion assay
Sub-confluent melanoma cells were cultured overnight in serum-free medium. Cells (4.2×104) in serum-free medium were placed inside 8.0μm pore-size cell culture inserts (BD Biosciences, Franklin Lakes, NJ). Cells were allowed to migrate for twenty-four hours towards an attractant of full-serum medium. Chamber filters were fixed in buffered formalin and stained with crystal violet. The cells in the inner chamber were removed. Images in triplicate from each filter were taken with a Nikon Ti-Eclipse inverted microscope and stained cells were counted manually utilizing the NIS-Elements Software package. Images used for quantitation were taken at 10× magnification. The average count from control filters triplicate counts from triplicate independent experiments was normalized to 100% migration. All counts were converted to percent migration in comparison to the control 100% migration. For invasion studies, the migration assay was modified with coating of the inner chamber with 0.75mg/ml Matrigel (BD Biosciences) for one hour prior to addition of cells.
Colony formation assay
Mutant NRAS cells (4 × 103) were plated per 6-well plate in complete medium with or without inhibitors, which were replenished every two days. After 7 days, cells were stained with crystal violet in formalin, plates were imaged by scanner, and colonies were imaged on a Nikon Eclipse Ti inverted microscope with NIS-Elements AR 3.00 software.
3D collagen gels and apoptosis assay
Collagen gels were cast and cells were isolated and stained with annexin V-APC as previously described (19). Apoptosis was analyzed by flow cytometry on FACSCalibur flow cytometer (BD Biosciences). Data were analyzed by FlowJo software (Tree Star Inc., Ashland, OR).
Reagents
AZD6244 was purchased from Selleck Chemicals LLC (Houston, TX). BX795 was purchased from InvivoGen (San Diego, CA). AZ13102909 was kindly provided by Dr. Claudio Chuaqui and AstraZeneca (London, UK).
RESULTS
TBK1 is expressed and activated in a subset of mutant NRAS melanoma cells lines
TBK1 is an atypical IκB kinase family member that plays an important role in KRAS-mediated oncogenesis through its phosphorylation of AKT at the Thr308 and Ser473 sites (17, 18). Given its role in mutant KRAS-mediated oncogenesis, we examined the potential role of TBK1 in the malignant properties of mutant NRAS melanoma. We initially analyzed the expression level of TBK1 in a panel of mutant NRAS melanoma cell lines from different stages of melanoma progression and NRAS mutation (Supplementary Table 1). By Western blot analysis, TBK1 was ubiquitously expressed in both mutant NRAS and mutant BRAF cell lines examined, albeit to different levels (Figure 1A). Autophosphorylation of TBK1 at Ser172 is required for its activation (21). Levels of phosphorylated TBK1 at Ser172 were detectable in several cell lines and, in the mutant NRAS panel, correlated with high levels of NRAS. IKKε is another member of the atypical IκB kinase family that shares 67% amino acid identity with TBK1 within the kinase domain (14). Since IKKε has been implicated in tumorigenesis, particularly breast, ovarian and pancreatic cancer (22), we analyzed levels of IKKε and found that its expression level was barely detectable in mutant NRAS melanoma cell lines compared to RAW 264.7 macrophage cells (Supplementary Figure 1).
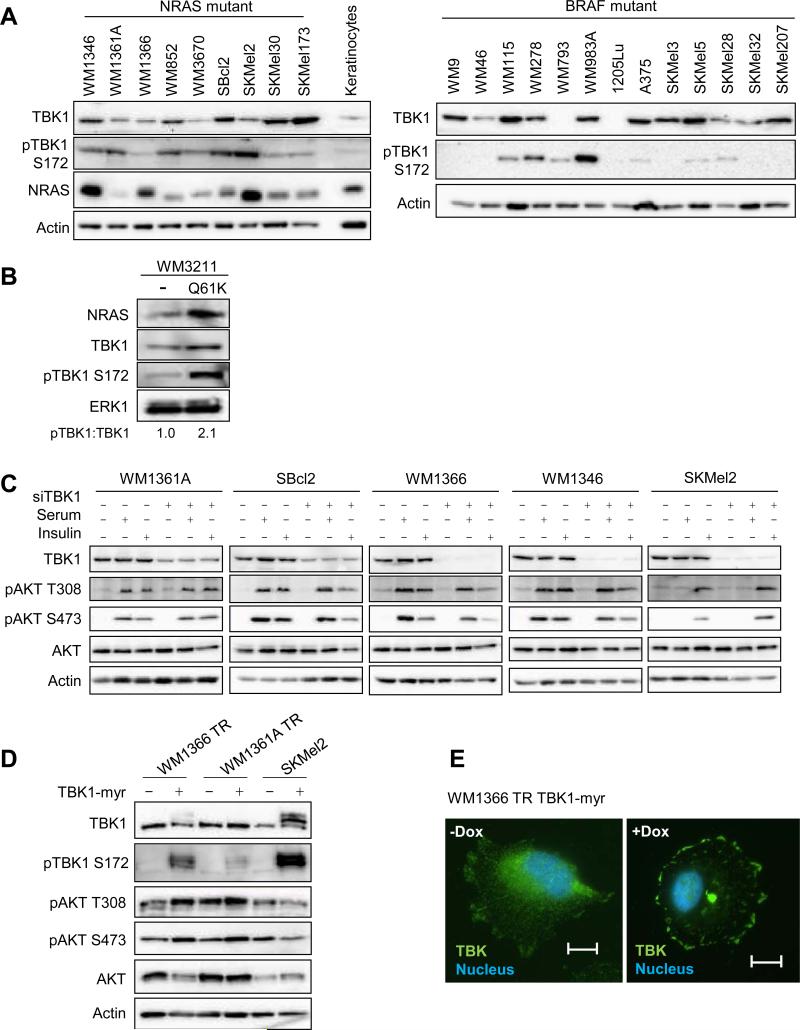
Figure 1. TBK1 is expressed in mutant NRAS cell lines.
(A) Cell lysates from mutant NRAS melanoma cells, a primary human keratinocyte culture, and mutant BRAF melanoma cells were analyzed by Western blot for TBK1, phospho-TBK1, NRAS and actin (loading control). (B) WM3211 melanoma cells wild-type for BRAF and NRAS were non-transduced (−) or transduced with constitutively active NRASQ61K. After selection, cells were lysed and lysates analyzed by Western blot for the proteins indicated. (C) Mutant NRAS melanoma cell lines were transfected with non-targeting or TBK1-targeting siRNA for 72 hours. Cells were serum-starved overnight and treated with serum-free medium, full serum medium, or 1 μM insulin for 20 min. Cells were lysed and lysates analyzed by Western blot analysis. (D) WM1366 and WM136A cells with a tetracycline-inducible system (TR) expressing a TBK1-myristoylated construct (myr) were treated with or without doxycycline for 48 hours; and parental SKMel2 and SKMel2 constitutively expressing TBK1-myr cells were lysed. Lysates were analyzed by Western blot analysis. (E) Immunofluorescence image of WM1366 TR-TBK1-myr with or without doxycycline for 24 hours stained for DAPI (blue) and TBK1 (green). Scale bar = 25 μm.
Despite previous evidence that KRAS regulates TBK1, it is unknown whether NRAS similarly activates TBK1. To test this, we utilized a lentiviral transduction system to express modest levels of mutant NRASQ61K in WM3211 cells, which are wild-type for NRAS and BRAF (23). Expression of mutant NRAS led to a marked increase in the phosphorylation of endogenous TBK1 (Figure 1B). To analyze TBK1 signaling to AKT, we depleted TBK1 in five mutant NRAS melanoma cell lines, and serum-starved cells before stimulation with either serum or insulin. AKT phosphorylation at both Thr308 and Ser473 was low in unstimulated cells (Figure 1C). TBK1 depletion slightly decreased AKT activation in SBcl2, WM1366 and SKMel2 cells in response to insulin stimulation but no effect was observed in WM1361A and WM1346 cells. Furthermore, TBK1 knockdown had minimal effects on serum-stimulated AKT phosphorylation in the five cell lines analyzed.
Previous efforts to generate active forms of TBK1 by introducing phosphomimetic mutants, such as S172D and S172E, have been unsuccessful and have actually decreased TBK1 activity (24). Since subcellular fractionation has revealed that TBK1 is constitutively localized on the membrane (18), we generated a TBK1 construct with an epitope at the N-terminus that can be myristoylated. Myristoylation is an irreversible, co-translational protein modification that targets proteins to the membrane and therefore enhances signaling (25). The TBK1-myristoylated (myr) construct was expressed and led to a robust increase in the autophosphorylation of TBK1 in WM1366 TR and SKMel2 cells (Figure 1D), whereas TBK1-myr expression/phosphorylation was low in WM1361A TR cells. Immunofluorescence showed that expression of TBK1-myr leads to increased staining of TBK1 localized at the plasma membrane (Figure 1E). As with TBK1 depletion, TBK1 activation had a marginal effect on AKT phosphorylation, with a slight increase in WM1366 TR and WM1361A TR but no effect in SKMel2 (Figure 1D).
TBK1 affects cell migration and invasion in mutant NRAS melanoma cells
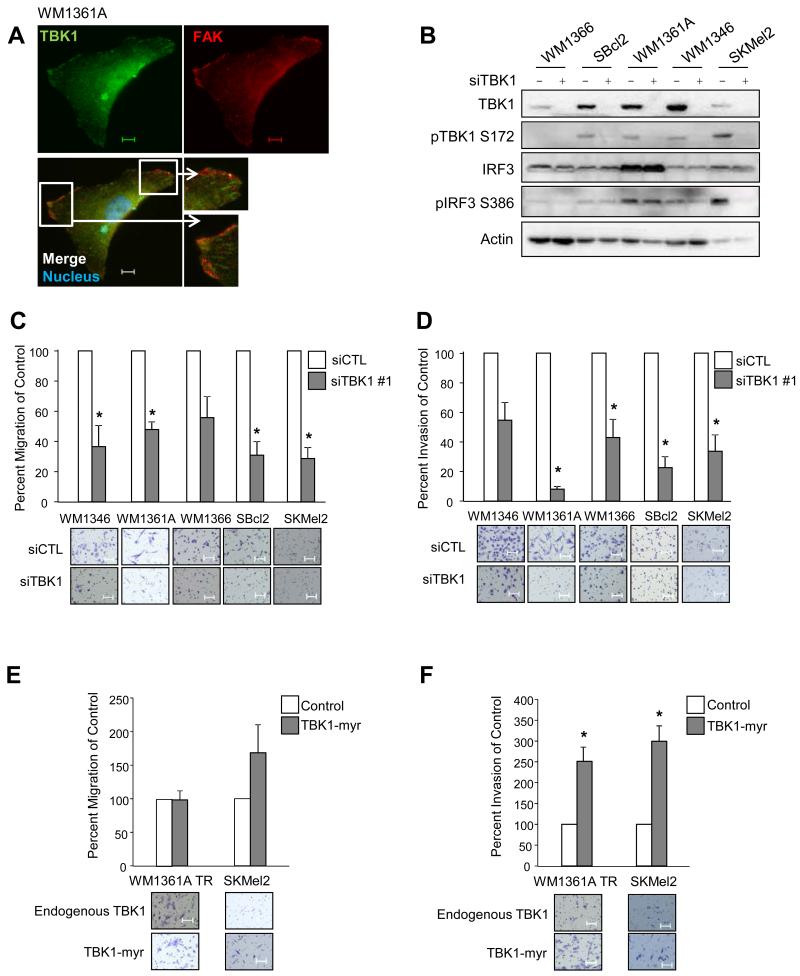
Through our immunofluorescence studies, we noticed that endogenous TBK1 localized to the edge of the cell and colocalizes, at least in part, with focal adhesion kinase (FAK) (Figure 2A). FAK is a cytoplasmic tyrosine kinase that localizes to focal adhesions, areas where cells interact with the extracellular matrix (26), and is required for migration and invasion of several different cancers, including melanoma (27). In addition, TBK1 is known to be recruited to the exocyst, a heterooctameric complex that is directly employed for maintenance of epithelial cell polarity, cell motility and cytokinesis (14). Given its colocalization with FAK and its known relationship to the exocyst complex, we tested the role of TBK1 in cell migration and invasion. We utilized siRNA to effectively knock down TBK1 in five mutant NRAS melanoma lines (Figure 2B). Upon TBK1 depletion in mutant NRAS cells, there was a decrease in transwell migration towards full serum medium (Figure 2C). Furthermore, in Matrigel-coated transwell chamber assays, TBK1 knockdown effectively inhibited invasion of mutant NRAS melanoma cells (Figure 2D). These findings were recapitulated with a second independent siRNA targeting TBK1 (Supplementary Figure 3A, B). The expression of TBK1-myr showed a trend towards an increase in migration in SKMel2 but this was not statistically significant (Figure 2E). In both WM1361A TR and SKMel2, the expression of TBK1-myr led to a significant increase in invasion (Figure 2F). Transduction of WM1366 TR cells with the TBK1-myr lentivirus did not enhance either migration or invasion likely due to less efficient expression (data not shown and Figure 1D). Overall, these results indicate a novel role for TBK1 in cancer cell migration and invasion.
Figure 2. TBK1 promotes migration and invasion of melanoma cells.
(A) Immunofluorescence image of WM1361A stained for DAPI (blue), FAK (red) and TBK1 (green). Scale bar = 25 μm. (B) WM1366, SBcl2, WM1346, WM1361A and SKMel2 cells were transfected with non-targeting or TBK1-targeting siRNA for 72 hours and lysates were analyzed by Western blotting. (C) Mutant NRAS cells were transfected as in (B). Cells were allowed to migrate through Boyden chambers toward an attractant of full serum medium for 24 hours. Counts taken (in triplicate fields of view) from the control siRNA (average set at 100% migration) were used to calculate percent migration (n=3; errors bars, S.E.; *, p<0.05). Representative 10× images of migrated cells are shown. Scale bar = 100 μm. (D) Mutant NRAS cells were transfected as in (B). Cells were allowed to invade through Boyden chambers coated with Matrigel toward an attractant of full serum medium for 24 hours. Counts taken (in triplicate fields of view) from the control siRNA (average set at 100% invasion) were used to calculate percent invasion (n=3; errors bars, S.E.; *, p<0.05). Representative 10× images of migrated cells are shown. Scale bar = 100 μm. (E) WM1366 TR TBK1-myr and WM136A TR TBK1-myr cells were treated with or without doxycycline for 24 hours; and parental SKMel2 and SKMel2 constitutively expressing TBK1-myr were seeded for migration assay as in (C). (F) WM1366 TR TBK1-myr and WM136A TR TBK1-myr cells were treated with or without doxycycline for 24 hours; and parental SKMel2 and SKMel2 constitutively expressing TBK1-myr were seeded for invasion assay as in (D).
Knockdown of TBK1 combined with AZD6244 enhances apoptosis in AZD6244-resistant cell lines in 3D culture
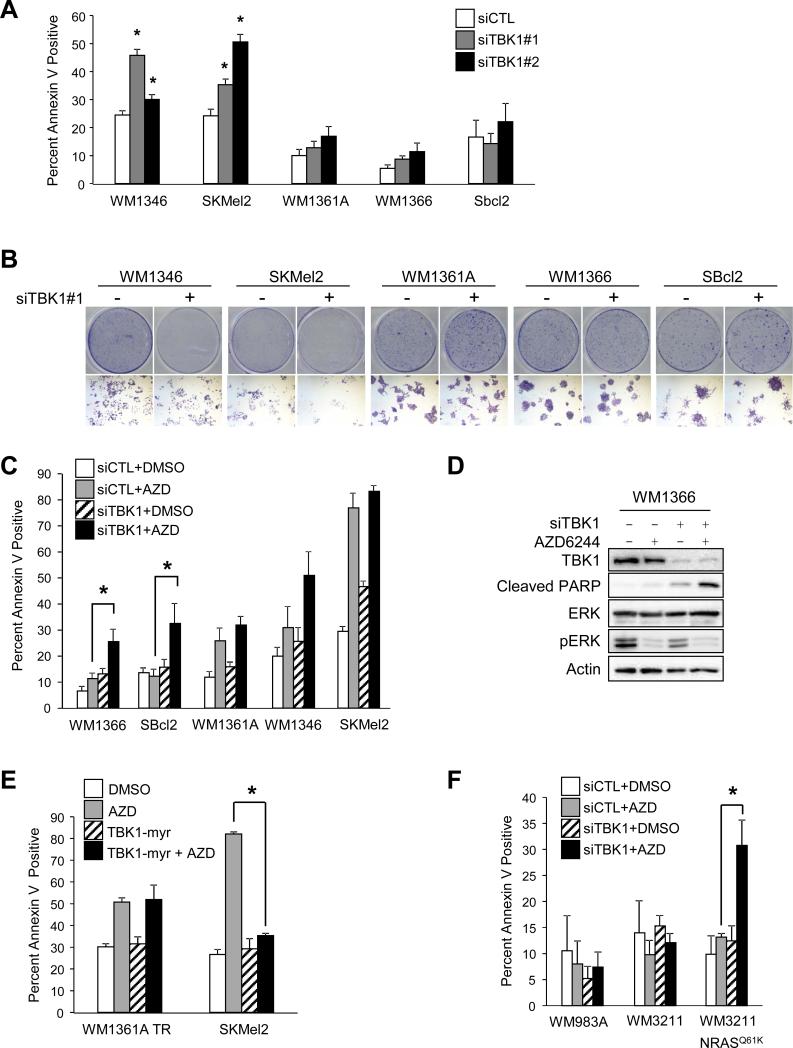
Previous groups have identified a role for TBK1 in promoting the growth and proliferation of KRAS-transformed cancer cells (16, 17). After depleting TBK1 with siRNA, we observed a significant increase in annexin V staining in some but not all mutant NRAS melanoma cell lines (Figure 3A). Similarly, TBK1 knockdown inhibited colony growth in 2D assays (Figure 3B). To further examine the effects of TBK1 on apoptosis, we utilized a 3D collagen culture system that better mimics the dermal microenvironment, where melanomas and cutaneous metastases reside (28), and is thought to be a better medium for the investigation of anti-cancer drugs (29). In contrast to 2D culture assays, TBK1 depletion alone in 3D cultures did not have a dramatic effect on annexin V staining (Figure 3C). MEK inhibitors have been used in clinical trials for mutant NRAS melanoma patients, but with mixed results (9, 10); hence, we tested whether combining TBK1 depletion would enhance the effects of MEK inhibitors. Targeting MEK with the allosteric MEK inhibitor, AZD6244, increased apoptosis in two of the five mutant NRAS cell lines examined. The combination of TBK1 depletion and AZD6244 significantly increased annexin V staining in two cell lines that were non-responsive to either alone: WM1366 and SBcl2 (Figure 3C). These data were supported by the observation of enhanced PARP cleavage in WM1366 cells in 3D culture with the combination of TBK1 depletion and AZD6244 treatment (Figure 3D).
Figure 3. Knockdown of TBK1 enhances apoptosis in combination with AZD6244 specifically in mutant NRAS cell lines.
(A) Mutant NRAS cells were transfected with non-targeting or TBK1-targeting siRNA #1 and #2, and, after 72 hours in 2D conditions, were analyzed for annexin V staining by flow cytometry (n=3; errors bars, S.E.; *, p<0.05). (B) Mutant NRAS cells were transfected with non-targeting or TBK1-targeting siRNA and plated at low density for a total of 7 days. Full-sized image, top, and 4× magnification, bottom. (C) Mutant NRAS cells were transfected as in (A), cultured in 3D collagen in the presence or absence of AZD6244 (3.3 μM). After 24 hours, cells were extracted and analyzed for annexin V staining by flow cytometry (n=3; errors bars, S.E.; *, p<0.05). (D) WM1366 cells were transfected as in (A) and cultured in 3D collagen as in (C). After 24 hours, cells were lysed and lysates analyzed by Western blotting. (E) WM1361A TR TBK1-myr pretreated with or without doxycycline, SKMel2 and SKMel2 TBK1-myr were cultured in 3D collagen in the presence or absence of AZD6244 (3.3 μM). After 48 hours, cells were released from the gel and analyzed for annexin V staining by flow cytometry (n=3; errors bars, S.E.; *, p<0.05). (F) WM3211, WM3211 transduced with NRASQ61K, and WM983A cells were transfected as in (A), then cultured and treated in 3D collagen as in (C). After 24 hours, cells were extracted and analyzed for annexin V staining by flow cytometry (n=3; errors bars, S.E.; *, p<0.05).
Given that TBK1 depletion cooperates with MEK inhibition to promote apoptosis, we analyzed the ability of activated TBK1 to protect cells from AZD6244-induced apoptosis. The expression of TBK1-myr in the AZD6244-sensitive SKMel2 cell line reduced AZD6244-induced apoptosis (Figure 3F). The expression of TBK1-myr in WM1361A TR did not show a similar response, which may be due to the lower level of activated TBK1 in this cell line (Figure 1D). To determine if the effect of TBK1 and MEK inhibition is specific to mutant NRAS melanoma cells, we utilized the WM3211, WM3211-NRASQ61K, and WM983A melanoma cell lines. WM983A is a mutant BRAF/wild-type NRAS melanoma cell line with very high levels of active TBK1 (Figure 1A). In 3D culture, the depletion of TBK1 cooperated with AZD6244 to enhance apoptosis in WM3211-NRASQ61K but not in WM3211 or WM983A (Figure 3G). In summary, although targeting TBK1 alone has limited effects on melanoma apoptosis in 3D culture, it cooperates with AZD6244 to promote apoptosis in MEK inhibitor resistant lines.
BX795 in combination with AZD6244 enhances apoptosis in AZD6244-resistant lines in 3D culture
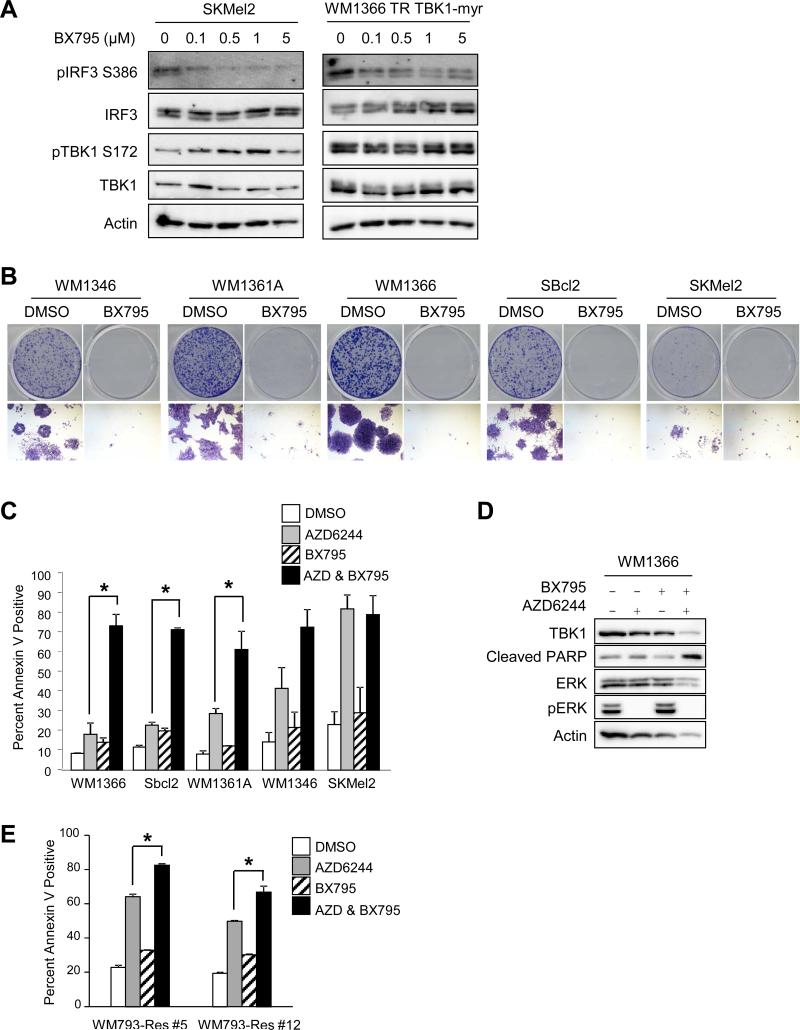
To pursue targeting TBK1 by pharmacological inhibition, we used BX795, an ATP competitive inhibitor of TBK1 (30, 31). BX795 has significant activity towards both TBK1 and PDK1, with an IC50 of 6 nM for TBK1 and an IC50 of 111 nM for PDK1 (30). To determine the ability of BX795 to inhibit TBK1 activity, we analyzed levels of IRF3 Ser386 phosphorylation, a TBK1-specific phosphorylation site (32). In SKMel2 and WM1366 TR expressing TBK1-myr, BX795 efficiently inhibited IRF3 phosphorylation at concentrations of 500 nM and above (Figure 4A). By contrast, BX795 did not dramatically alter TBK1 autophosphorylation. In 2D cultures, BX795 effectively inhibited the growth of all mutant NRAS cell lines (Figure 4B). In 3D collagen survival assays, BX795 treatment alone elicited only a minor increase in annexin V staining (Figure 4C). As noted before, MEK inhibition alone was more effective at inducing apoptosis in WM1346, SKMEL2 and, to a lesser extent, WM1361A cells compared to WM1366 and SBcl2. Notably, the addition of BX795 with AZD6244 significantly increased apoptosis in WM1366, WM1361A and SBcl2 cells, the cell lines most resistant to MEK inhibition. This increase in apoptosis was supported by increased PARP cleavage following treatment with the BX795 and AZD6244 combination in WM1366 cells (Figure 4D).
Figure 4. BX795 in combination with AZD6244 enhances apoptosis in MEK inhibitor-resistant mutant NRAS melanoma cells in 3D.
(A) SKMel2 and doxycycline-induced WM1366 TR TBK1-myr cells were treated with increasing concentrations of the TBK1 inhibitor, BX795, for 24 hours. Cells were lysed and lysates analyzed by Western blotting. (B) Mutant NRAS cells were plated at low density and treated with DMSO or BX795 (1 μM) for 1 week. Full-sized image, top, and 4× magnification, bottom. (C) WM1366, SBcl2, WM1346, WM1361A, and SKMel2 cells were cultured in 3D collagen in the presence or absence of BX795 (1 μM) and/or AZD6244 (3.3 μM). After 48 hours, cells were extracted and analyzed for annexin V staining by flow cytometry (n=3; errors bars, S.E.; *, p<0.05). (D) WM1366 cells were placed in 3D collagen and treated as in (C). After 24 hours, cells were lysed and lysates analyzed by Western blotting. (E) WM793-Res #5 and WM793-Res #12 cells grown in 5 μM PLX4720 were cultured in 3D collagen and treated as in (C). After 48 hours, cells were extracted and analyzed for annexin V staining by flow cytometry (n=3; errors bars, S.E.; *, p<0.05).
NRAS mutations also mediate acquired resistance to RAF inhibition in mutant BRAF melanoma cells (7). Thus, we examined mutant BRAF cells that have acquired resistant to the RAF inhibitor, PLX4720, through a secondary mutation in NRAS (19). Two PLX4720-resistant lines, WM793-Res #5 and WM793-Res #12, both showed an increase in apoptosis in response to AZD6244, that was further enhanced when combined with BX795 (Figure 4E). These data suggest that in 3D dermal mimetic conditions, BX795 combines with MEK inhibitors to promote apoptosis in mutant NRAS melanoma cells.
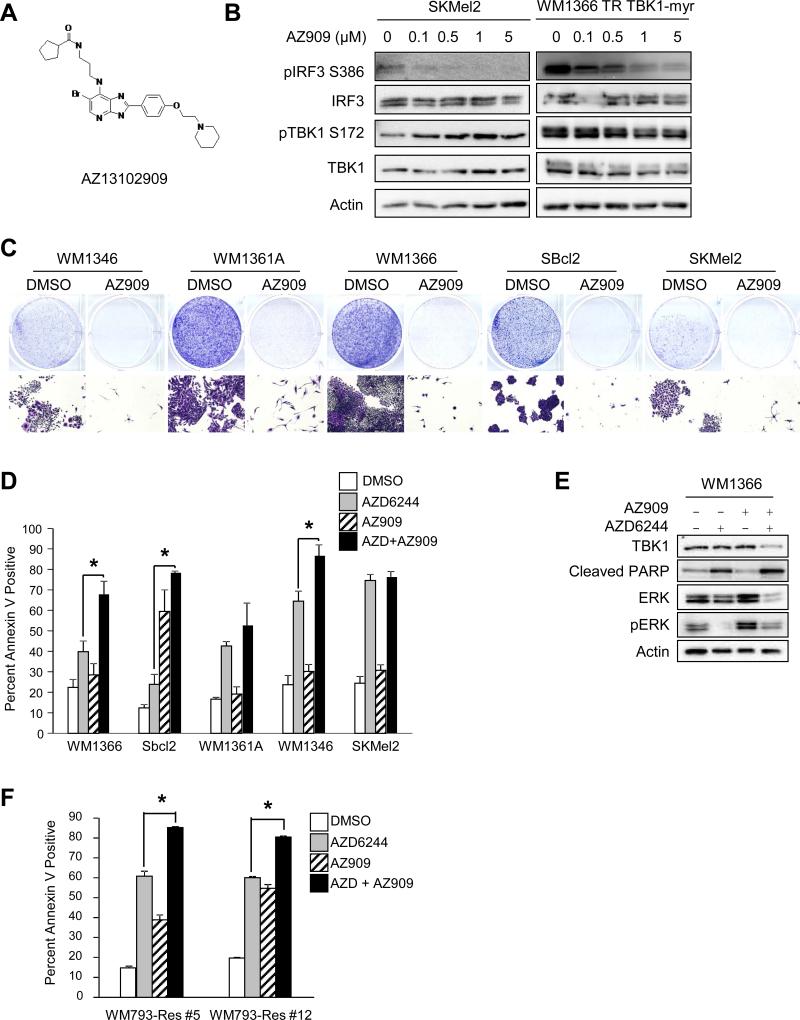
AZ909 combines with AZD6244 to enhance apoptosis in AZD6244-resistant lines in 3D culture
BX795 is a promiscuous inhibitor and has significant effects on PDK1, and therefore AKT signaling. Due to the effects of BX795 on AKT signaling, we used an ATP-binding site TBK1 inhibitor, AZ13102909 (AZ909, AstraZeneca), with a different selectivity profile. AZ909 has an IC50 of 5 nM against TBK1 and an IC50 of 100-1000 fold greater for other related kinases (Dr. Claudio Chaqui, personal communication). The structure of AZ909 is very similar to those of previously published azabenzimidazole derivatives (33) (Figure 5A). Treatment of SKMel2 cells with AZ909 resulted in inhibition of phosphorylation of IRF3 at 100 nM, whereas 1 μM of AZ909 was required to inhibit IRF3 phosphorylation in WM1366 TR cells expressing TBK1-myr (Figure 5B). In mutant NRAS cells, treatment with 1 μM of AZ909 had no effect on PDK1 and IKKε, and weakly effected inhibited aurora B phosphorylation (Supplementary Figure 3). In 2D cultures, AZ909 effectively inhibited the growth of all mutant NRAS cell lines (Figure 5C); however, as was noted with BX795, it did not induce significant annexin V staining in cells in 3D collagen with the exception of SBcl2. Notably, the combination of AZ909 (1 μM) and AZD6244 (3.3 μM) cooperated to significantly enhance apoptosis in several mutant NRAS cell lines, namely, WM1366, SBcl2, and WM1346 (Figure 5D, 5E). In the PLX4720-resistant WM793-Res#5 and WM793-Res#12 cells, AZD6244 and AZ909 alone elicited an increase in apoptosis but the combination of AZ909 and AZD6244 cooperated to significantly enhance apoptosis levels (Figure 5F).
Figure 5. AZ245 in combination with AZD6244 enhances apoptosis in MEK inhibitor-resistant lines in 3D.
(A) Structure of the TBK1 inhibitor AZ13102909 (AZ909). (B) SKMel2 and doxycycline-induced WM1366 TR TBK1-myr cells were treated with increasing concentrations of AZ909 for 24 hours. Cells were lysed and lysates analyzed by Western blotting. (C) Mutant NRAS cells were plated at low density and treated with DMSO or AZ909 (1 μM) for 1 week. Full-sized image, top, and 4× magnification, bottom. (D) Mutant NRAS cells were cultured in 3D collagen in the presence or absence of AZ909 (1 μM) and/or AZD6244 (3.3 μM). After 48 hours, cells were extracted and analyzed for annexin V staining by flow cytometry (n=3; errors bars, S.E.; *, p<0.05). (E) WM1366 cells were placed in 3D collagen and treated as in (D). After 48 hours, cells were lysed and lysates analyzed by Western blotting. (F) WM793-Res #5 and WM793-Res #12 cells grown in 5 μM PLX4720 were cultured in 3D collagen and treated as in (D). After 48 hours, cells were extracted and analyzed for annexin V staining by flow cytometry (n=3; errors bars, S.E.; *, p<0.05).
We examined the levels of the apoptotic proteins Bim-EL, Mcl-1, and Bmf in cells cultured in 3D conditions treated with TBK1 and MEK inhibitors. The levels of Mcl-1 did not change and Bim-EL increased with AZD6244 treatment alone and in combination with TBK1 knockdown or AZ909 treatment (Supplementary Figure 4A, B). The levels of Bmf were increased in cells treated with the combination of AZ909 and AZD6244 but this was not seen with the combination of siTBK1 and AZD6244. Due to this difference, we evaluated broader apoptotic pathways with an apoptosis RT2 Profiler PCR Array (Supplementary Figure 4C). While the most variation in apoptotic genes was seen between the DMSO and AZD6244-treated groups, some genes were differentially regulated (e.g., Nod1, TNFRSF25, and TRAF3) with the combination of AZD6244 and TBK1 knockdown. In sum, these data implicate that targeting TBK1 enhances the susceptibility of AZD6244-resistant mutant NRAS melanoma cells to apoptosis.
DISCUSSION
NRAS is a driver mutation in fifteen to twenty percent of melanomas. Despite it being the second-most common subset in melanoma, mutant NRAS melanomas remain relatively understudied and the treatment options are limited (34). In the past few years, several targeted therapies have been approved for the treatment of mutant BRAF melanoma patients. These include the RAF inhibitors, vemurafenib and dabrafenib, and the RAF/MEK inhibitor combination of dabrafenib plus trametinib. In contrast, no such targeted therapeutic options are FDA-approved for mutant NRAS melanoma patients. To further understand the signaling networks in mutant NRAS melanomas, we focused on TBK1, an atypical IκB kinase family member that acts downstream of the RAS effector, RalGEF.
TBK1 has been implicated in several cancers: non-small cell lung cancer (16), pancreatic cancer cells (17), breast cancer (35), and prostate cancer (36). Given this, there has been an interest in developing compounds that target TBK1. To our knowledge, our study represents the first analysis of TBK1 in the malignant traits of melanoma. TBK1 is expressed widely in melanoma cells regardless of genotype but its phosphorylated state was variable. We focused our effort on the subset of melanomas that harbor NRAS mutations since mutant NRAS expression was sufficient to promote TBK1 autophosphorylation. Our data indicate that TBK1 has a limited role in AKT signaling in mutant NRAS melanoma cells. While one group has found that TBK1 depletion in KRAS transformed cells did not change phospho-AKT levels (16), others have found that it leads to a decrease in phosphorylation (17). The mechanism of TBK1 activation of AKT is believed to be through its recruitment to the exocyst complex and subsequent interaction with AKT1 and AKT2, which interact with distinct exocyst subunits (17). In melanoma, reliance on other AKT isoforms, e.g. AKT3 (37), may account for the contrast in findings of AKT regulation by TBK1.
To examine the effects of constitutively active TBK1, we generated a novel TBK1 construct with a tag for myristoylation (TBK1-myr). Unlike previous attempts to generate a phosphomimetic construct of TBK1 (24), the expression of TBK1-myr led to a robust increase in levels of phosphorylated TBK1. Recent studies have shown that TBK1 promotes tumorigenesis through its modulation of cytokines, specifically CCL5, IL-6, and IL-8 (38, 39). While TBK1-myr expression did not dramatically affect signaling through AKT, it did lead to an increase in secretion of CCL5 (data not shown).
Despite its highly metastatic nature, little is known about the regulators of migration and invasion in mutant NRAS melanoma. We observed that TBK1 is localized to areas of focal adhesions. Furthermore, we found that TBK1 depletion decreased migration and invasion. Conversely, the expression of TBK1-myr in some cell lines led to an increase in migratory and invasive properties. Consistent with our findings, other groups have recently implicated TBK1 in glioblastoma and lung carcinoma cell migration, as assessed by wound healing assays (40, 41).
To analyze effects on cell growth, we tested the effects of TBK1 depletion and pharmacological inhibitors in 2D and 3D collagen. Cells in 3D culture regulate signaling differently from those in 2D conditions (42, 43) and 3D models have been suggested to be a better medium for the investigation of anti-cancer drugs than 2D monolayers (29). Others have utilized 3D collagen cultures to mimic the dermal microenvironment of invasive primary melanomas and cutaneous metastases (28). In 2D cultures, we found a significant increase in apoptosis with targeting TBK1; by contrast, the effect in 3D cultures was limited. This prompted us to combine TBK1 targeting with MEK inhibitors in 3D to block two major RAS effector pathways. MEK inhibitors have been noted to elicit highly variable effects in mutant NRAS melanoma cells (44). We noted that the combination of TBK1 targeting and MEK inhibition promoted apoptosis, particularly in those mutant NRAS cells lines that are resistant to MEK inhibition. This observation was made using molecular knockdowns, as well as pharmacological inhibitors, underscoring that it is likely a TBK1 selective effect. Notably, we utilized a new inhibitor, AZ909, that has selectivity toward TBK1 versus PDK1. While we sought the use of TBK1 selective inhibitors for our line of investigation, we note that more broad spectrum TBK1-PDK1 inhibitors may be useful in other aspects. For example, inhibition of PDK1, through both genetic modification and pharmacological inhibition, attenuated the growth and metastatic potential of BRAFV600E in a PTEN and CDKN2A null background melanomas (45).
In summary, melanoma is renowned for its tendency for invasion and its resistance to apoptosis. In the understudied mutant NRAS subset of cutaneous melanoma, we identify a role for TBK1 in migration and invasion and show that targeting TBK1 cooperates with MEK inhibition to promote apoptosis. Since the effects of TBK1 in mutant NRAS melanomas do not appear to be mediated by AKT, future work should examine the downstream effectors of TBK1 in this context. Furthermore, it may be worth exploring the role of TBK1 in other melanoma backgrounds. In addition to BRAF and NRAS mutations, the genomic landscape of melanoma has been updated to include the loss of NF1, which encodes the RAS-GTPase activating protein neurofibromin that is mutated in ~10% of melanomas (46). TBK1 may have a role in NF1 mutant melanomas or in those that are triple negative for BRAF, NRAS, and NF1 mutations.
Supplementary Material
IMPLICATIONS.
TBK1 promotes the malignant properties of NRAS mutant melanoma and its targeting, in combination with MEK, promotes apoptosis; thus, providing a potential novel targeted therapeutic option.
ACKNOWLEDGEMENTS
We thank Dr. Claudio Chaqui, Dr. Haixia Wang and AstraZeneca for providing AZ13102909. We are grateful to Dr. Meenhard Herlyn (Wistar Institute, Philadelphia, PA) and Dr. David Solit (Memorial Sloan-Kettering Cancer Center, New York, NY) for supplying the WM and SKMel melanoma cell lines, respectively. We thank Dr. Fred Kaplan for initial studies and Sheera Rosenbaum for technical assistance.
GRANT SUPPORT
This work was supported by the National Institutes of Health Grant F31-CA174331 (H.L. Vu) and the Melanoma Research Foundation (A.E. Aplin). The Kimmel Cancer Center is funded by National Cancer Institute Support Grant 1P30CA56036.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
There are no potential conflicts of interest.
BIBLIOGRAPHY
- 1.Howe HL, Wu X, Ries LA, Cokkinides V, Ahmed F, Jemal A, et al. Annual report to the nation on the status of cancer, 1975-2003, featuring cancer among U.S. Hispanic/Latino populations. Cancer. 2006;107:1711–42. doi: 10.1002/cncr.22193. [DOI] [PubMed] [Google Scholar]
- 2.Miller AJ, Mihm MC., Jr. Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 3.Russo AE, Torrisi E, Bevelacqua Y, Perrotta R, Libra M, McCubrey JA, et al. Melanoma: molecular pathogenesis and emerging target therapies (Review). Int J Oncol. 2009;34:1481–9. doi: 10.3892/ijo_00000277. [DOI] [PubMed] [Google Scholar]
- 4.Soengas MS, Lowe SW. Apoptosis and melanoma chemoresistance. Oncogene. 2003;22:3138–51. doi: 10.1038/sj.onc.1206454. [DOI] [PubMed] [Google Scholar]
- 5.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–7. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 6.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adjei AA, Cohen RB, Franklin W, Morris C, Wilson D, Molina JR, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26:2139–46. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkwood JM, Bastholt L, Robert C, Sosman J, Larkin J, Hersey P, et al. Phase II, open-label, randomized trial of the MEK1/2 inhibitor selumetinib as monotherapy versus temozolomide in patients with advanced melanoma. Clin Cancer Res. 2012;18:555–67. doi: 10.1158/1078-0432.CCR-11-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 12.Mishra PJ, Ha L, Rieker J, Sviderskaya EV, Bennett DC, Oberst MD, et al. Dissection of RAS downstream pathways in melanomagenesis: a role for Ral in transformation. Oncogene. 2010;29:2449–56. doi: 10.1038/onc.2009.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zipfel PA, Brady DC, Kashatus DF, Ancrile BD, Tyler DS, Counter CM. Ral activation promotes melanomagenesis. Oncogene. 2010;29:4859–64. doi: 10.1038/onc.2010.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–70. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 15.Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–40. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- 16.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou YH, Torres M, Ram R, Formstecher E, Roland C, Cheng T, et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol Cell. 2011;41:458–70. doi: 10.1016/j.molcel.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie X, Zhang D, Zhao B, Lu MK, You M, Condorelli G, et al. IkappaB kinase epsilon and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proc Natl Acad Sci USA. 2011;108:6474–9. doi: 10.1073/pnas.1016132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan FM, Kugel CH, 3rd, Dadpey N, Shao Y, Abel EV, Aplin AE. SHOC2 and CRAF mediate ERK1/2 reactivation in mutant NRAS-mediated resistance to RAF inhibitor. J Biol Chem. 2012;287:41797–807. doi: 10.1074/jbc.M112.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abel EV, Aplin AE. FOXD3 is a mutant B-RAF-regulated inhibitor of G(1)-S progression in melanoma cells. Cancer Res. 2010;70:2891–900. doi: 10.1158/0008-5472.CAN-09-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei CQ, Zhong B, Zhang Y, Zhang J, Wang S, Shu HB. Glycogen synthase kinase 3beta regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity. 2010;33:878–89. doi: 10.1016/j.immuni.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 22.Verhelst K, Verstrepen L, Carpentier I, Beyaert R. IkappaB kinase epsilon (IKKepsilon): a therapeutic target in inflammation and cancer. Biochem Pharmacol. 2013;85:873–80. doi: 10.1016/j.bcp.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodman SE, Trent JC, Stemke-Hale K, Lazar AJ, Pricl S, Pavan GM, et al. Activity of dasatinib against L576P KIT mutant melanoma: molecular, cellular, and clinical correlates. Mol Cancer Ther. 2009;8:2079–85. doi: 10.1158/1535-7163.MCT-09-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kishore N, Huynh QK, Mathialagan S, Hall T, Rouw S, Creely D, et al. IKK-i and TBK-1 are enzymatically distinct from the homologous enzyme IKK-2: comparative analysis of recombinant human IKK-i, TBK-1, and IKK-2. J Biol Chem. 2002;277:13840–7. doi: 10.1074/jbc.M110474200. [DOI] [PubMed] [Google Scholar]
- 25.Boutin JA. Myristoylation. Cell Signal. 1997;9:15–35. doi: 10.1016/s0898-6568(96)00100-3. [DOI] [PubMed] [Google Scholar]
- 26.Schaller MD, Borgman CA, Parsons JT. Autonomous expression of a noncatalytic domain of the focal adhesion-associated protein tyrosine kinase pp125FAK. Mol Cell Biol. 1993;13:785–91. doi: 10.1128/mcb.13.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hess AR, Postovit LM, Margaryan NV, Seftor EA, Schneider GB, Seftor RE, et al. Focal adhesion kinase promotes the aggressive melanoma phenotype. Cancer Res. 2005;65:9851–60. doi: 10.1158/0008-5472.CAN-05-2172. [DOI] [PubMed] [Google Scholar]
- 28.Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006;5:1136–44. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 29.Fallica B, Maffei JS, Villa S, Makin G, Zaman M. Alteration of cellular behavior and response to PI3K pathway inhibition by culture in 3D collagen gels. PloS One. 2012;7:e48024. doi: 10.1371/journal.pone.0048024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldman RI, Wu JM, Polokoff MA, Kochanny MJ, Dinter H, Zhu D, et al. Novel small molecule inhibitors of 3-phosphoinositide-dependent kinase-1. J Biol Chem. 2005;280:19867–74. doi: 10.1074/jbc.M501367200. [DOI] [PubMed] [Google Scholar]
- 31.Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct 23 upstream kinase mediates Ser-172 phosphorylation and activation. The Journal of biological chemistry. 2009;284:14136–46. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu C, Sankaran B, Chaton CT, Herr AB, Mishra A, Peng J, et al. Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure. 2013;21:1137–48. doi: 10.1016/j.str.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang T, Block MA, Cowen S, Davies AM, Devereaux E, Gingipalli L, et al. Discovery of azabenzimidazole derivatives as potent, selective inhibitors of TBK1/IKKepsilon kinases. Bioorg Med Chem Lett. 2012;22:2063–9. doi: 10.1016/j.bmcl.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 34.Fedorenko IV, Gibney GT, Smalley KS. NRAS mutant melanoma: biological behavior and future strategies for therapeutic management. Oncogene. 2013;32:3009–18. doi: 10.1038/onc.2012.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang KM, Jung Y, Lee JM, Kim W, Cho JK, Jeong J, et al. Loss of TBK1 Induces Epithelial-Mesenchymal Transition in the Breast Cancer Cells by ERalpha Downregulation. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-0891. [DOI] [PubMed] [Google Scholar]
- 36.Kim JK, Jung Y, Wang J, Joseph J, Mishra A, Hill EE, et al. TBK1 regulates prostate cancer dormancy through mTOR inhibition. Neoplasia. 2013;15:1064–74. doi: 10.1593/neo.13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–10. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Z, Aref AR, Cohoon TJ, Barbie TU, Imamura Y, Yang S, et al. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-13-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Czabanka M, Korherr C, Brinkmann U, Vajkoczy P. Influence of TBK-1 on tumor angiogenesis and microvascular inflammation. Front Biosci. 2008;13:7243–9. doi: 10.2741/3225. [DOI] [PubMed] [Google Scholar]
- 40.Stellzig J, Chariot A, Shostak K, Ismail Goktuna S, Renner F, Acker T, et al. Deregulated expression of TANK in glioblastomas triggers pro-tumorigenic ERK1/2 and AKT signaling pathways. Oncogenesis. 2013;2:e79. doi: 10.1038/oncsis.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu W, Huang YJ, Liu C, Yang YY, Liu H, Cui JG, et al. Inhibition of TBK1 attenuates radiation-induced epithelial-mesenchymal transition of A549 human lung cancer cells via activation of GSK-3beta and repression of ZEB1. Lab Invest. 2014 doi: 10.1038/labinvest.2013.153. [DOI] [PubMed] [Google Scholar]
- 42.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–12. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 43.Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J Cell Biol. 2003;163:583–95. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scortegagna M, Ruller C, Feng Y, Lazova R, Kluger H, Li JL, et al. Genetic inactivation or pharmacological inhibition of Pdk1 delays development and inhibits metastasis of Braf::Pten melanoma. Oncogene. 2013 doi: 10.1038/onc.2013.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.