Abstract
Isocitrate dehydrogenase 1 gene mutations are found in most World Health Organization grade II and III gliomas and secondary glioblastomas. Isocitrate dehydrogenase 1 mutations are known to have prognostic value in high-grade gliomas. However, their prognostic significance in low-grade gliomas remains controversial. We determined the predictive and prognostic value of isocitrate dehydrogenase 1 status in low-grade gliomas. The association of isocitrate dehydrogenase 1 status with clinicopathological and genetic factors was also evaluated. Clinical information and genetic data including isocitrate dehydrogenase 1 mutation, O 6-methylguanine DNA methyltransferase promoter methylation, 1p/19q chromosome loss, and TP53 mutation of 417 low-grade gliomas were collected from the Chinese Glioma Genome Atlas database. Kaplan–Meier and Cox proportional hazards regression analyses were performed to evaluate the prognostic effect of clinical characteristics and molecular biomarkers. Isocitrate dehydrogenase 1 mutation was identified as an independent prognostic factor for overall, but not progression-free, survival. Notably, isocitrate dehydrogenase 1 mutation was found to be a significant prognostic factor in patients with oligodendrogliomas, but not in patients with astrocytomas. Furthermore, O 6-methylguanine DNA methyltransferase promoter methylation (p = 0.017) and TP53 mutation (p < 0.001), but not 1p/19q loss (p = 0.834), occurred at a higher frequency in isocitrate dehydrogenase 1-mutated tumors than in isocitrate dehydrogenase 1 wild-type tumors. Younger patient age (p = 0.041) and frontal lobe location (p = 0.010) were significantly correlated with isocitrate dehydrogenase 1 mutation. Chemotherapy did not provide a survival benefit in patients with isocitrate dehydrogenase 1-mutated tumors. Isocitrate dehydrogenase 1 mutation was an independent prognostic factor in low-grade gliomas, whereas it showed no predictive value for chemotherapy response. Isocitrate dehydrogenase 1 mutation was highly associated with O 6-methylguanine DNA methyltransferase promoter methylation and TP53 mutation.
Introduction
Low-grade gliomas (LGGs) are the most common primary brain tumors and are comprised of specific histological subtypes, including astrocytomas, oligodendrogliomas, and oligoastrocytomas. LGGs have been classified as grade II tumors based on the histopathological and clinical criteria established by the World Health Organization (WHO) [1]. Although histopathology has been considered the gold standard for the pathological classification of brain tumors, it has become increasingly clear that these criteria still have limitations. Prognosis has been shown to vary widely in patients with the same histologic subtype, which may due to the significant genetic variation among brain tumors. Therefore, identification of molecular characteristics is essential for tumor diagnosis and outcome prediction. Low-grade diffuse gliomas (WHO grade II) are well-differentiated and slow-growing tumors that diffusely infiltrate surrounding brain structures. However, these tumors show a consistent tendency to recur even after surgical resection. As the management of patients with LGGs remains controversial, the development of markers that unfailingly predict tumor performance would be beneficial in treatment planning.
Recent genome-wide mutational analyses have demonstrated the presence of isocitrate dehydrogenase 1 (IDH1) mutations in more than 70% of WHO grade II and III astrocytomas, oligodendrogliomas, and secondary glioblastomas (GBMs) [2, 3], whereas fewer than 5% of primary GBMs harbor this mutation [4]. In addition, accumulating research has confirmed that IDH1 mutation occurs early in gliomagenesis [5], which suggests that this genetic event drives tumor progression. Therefore, IDH1 status is a useful biomarker in assisting molecular-based classification [6]. However, to date, the usefulness of IDH1 mutation as a predictive marker for treatment response and survival outcome in patients with LGGs is still unknown. Studies investigating the role of IDH1 mutation in predicting chemotherapy response in LGGs have produced conflicting results. A few studies have suggested that IDH1 mutation is associated with better outcome and sensitivity to temozolomide; however, evidence of its predictive value for response to alkylating agents, 1-(2-chloroethyl)-3-cyclohexyl-L-nitrosourea, and vincristine chemotherapy is lacking [7–9].
Many previous studies have suggested that the prevalence of IDH1 mutations is particularly high in LGGs with 1p/19q deletion [10], TP53 mutation [5], and O 6-methylguanine-DNA methyltransferase (MGMT) gene promoter methylation [11], which has been shown to play a role in predicting survival in patients with newly diagnosed GBM [11]. However, the clinical and histopathological characteristics associated with IDH1 status in LGGs have not yet been systematically elucidated.
Our previous study demonstrated that IDH1 mutation was a prognostic factor and correlated with various clinicopathological parameters in primary GBM [12]. In the present study, we focused on the predictive and prognostic value of IDH1 mutations for survival and treatment response in a large series of patients with LGGs (n = 417). We also examined the association of IDH1 status with clinicopathological parameters including age, gender, tumor location, histology, MGMT promoter methylation, TP53 mutation, and 1p/19q chromosome deletion.
Materials and Methods
Study cohort
A total of 417 clinical patients with LGGs from the Chinese Glioma Genome Atlas (CGGA) database were analyzed. All tumors were pathologically diagnosed as WHO grade II gliomas. Clinical information including patient gender, age at the time of diagnosis, tumor location, preoperative Karnofsky Performance Status (KPS) score, extent of resection, adjuvant chemotherapy and radiotherapy, and the recorded date of disease progression or death were systematically reviewed. This study was approved by the Ethics Committee of Beijing Tiantan Hospital, and written informed consent was obtained from all patients included in this study.
DNA extraction
Materials were selected for DNA extraction after careful examination of corresponding hematoxylin and eosin-stained sections. All selected samples contained at least 80% of vital tumor. Genomic DNA was extracted from frozen tumor tissues using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s protocol. RNA sequencing was performed as described in our previous study [12].
DNA pyrosequencing for IDH1 mutation
The genomic region spanning wild-type R132 of IDH1 was analyzed by pyrophosphate sequencing using the following primers: (forward) 5'-GCTTGTGAGTGGATGGGTAAAAC-3' and (reverse) 5'-biotin-TTGCCAACATGACTTACTTGATC-3'. Polymerase chain reaction (PCR) was performed using the ABI PCR System 9700 (Applied Biosystems). Polymerase chain reaction amplification was performed in a 40 μl reaction volume containing 1 μl each of forward and reverse primer (10 μM), 4 μl 10× buffer, 3.2 μl dNTPs (2.5 μM), 2.5 U hotstart Taq (Takara), and 2 μl DNA (10 μM). The PCR conditions were as follows: 95° for 3 min; 50 cycles of 95°C for 15 s, 56°C for 20 s, and 72°C for 30 s; and 72°C for 5 min. After extraction from the amplified product, single-stranded DNA was subjected to bisulfate modification using the EpiTect Bisulfite Kit (Qiagen) and pyrosequencing using the PyroMark Q96 ID System (Qiagen) with the primer 5'- TGGATGGGTAAAACCT-3'.
DNA pyrosequencing for MGMT promoter methylation
Bisulfite modification of DNA was performed using the EpiTect Kit. The following primers were used to amplify the MGMT promoter region: (forward) 5'-GTTTYGGATATGTTGGGATA-3' and (reverse) 5'-biotin-ACCCAAACACTCACCAAATC-3'. PCR amplification was performed in a 40 μl reaction volume containing 0.5 μl each of the forward and reverse primers (10 μM), 4 μl 10× buffer, 3.2 μl dNTPs (2.5 μM), 2.5 U hotstart Taq, and 2 μl bisulfite-treated DNA (10 μM). The PCR conditions were as follows: 95°C for 3 min; 40 cycles of 95°C for 15 s, 52°C for 30 s, and 72°C for 30 s; and 72°C for 5 min. Single-stranded DNA was extracted from the amplified product using the QIAamp DNA Mini Kit and subjected to pyrosequencing using the PyroMark Q96 ID System with the primer 5'-GGATATGTTGGGATAGT-3' according to the manufacturer’s instructions. The methylation values acquired were averaged across the seven CpG loci tested within the MGMT promoter. LGG samples with an average methylation > 10% were considered MGMT promoter methylated.
Detection of TP53 mutation and 1p/19q chromosome loss
Mutation scanning of TP53 exons 5–8 was done using the following primers: 5'-AGGCCCTTAGCCTCTGTAAGC-3' (sense) and 5'-M13-CTGCTCAGATAGCGATGGTG-3' (antisense) for exon 5; 5'-M13-AGAAATCGGTAAGAGGTGGGC-3' (sense) and 5'-CATCCTGGCTAACGGTGAAAC-3' (antisense) for exon 6; 5'-M13-TTGGGCAGTGCTAGGAAAGAG-3' (sense) and 5'-GTTGGGAGTAGATGGAGCCTG-3' (antisense) for exon 7; 5'-TTGTCTTTGAGGCATCACTGC-3' (sense) and 5'-M13-GGAGCACTAAGCGAGGTAAGC-3' (antisense) for exon 8. PCR products were subsequently analyzed using Sanger sequencing. 1p/19q loss was detected by fluorescence in situ hybridization using LSI probe sets 1p36/1q25 and 19q13/19p13 (spectrum orange-labeled 1p36 and 19q13 probes; spectrum green-labeled 1q25 and 19p13 probes; Vysis) and evaluated in at least 200 non-overlapping nuclei with intact morphology.
Statistical analysis
Progression-free survival (PFS) was calculated from the date of diagnosis to the date of recurrence or last follow-up. Overall survival (OS) was defined as the time from primary surgery to death. PFS and OS estimates were obtained using the Kaplan–Meier method and compared using the log-rank test. A p value ≤ 0.05 was considered significant. Cox proportional hazards regression was used to calculate the hazard ratios and 95% confidence intervals for the association of IDH1 mutation, 1p/19q deletion, TP53 mutation, MGMT promoter methylation, and clinical factors with prognosis.
Results
IDH1 mutation status
Of the 417 grade II gliomas examined, mutations at codon 132 of the IDH1 gene were detected in 309 tumors (74%) including 304 R132H mutations (arginine to histidine substitution [CGT to CAT]) and 5 R132G mutations (arginine to glycine substitution [CGT to GGT]), which resulted in amino acid sequence alterations.
Association of IDH1 status with survival
The study cohort consisted of 417 patients with grade II gliomas. Of the 417 patients, 309 and 108 patients had IDH1-mutated and wild-tumors, respectively. OS was significantly longer in patients with IDH1-mutated tumors than in patients with IDH1 wild-type tumors (log-rank test, p = 0.015; Fig 1A). Although PFS was longer in patients with IDH1-mutated tumors than in patients with IDH1 wild-type tumors, this difference was not significant (log-rank test, p = 0.095; Fig 1B). Histological subtype influenced the prognostic effect of IDH1 mutation (log-rank test: OS p = 0.005, PFS p = 0.008; Fig 1C and 1D). IDH1 mutation was associated with better OS in patients with oligoastrocytomas or oligodendrogliomas (p = 0.047; Fig 1E), but not in patients with astrocytomas (p = 0.124; Fig 1F).
Fig 1. Kaplan-Meier Analysis of Overall Survival and Progression-free Survival in Patients with IDH1-mutated and Wild-type Gliomas.
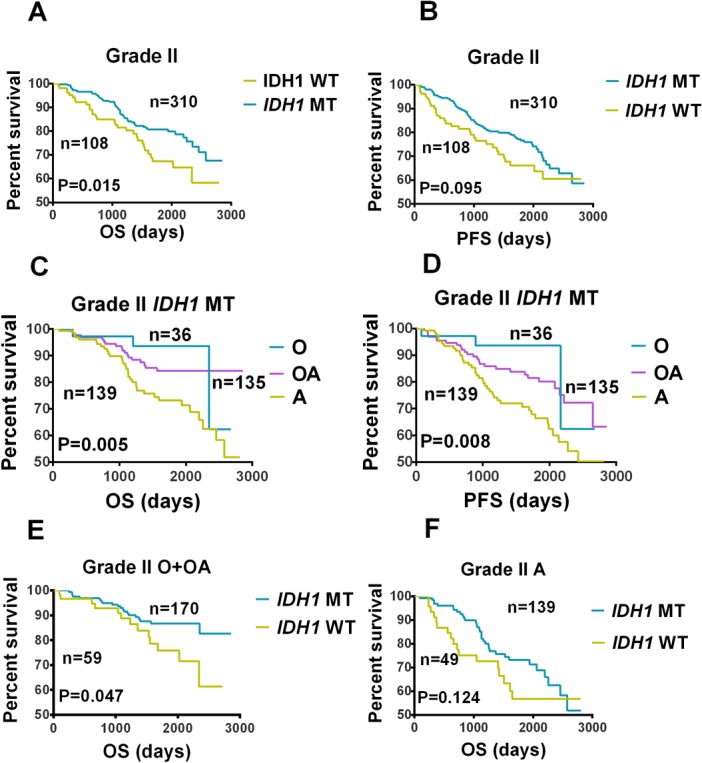
Comparison of (A) overall survival (OS) and (B) progression-free survival (PFS) between patients with IDH1-mutated (IDH1 MT) and wild-type (IDH1 WT) grade II gliomas. Comparison of (C) OS and (D) PFS among patients with IDH1 MT grade II oligodendrogliomas (O), oliogoastrocytomas (OA), and atrocytomas (A). (E) A comparison of OS between patients with IDH1 MT or IDH1 WT grade II O or OA. (F) A comparison of OS between patients with IDH1 MT or IDH1 WT grade II As.
The univariate regression analysis demonstrated that preoperative KPS score, chemotherapy, TP53 mutation, 1p/19q loss, MGMT promoter methylation, and IDH1 mutation were significantly associated with OS (Table 1), whereas preoperative KPS score, 1p/19q loss, and chemotherapy were associated with PFS (S1 Table). In the multivariate regression analysis, preoperative KPS score and IDH1 mutation were identified as independent prognostic factors for OS (Table 1 and S3 Table), whereas only preoperative KPS score was shown to be an independent prognostic factor for PFS (S1 Table).
Table 1. Overall survival of grade II gliomas (n = 417).
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% Cl | P-value | |
| Gender | 0.825 | 0.529–1.287 | 0.397 | |||
| Age | 1.012 | 0.990–1.035 | 0.290 | |||
| Preoperative KPS | 0.968 | 0.952–0.984 | <0.001 | 0.905 | 0.852–0.961 | 0.001 |
| IDH1mutation | 0.607 | 0.384–0.958 | 0.032 | 0.253 | 0.087–0.735 | 0.012 |
| TP53 mutation | 1.676 | 1.028–2.735 | 0.039 | 1.989 | 0.684–5.788 | 0.207 |
| 1p/19q loss | 0.596 | 0.357–0.996 | 0.048 | 0.675 | 0.171–2.666 | 0.575 |
| Chemotherapy | 2.515 | 1.409–4.489 | 0.002 | 0.723 | 0.266–1.967 | 0.526 |
| MGMT promoter methylation | 1.039 | 0.385–2.804 | 0.939 | |||
| Extent of resection | 0.932 | 0.684–1.271 | 0.656 | |||
| Radiotherapy | 1.096 | 0.466–2.582 | 0.833 | |||
Association of IDH1 status with clinicopathological parameters
Next, we sought to ascertain the correlation of IDH1 status with clinicopathological and molecular pathology features (Table 2). The IDH1 mutation group consisted of 181 males and 128 females with a median age of 37 (range, 18–66) years, whereas the IDH1 wild-type group consisted of 73 males and 34 females with a median age of 41 (range, 14–72) years. The median OS was 56 (range, 3–95) months and 49 (range, 3–93) months in the mutation and wild-type groups, respectively. IDH1 mutation was associated with younger age (chi square test, p = 0.041). Median KPS score (chi square test, p = 0.200) and gender (chi square test, p = 0.078) were not significantly associated with IDH1 status. Similarly, IDH1 status did not differ according to histological subtype. A total of 224 (72%) and 64 (59%) IDH1-mutated and IDH1 wild-type tumors were located in the frontal lobe, respectively (chi square test, p = 0.010). The Cancer Genome Atlas subtype was more favorable in the IDH1 mutation group compared with the IDH1 wild-type group (chi square test: proneural p = 0.001 and neural p = 0.022). Extent of resection was not significantly different between the two groups (chi square test, p = 0.246).
Table 2. Clinical and genetic features of WHO grade II gliomas (n = 417).
| Characteristics | IDH1 mutation (%) | IDH1 wide-type (%) | P-value |
|---|---|---|---|
| No. of cases | 309(74) | 108(26) | |
| Gender (F/M) | 128(41)/181 | 34(31)/73 | 0.078 |
| Age (<40 / ≥40yrs) | 187/125 | 52/55 | 0.041 |
| Histopathology | |||
| Astrocytomas | 139(45) | 49(45) | 0.945 |
| Oligodendrogliomas | 36(12) | 11(10) | 0.678 |
| Oligoastrocytomas | 134(43) | 48(44) | 0.846 |
| Tumor location | |||
| Frontal lobe | 224(72) | 64(59) | 0.010 |
| Temporal lobe | 105(34) | 45(42) | 0.152 |
| Parietal lobe | 23(7) | 8(7) | 0.288 |
| Insula | 63(20) | 23(21) | 0.841 |
| Occipital lobe | 6(2) | 6(6) | 0.110 |
| Others | 20(6) | 12(11) | 0.119 |
| Extent of resection | |||
| GTR (%) | 109(38)/176 | 44(45)/54 | 0.246 |
| Molecular biomarkers | |||
| MGMT promoter methylation (high/low) | 42(65)/23 | 6(33)/12 | 0.017 |
| 1p/19q loss (Yes/No) | 89(30)/208 | 32(31)/71 | 0.834 |
| Mutant TP53 (Yes/No) | 81(36)/145 | 7(9)/73 | <0.001 |
Molecular features
MGMT promoter methylation status could be determined in 83 tumors. MGMT promoter methylation was present in 48 (58%) tumors (Table 2). Strikingly, of the 48 tumors with MGMT promoter methylation, 42 (87%) were IDH1-mutated tumors, whereas only 6 (13%) were IDH1 wild-type tumors (chi square test, p = 0.017). The proportion of patients with TP53 mutations was higher in the IDH1 mutation group than in the IDH1 wild-type group (36% [81/226] vs. 9% [7/80]; chi square test, p < 0.001). The frequency of 1p/19q deletion was not significantly different between the IDH1 mutation and wild-type groups (30% vs. 31%; chi square test, p = 0.834). Furthermore, in the IDH1 wild-type group, OS and PFS were significantly longer in patients who underwent radiotherapy than those who underwent radiotherapy plus chemotherapy (p = 0.002 and p = 0.003, respectively). In contrast, OS and PFS were not significantly different between patients who received radiotherapy or radiotherapy plus chemotherapy in the IDH1 mutation group (p = 0.194 and p = 0.137, respectively; Fig 2).
Fig 2. Kaplan-Meier Analysis of Overall Survival and Progression-free Survival According to IDH1 Status and Adjuvant Treatment.
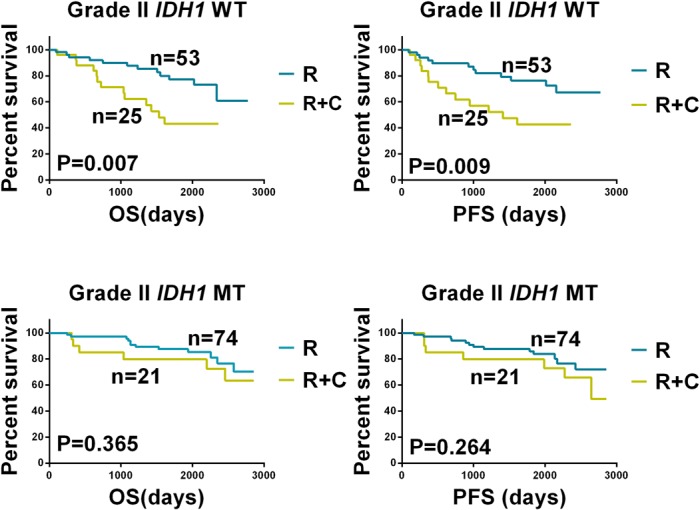
A comparison of (A) overall survival (OS) and (B) progression-free survival (PFS) between patients with IDH1 wild-type (IDH1 WT) grade II gliomas who received radiotherapy (RT) or radiotherapy plus chemotherapy (RT+CT). A comparison of (C) OS and (D) PFS between patients with IDH1-mutated (IDH1 MT) grade II gliomas who received RT or RT+CT. For the IDH1 WT group, PFS and OS were longer in patients who underwent RT compared with those who underwent RT+CT (p = 0.002). For the IDH1 MT group, PFS and OS were not significantly different between patients who underwent RT or RT+CT (p = 0.194).
Discussion
Using large-scale sequencing, nearly 12% of GBMs were found to harbor IDH1 mutations [13]. Approximately 70% of IDH1 mutations were found in grade II and III gliomas and secondary GBMs [5]. Yan et al.[3] reported that more than 70% of WHO grade II and III astrocytomas and oligodendrogliomas had mutations in amino acid position 132 of IDH1. IDH1 mutations also have been shown to occur predominantly in younger patients [5]. Several previous studies have demonstrated the important prognostic role of IDH1 mutations in patients with high-grade gliomas [12, 14]. Sanson et al. [15] reported that IDH1 mutation was associated with longer survival in the univariate and multivariate analyses of 404 gliomas, including 100 grade II gliomas [15]. A study of 139 LGGs demonstrated an association between IDH1 mutation and OS [16]. In contrast, a study with a larger sample size (n = 360) suggested that IDH1 mutation does not have prognostic value in LGGs [17]. Therefore, the predictive role of IDH1 mutation in WHO grade II gliomas is controversial and remains to be established. We investigated the frequency and prognostic impact of IDH1 mutations in a large LGG dataset. Our results demonstrated that IDH1 mutations occur at a high frequency in WHO grade II astrocytomas and oligodendrogliomas. Furthermore, IDH1 mutation was found to be a robust predictor of patient survival, which corroborates prior reports [8, 15, 16].
1p/19q loss, TP53 mutation, and MGMT promoter methylation have been investigated as potential prognostic predictors in glioma. Most previous studies have suggested that OS is longer in LGG patients with combined 1p and 19q loss [18]. However, the prognostic role of TP53 mutations has remained controversial, and no consistent association of TP53 mutation with survival outcome has been reported. In a cohort study of 159 patients with LGGs, PFS, but not OS, was significantly shorter in patients with TP53-mutated tumors than in those with TP53 wild-type tumors [19]. Another study of 122 glioma cases demonstrated that TP53 mutation is a predictor of shorter survival in patients with low-grade diffuse gliomas [20]. Moreover, evaluation of the prognostic role of MGMT promoter methylation is complicated by the influence of clinical factors, such as age and histopathology [21]. Our study underscores the prognostic relevance of 1p/19q loss and TP53 mutation, but not MGMT promoter methylation, in LGGs [16]. Notably, our study is the first to demonstrate the prognostic significance of IDH1 mutation in patients with oligodendroglioma or oligoastrocytoma, but not in patients with astrocytoma.
Several clinical parameters and genetic factors have been shown to be related to IDH1 status. Parsons et al. [13] found that mean age was significantly lower in IDH1-mutated GBMs than in those with IDH1 wild-type GBMs. In the present study, we showed that patients with LGGs carrying IDH1 mutations were significantly younger than those with IDH1 wild-type LGGs. In line with previous reports, we found that IDH1 mutation is correlated with MGMT promoter methylation. A high level of MGMT promoter methylation was detected in 65% of patients in the IDH1 mutation group, whereas only 33% of patients in the IDH1 wild-type group had a high level of MGMT promoter methylation. This finding indicates that MGMT hypermethylation is highly associated with IDH1 mutation. Furthermore, Watanabe et al. [5] demonstrated the copresence of IDH1 and TP53 mutations in 63% of low-grade astrocytomas, whereas IDH1 mutation plus 1p/19q loss was present in most (64%) oligodendrogliomas. In concordance with previous reports [3, 22], we observed a highly significant correlation between TP53 mutation and IDH1 mutation. Interestingly, the frequency of 1p/19q loss was not significantly different between the IDH1 mutation and wild-type groups. To date, few studies have focused on the association between tumor location and IDH1 mutation. Yan et al. [12] and Zhang et al. [14] reported that IDH1-mutated primary GBMs and anaplastic astrocytomas mainly involved the frontal lobe. Similarly, we found that a higher proportion of IDH1-mutated LGGs were located in the frontal lobe compared with wild-type LGGs.
GBM patients with MGMT promoter methylation have been reported to have a higher response rate to temozolomide [23]; however, whether IDH1 mutation can predict outcome to this specific treatment is unknown. Previously, a Dutch study [24] revealed that IDH1 mutation was a good predictor of OS but not response to temozolomide after radiotherapy in patients with low-grade astrocytomas, which is in agreement with our findings. Furthermore, in the IDH1 mutation group, we found that survival outcome was not different between patients who did and did not receive chemotherapy (primarily temozolomide). However, in the wild-type IDH1 group, patients who underwent radiotherapy plus chemotherapy had a shorter survival than those who only underwent radiotherapy. Despite the wide clinical application of chemotherapy in GBM patients, our results suggest that chemotherapy is unsuitable for patients without IDH1 mutations. This may be because the toxic effects of temozolomide are even more apparent in LGGs. Our study suggests that wild-type IDH1 status is associated with good response to radiotherapy but not to radiotherapy plus chemotherapy. This association and its underlying mechanisms require further investigation.
Conclusions
IDH1 mutation was an independent prognostic factor in LGGs; however, it had no predictive value for chemotherapy response. IDH1 mutation was highly associated with MGMT promoter methylation and TP53 mutation.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Yu-ling Yang for tissue sample collection and clinical data retrieval.
Data Availability
Relevant data owned by the author are included in the paper and its Supporting Information. Additional clinical data are owned by the Chinese Genome Atlas. Interested researchers can submit an application for the data from the Chinese Genome Atlas website (http://www.cgga.org.cn/).
Funding Statement
This work was supported by grants from the Beijing Science and Technology Plan (No. Z131100006113018) and National Science Foundation of China (No. 91229121 and No. 81201993). The funders had the key role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116: 597–602. 10.1007/s00401-008-0455-2 [DOI] [PubMed] [Google Scholar]
- 3. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360: 765–73. 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ichimura K, Pearson DM, Kocialkowski S, Backlund LM, Chan R, Jones DT, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. 2009;11: 341–7. 10.1215/15228517-2009-025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174: 1149–53. 10.2353/ajpath.2009.080958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gupta R, Webb-Myers R, Flanagan S, Buckland ME. Isocitrate dehydrogenase mutations in diffuse gliomas: clinical and aetiological implications. J Clin Pathol. 2011;64: 835–44. 10.1136/jclinpath-2011-200227 [DOI] [PubMed] [Google Scholar]
- 7. SongTao Q, Lei Y, Si G, YanQing D, HuiXia H, XueLin Z, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012;103: 269–73. 10.1111/j.1349-7006.2011.02134.x [DOI] [PubMed] [Google Scholar]
- 8. Houillier C, Wang X, Kaloshi G, Mokhtari K, Guillevin R, Laffaire J, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75: 1560–6. 10.1212/WNL.0b013e3181f96282 [DOI] [PubMed] [Google Scholar]
- 9. van den Bent MJ, Dubbink HJ, Marie Y, Brandes AA, Taphoorn MJ, Wesseling P, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16: 1597–604. 10.1158/1078-0432.CCR-09-2902 [DOI] [PubMed] [Google Scholar]
- 10. Maintz D, Fiedler K, Koopmann J, Rollbrocker B, Nechev S, Lenartz D, et al. Molecular genetic evidence for subtypes of oligoastrocytomas. J Neuropathol Exp Neurol. 1997;56: 1098–104. [DOI] [PubMed] [Google Scholar]
- 11. Gorlia T, van den Bent MJ, Hegi ME, Mirimanoff RO, Weller M, Cairncross JG, et al. Nomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981-22981/CE.3. Lancet Oncol. 2008;9: 29–38. [DOI] [PubMed] [Google Scholar]
- 12. Yan W, Zhang W, You G, Bao Z, Wang Y, Liu Y, et al. Correlation of IDH1 mutation with clinicopathologic factors and prognosis in primary glioblastoma: a report of 118 patients from China. PLoS One. 2012;7: e30339 10.1371/journal.pone.0030339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321: 1807–12. 10.1126/science.1164382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang CB, Bao ZS, Wang HJ, Yan W, Liu YW, Li MY, et al. Correlation of IDH1/2 mutation with clinicopathologic factors and prognosis in anaplastic gliomas: a report of 203 patients from China. J Cancer Res Clin Oncol. 2014;140: 45–51. 10.1007/s00432-013-1519-9 [DOI] [PubMed] [Google Scholar]
- 15. Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27: 4150–4. 10.1200/JCO.2009.21.9832 [DOI] [PubMed] [Google Scholar]
- 16. Hartmann C, Hentschel B, Tatagiba M, Schramm J, Schnell O, Seidel C, et al. Molecular markers in low-grade gliomas: predictive or prognostic? Clin Cancer Res. 2011;17: 4588–99. 10.1158/1078-0432.CCR-10-3194 [DOI] [PubMed] [Google Scholar]
- 17. Kim YH, Nobusawa S, Mittelbronn M, Paulus W, Brokinkel B, Keyvani K, et al. Molecular classification of low-grade diffuse gliomas. Am J Pathol. 2010;177: 2708–14. 10.2353/ajpath.2010.100680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith JS, Perry A, Borell TJ, Lee HK, O'Fallon J, Hosek SM, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18: 636–45. [DOI] [PubMed] [Google Scholar]
- 19. Peraud A, Kreth FW, Wiestler OD, Kleihues P, Reulen HJ. Prognostic impact of TP53 mutations and P53 protein overexpression in supratentorial WHO grade II astrocytomas and oligoastrocytomas. Clin Cancer Res. 2002;8: 1117–24. [PubMed] [Google Scholar]
- 20. Okamoto Y, Di Patre PL, Burkhard C, Horstmann S, Jourde B, Fahey M, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108: 49–56. [DOI] [PubMed] [Google Scholar]
- 21. Boots-Sprenger SH, Sijben A, Rijntjes J, Tops BB, Idema AJ, Rivera AL, et al. Significance of complete 1p/19q co-deletion, IDH1 mutation and MGMT promoter methylation in gliomas: use with caution. Mod Pathol. 2013;26: 922–9. 10.1038/modpathol.2012.166 [DOI] [PubMed] [Google Scholar]
- 22. Gorovets D, Kannan K, Shen R, Kastenhuber ER, Islamdoust N, Campos C, et al. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin Cancer Res. 2012;18: 2490–501. 10.1158/1078-0432.CCR-11-2977 [DOI] [PubMed] [Google Scholar]
- 23. Wick W, Platten M, Meisner C, Felsberg J, Tabatabai G, Simon M, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13: 707–15. 10.1016/S1470-2045(12)70164-X [DOI] [PubMed] [Google Scholar]
- 24. Dubbink HJ, Taal W, van Marion R, Kros JM, van Heuvel I, Bromberg JE, et al. IDH1 mutations in low-grade astrocytomas predict survival but not response to temozolomide. Neurology. 2009;73: 1792–5. 10.1212/WNL.0b013e3181c34ace [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Relevant data owned by the author are included in the paper and its Supporting Information. Additional clinical data are owned by the Chinese Genome Atlas. Interested researchers can submit an application for the data from the Chinese Genome Atlas website (http://www.cgga.org.cn/).


