Abstract
Ectopic lipid accumulation has been observed in fish fed a high-lipid diet. However, no information is available on the mechanism by which dietary lipid levels comprehensively regulate lipid transport, uptake, synthesis and catabolism in fish. Therefore, the present study aimed to gain further insight into how dietary lipids affect lipid deposition in the liver of large yellow croaker(Larimichthys crocea). Fish (150.00±4.95 g) were fed a diet with a low (6%), moderate (12%, the control diet) or high (18%) crude lipid content for 10 weeks. Growth performance, plasma biochemical indexes, lipid contents and gene expression related to lipid deposition, including lipoprotein assembly and clearance, fatty acid uptake and triacylglycerol synthesis and catabolism, were assessed. Growth performance was not significantly affected. However, the hepato-somatic and viscera-somatic indexes as well as plasma triacylglycerol, non-esterified fatty acids and LDL-cholesterol levels were significantly increased in fish fed the high-lipid diet. In the livers of fish fed the high-lipid diet, the expression of genes related to lipoprotein clearance (LDLR) and fatty acid uptake (FABP11) was significantly up-regulated, whereas the expression of genes involved in lipoprotein assembly (apoB100), triacylglycerol synthesis and catabolism (DGAT2, CPT I) was significantly down-regulated compared with fish fed the control diet, and hepatic lipid deposition increased. In fish fed the low-lipid diet, the expression of genes associated with lipoprotein assembly and clearance (apoB100, LDLR, LRP-1), fatty acid uptake (CD36, FATP1, FABP3) and triacylglycerol synthesis (FAS) was significantly increased, whereas the expression of triacylglycerol catabolism related genes (ATGL, CPT I) was reduced compared with fish fed the control diet. However, hepatic lipid content in fish fed the low-lipid diet decreased mainly due to low dietary lipid intake. In summary, findings of this study provide molecular insight into the role of lipid deposition in the liver in response to different dietary lipid contents.
Introduction
High-lipid diets have increasingly been used for cost-effective farming in aquaculture in recent years. This farming system is based upon the notion that dietary protein can be reduced when more energy is supplied in the form of dietary lipids [1,2]. However, high-lipid diets often lead to ectopic lipid accumulation in the tissues of farmed fish, including the liver and abdominal adipose tissue, and induce metabolic disturbances [3–5]. Lipid deposition is a complex process involving lipid transport, uptake, synthesis and catabolism. In the liver, the lipid stores include fatty acids (FAs) from three sources: diet (delivered via chylomicron remnants), de novo lipogenesis and circulating non-esterified fatty acids (NEFAs) [6].
Lipid transport in fish is similar to that in mammals, mainly occurring via lipoproteins (LPs). The process consists of two loops: an exogenous loop and an endogenous loop [7,8]. The exogenous pathway transports dietary lipids absorbed by the intestine via chylomicrons (CMs). The endogenous pathway refers to the hepatic secretion of lipids via very low-density LP (VLDL) and the metabolism of VLDL to intermediate-density LP (IDL) and low-density LP (LDL). In mammals, triacylglycerol (TAG) in CMs and VLDL are rapidly hydrolyzed by lipoprotein lipase (LPL) and taken up by peripheral tissues, and the hydrolyzed LP remnants are removed from the plasma via LDL receptor (LDLR) and LDL receptor-related protein-1 (LRP-1)mainly by the liver but also partially by peripheral tissues [9–12]. Additionally, scavenger receptor class B type I (SRBI) is also closely related to CM and VLDL metabolism. Although LP receptors play an important role in lipid transport, few studies have specifically targeted LP receptors in fish. Long-chain FAs (LCFAs) released from TAG in LPs and other sources have been widely demonstrated to cross the plasma membrane into different tissues via a protein-mediated mechanism [13–15]. In adipocytes, protein-mediated LCFA uptake accounts for approximately 90% of cellular acquisition [16]. A number of membrane FA transporters have been identified, including FA translocase (cluster of differentiation, CD36), a family of FA transport proteins (FATP1-6) and plasma membrane-associated FA binding proteins (FABPpm). Subsequently, FAs transported across the membrane are transferred through the cytosol via FA binding proteins (FABPs) for oxidation or storage. FABPs are ubiquitously expressed in vertebrate tissues, with distinct expression patterns being observed for individual FABP and are named after the tissues in which they were discovered or are mainly expressed, such as basic liver-type FABP (FABP10), heart-type FABP (FABP3) and adipocyte-type FABP (FABP4) [17]. In fish, FABP10 mRNA is highly expressed in the liver, whereas FABP3 is abundantly expressed in muscle. Although FABP4 has not been described, FABP11 may represent the fish lipid storage tissue FABP homologue [18,19]. It should be noted that most tissues express various FABP types because no FABP is specific to a given tissue. The function and regulation of FA transporters and FABPs have been studied under various nutritional and hormonal conditions in fish [13–15,18,20–22]. However, these studies primarily focused on the regulation of hormonal stimulation. In addition to lipid transport and uptake, TAG synthesis and catabolism also influence lipid deposition, and the regulation of these processes has been extensively demonstrated in fish [23,24].
The large yellow croaker (Larimichthys crocea) is a commercially important carnivorous species for marine culture given its high market value in China. To our knowledge, no information is available on the mechanism by which dietary lipid levels comprehensively regulate lipid transport, uptake, synthesis and catabolism in fish. Thus, the aim of this study was to investigate the effect of dietary lipid levels on lipid deposition in the liver of large yellow croaker and to determine the mechanism by which dietary lipids regulate lipid deposition at the transcriptional level.
Materials and Methods
Ethics statement
The present study was performed in strict accordance with the Standard Operation Procedures (SOPs) of the Guide for the Use of Experimental Animals of Ocean University of China. All animal care and use procedures were approved by the Institutional Animal Care and Use Committee of Ocean University of China (Permit Number: 20001001). Fish were anesthetized with eugenol (1:10,000) (Shanghai Reagent Corp., Shanghai, China) to minimize suffering before being assigned to cages and sampling.
Diet and feeding trial
Three isoproteic (42% crude protein) diets were formulated to contain low (6%), moderate (12%) and high (18%) crude lipid levels by gradually adding fish oil, and wheat starch was used to adjust the level of fish oil. The diet with 12% crude lipid was used as the control, which has been proven to be appropriate for the growth of this fish [25]. The formulation and approximate composition of the diets are presented in Table 1, and the FA composition of the diets is shown in Table 2.
Table 1. Formulation and proximate composition of the experimental diets.
| Dietary lipid levels (%) | |||
|---|---|---|---|
| Low (6) | Moderate (12) | High (18) | |
| Ingredients (g/100 g) | |||
| Fish meal 1 | 39.0 | 39.0 | 39.0 |
| Soybean meal 1 | 20.0 | 20.0 | 20.0 |
| Wheat meal 1 | 23.3 | 23.3 | 23.3 |
| Wheat starch 1 | 12.0 | 6.0 | 0 |
| Fish oil 1 | 0 | 6.0 | 12.0 |
| Soybean lecithin 1 | 1.5 | 1.5 | 1.5 |
| Vitamin premix 2 | 2.0 | 2.0 | 2.0 |
| Mineral premix 3 | 2.0 | 2.0 | 2.0 |
| Attractant 4 | 0.1 | 0.1 | 0.1 |
| Mold inhibitor 5 | 0.1 | 0.1 | 0.1 |
| Proximate composition (g/100 g) | |||
| Moisture | 9.5 | 9.4 | 9.2 |
| Crude protein | 43.1 | 42.6 | 43.2 |
| Crude lipid | 6.1 | 11.5 | 17.8 |
1All of these ingredients were supplied by Great Seven Biotechnology Co., Ltd., China.
2Vitamin premix (mg or g/kg diet): cholecalciferol, 5 mg; retinol acetate, 32 mg; thiamin 25 mg;vitamin B12 (1%), 10 mg; riboflavin, 45 mg; pyridoxine HCl, 20 mg; ascorbic acid, 2000 mg; alpha-tocopherol (50%), 240 mg; vitamin K3, 10 mg; pantothenic acid, 60 mg; inositol, 800 mg; niacin acid, 200 mg; folic acid, 20 mg; biotin (2%), 60 mg; choline chloride (50%), 4000 mg;microcrystalline cellulose, 12.47 g.
3Mineral premix (mg or g/kg diet): CuSO4·5H2O, 10 mg; Ca (IO3)2·6H2O (1%), 60 mg; CoCl2·6H2O (1%), 50 mg; FeSO4·H2O, 80 mg; MgSO4·7H2O, 1200 mg; MnSO4·H2O, 45 mg; NaSeSO3·5H2O (1%), 20 mg; ZnSO4·H2O, 50 mg; CaH2PO4·H2O, 10 g; zeolite, 8.485 g.
4Attractants: glycine and betaine.
5Mold inhibitor: contained 50% calcium propionic acid and 50% fumaric acid.
Table 2. Fatty acid composition of the experimental diets (for total fatty acids).
| Dietary lipid levels (%) | |||
|---|---|---|---|
| Fatty acid (%) | Low (6) | Moderate (12) | High (18) |
| C14:0 | 4.90 | 6.17 | 6.17 |
| C16:0 | 20.81 | 20.43 | 19.52 |
| C18:0 | 3.27 | 2.92 | 2.51 |
| C20:0 | nd 1 | 1.72 | 2.26 |
| ΣSFA 2 | 28.97 | 31.23 | 30.46 |
| C16:n-9 | 8.61 | 8.83 | 8.68 |
| C18:n-9 | 14.76 | 19.18 | 22.31 |
| C22:n-9 | nd | 1.21 | 1.75 |
| ΣMUFA 3 | 23.36 | 29.22 | 32.74 |
| C18:2n-6 | 20.46 | 12.53 | 9.86 |
| C18:3n-6 | 0.79 | 1.43 | 1.46 |
| C20:4n-6 | 0.65 | 0.71 | 0.65 |
| Σn-6PUFA 4 | 21.90 | 14.67 | 11.97 |
| C18:3n-3 | 2.10 | 1.86 | 1.71 |
| C18:4n-3 | nd | 0.31 | 0.39 |
| C20:5n-3 (EPA) | 10.22 | 9.19 | 8.25 |
| C22:6n-3 (DHA) | 8.20 | 8.46 | 8.68 |
| Σn-3PUFA | 20.52 | 19.81 | 19.03 |
| n-3/n-6PUFA | 0.94 | 1.35 | 1.59 |
| Σn-3LC-PUFA 5 | 18.41 | 17.65 | 16.93 |
| DHA/EPA | 0.80 | 0.92 | 1.05 |
1nd: not detected.
2SFA: saturated fatty acids.
3MUFA: monounsaturated fatty acids.
4PUFA: polyunsaturated fatty acids.
5LC-PUFA: long-chain-polyunsaturated fatty acids.
Large yellow croaker of the Tai-chu race were obtained from a local farm (Aquatic Fingerlings Limited Company of Xiangshan Harbour, Ningbo, China) located at [121.752, 29.545]. The fish were reared in floating sea cages (3.0×3.0×3.0 m) for 2 weeks for acclimation to the experimental conditions. Before the feeding trial, fish were fasted for 24 h and weighed after being anaesthetized with eugenol (1:10,000) (Shanghai Reagent Corp., Shanghai, China). Fish of similar sizes with an initial weight of 150.00±4.95 g were randomly assigned to nine cages (1.5×1.5×2.0 m) with 40 fish per cage. Each diet was randomly distributed in triplicate cages. The fish were hand-fed to apparent satiation twice daily (05:00 and 17:00). During the 10-week feeding trail, the water temperature ranged from 21 to 28.5°C. In addition, the salinity ranged from 28 to 32‰ and the dissolved oxygen level ranged from 6.7 to 7.8 mg/L.
Sample collection
At the end of the trial, the fish were fasted for 24 h and anesthetized with eugenol (1:10,000). Then, the fish in each cage were weighed and counted. Five fish per cage were randomly selected for proximate analysis. Blood was sampled from the caudal veins of four fish per cage using ethylenediaminetetraacetic acid(EDTA)-containing Vacutainers (Huabo medical instrument Co., Ltd, Heze, Shandong province, China). Plasma was separated from the blood via centrifugation at 3000 rpm for 10 min at 4°Cand stored at -80°C until use. After blood sampling, the fish were dissected to calculate the hepato-somatic index (HSI) and viscera-somatic index (VSI). The liver was sampled for moisture and lipid contents analyse. For molecular analysis, the livers of three fish per cage were sampled and immediately transferred to liquid nitrogen and then stored at -80°C until analysis.
Plasma biochemical analysis
The plasma samples from each cage were pooled. Plasma TAG, total cholesterol, LDL-cholesterol (LDL-c) and high-density lipoprotein-cholesterol (HDL-c) concentrations were analyzed using commercial assay kits (Mindray Bio Medical Co., Ltd., Shenzhen, China)and a Mindray Auto Bio-chemical Analyzer (BS-400, Mindray, Shenzhen, China) according to the manufacturer’s instructions. TAG was measured via the glycerol lipase oxidase (GPO-PAP) method[26]. Total cholesterol, LDL-c and HDL-c were determined using the cholesterol oxidase method based on Schettler and Nussel [27], Okada et al. [28] and Gordon et al. [29], respectively. NEFA levels were measured using a Cu-NEFA coextraction-based colorimetric assay kit (Nanjing Jiancheng Bioengineering Inc., Nanjing, China) according to Falholt et al. [30].
Measurement of moisture and lipid contents
Whole body moisture content was analyzed by drying the samples to constant weight at 105°C. The moisture contents of the liver were determined via the freeze-drying method using vacuum-freezing drying equipment (Christ ALPHA, Germany). The crude lipid content of the whole body was measured through ether extraction using the Soxhlet method (Soxhlet Extraction System B-811, Switzerland). Approximately 100 mg of dried liver tissue was subjected to lipid extraction via the Folch method [31]. Briefly, the tissues were first homogenized with 6 mL of chloroform-methanol (2:1) and then centrifuged at 3000 rpm for 15 min to recover the liquid phase. The upper solvent was washed with 1.2 mL of a 1.6% CaCl2 solution. The lower chloroform phase containing lipids was first dried under nitrogen and then dried to constant weight at 75°C.
RNA extraction and real-time quantitative PCR
Total RNA was extracted from the liver using TRIzol reagent (Invitrogen, Carlsbad, CA,USA). Isolated RNA quantity and quality were determined via spectrophotometry using a NanoDrop spectrophotometer and on a 1.2% denaturing agarose gel, respectively. For cDNA synthesis, the TransScript One-step gDNA Removal and cDNA Synthesis Super Mix Kit (Transgen Biotech, Beijing, China) were used according to the manufacturer’s instructions. cDNA was diluted 5-fold using RNase- and DNase-free water. Real-time quantitative PCR was performed in a quantitative thermal cycler (Mastercycle ep realplex, Eppendorf, Hamburg, Germany). PCR measurements were performed in a total volume of 20μL, containing 0.4μL of each primer (10μM), 10μL of 2×TransStart Top green qPCR SuperMix (Transgen Biotech, Beijing, China) and 0.8 μL of cDNA. The following quantitative PCR program was employed: 95°C for 2 min, followed by 40 cycles of 95°C for 10 s, 60°C for 10 s and 72°C for 20 s. The primer sequences for each gene are listed in Table 3. At the end of PCR amplification, melting curve analysis was performed to confirm that only one PCR product was present. The fluorescence data acquired during the extension phase were normalized to β-actin via the delta-delta method [32]. Normalized gene expression for the control diet group (12% dietary lipid) was set at 1.
Table 3. Primer pair sequences and amplicon size of the genes used for real-time PCR.
| 5′-3′ primer sequence | ||||
|---|---|---|---|---|
| Gene | Forward | Reverse | Amplicon size (bp) | GenBank accession no. |
| apoAI | ttgctctcgcccttctcctg | cacgctgtccttgatctccttg | 179 | KM593125 |
| apoB100 | agagtgttgtccaggataaagatgc | cagggctcagggtctcagtc | 147 | KM593126 |
| MTP | atgtccaaaatgttctccatgtctg | atgtcaatagccaaccctccttg | 143 | KP027412 |
| LPL | gaattcaacgcggaaacacag | acgctcatagagggcagacac | 105 | JQ327827 |
| LDLR | acataagcgccggtgctgtt | tacgatgtcctctggctgattc | 95 | KM593127 |
| LRP-1 | tggactgggtggctggaaac | caatggcgtatggctcgtctatc | 126 | KM593128 |
| SRBI | acagatccagaaagacaacatcacg | gtagggcaacttctccatcatcac | 172 | KM593129 |
| CD36 | gagcatgatggaaaatggttcaaag | ctccagaaactccctttcaccttag | 159 | KM593122 |
| FATP1 | caaccagcaggacccattacg | catccatcaccagcacatcacc | 131 | KM593124 |
| FABP3 | ccaaacccaccactatcatctcag | gcaccatctttccctcctctattg | 171 | KM593123 |
| FABP10 | caatggaacatggcaggtttacg | tgattggcttgatgtccttggc | 107 | KM593131 |
| FABP11 | caggtgggcaatcggaccaa | ggctcgttgagcttgaacttga | 119 | KM593130 |
| FAS | cagccacagtgaggtcatcc | tgaggacattgagccagacac | 126 | JX456351 |
| DGAT2 | ttcggtgctttctgcaacttcg | aaggatggggaagcggaagt | 111 | KJ563922 |
| ATGL | ccatgcatccgtccttcaacc | Gagatccctaaccgcccact | 103 | HQ916211 |
| CPT I | gctgagcctggtgaagatgttc | Tccatttggttgaattgtttactgtcc | 159 | JX434612 |
| ACO | agtgcccagatgatcttgaagc | Ctgccagaggtaaccatttcct | 184 | JX456348 |
| β-actin | ctacgagggttatgccctgcc | Tgaaggagtaaccgcgctctgt | 107 | GU584189 |
apo: apolipoprotein; MTP: microsomal TAG transfer protein; LPL: lipoprotein lipase; LDLR: low-density lipoprotein receptor; LRP-1: lipoprotein receptor-related protein-1; SRBI: scavenger receptor class BI; CD36: cluster of differentiation 36; FATP1: fatty acid transport protein 1; FABP: fatty acid binding protein; FAS: fatty acid synthase; DGAT2: acyl-CoA: diacylglycerol acyltransferase 2; ATGL: adipose triglyceride lipase; CPT I: carnitine palmitoyltransferase I; ACO: acyl-CoA oxidase.
Statistical analysis
Statistical analysis was performed using SPSS 13.0 for Windows (SPSS Inc., Chicago, IL, USA). The data are presented as means with the standard error of the means (S.E.M.). The results were analyzed via one-way analysis of variance (ANOVA) followed by Tukey’s multiple-range test. When the homogeneity of variance was not satisfied, an independent-samples t test was performed to compare the differences. The level of significance was set at P<0.05.
Results
Survival rate, growth performance and somatic and plasma biochemical indexes
The survival rate, final weight, specific growth rate, feed intake and feed conversion ratio of the fish were not significantly affected by dietary lipid levels (P>0.05) (Table 4). Both HSI and VSI increased with increasing dietary lipid content. In particular, fish fed the high-lipid diet exhibited significantly increased HSI and VSI compared with fish fed the low-lipid and control diets (P<0.05) (Table 4). Plasma TAG, NEFA, and LDL-c levels significantly increased as the dietary lipid content increased from 6 to 18% (P<0.05), whereas no significant difference in plasma total cholesterol and HDL-c levels was observed among treatments (P>0.05).
Table 4. Effects of dietary lipid levels on the growth performance, somatic parameters and plasma biochemical indexes of large yellow croaker (Larimichthys crocea)*.
| Dietary lipid levels (%) | |||
|---|---|---|---|
| Low (6) | Moderate (12) | High (18) | |
| Survival rate (%) 1 | 81.58±2.63 | 93.86±3.16 | 85.96±3.16 |
| Final weight (g) | 281.55±4.47 | 280.17±3.85 | 269.3±3.13 |
| Specific growth rate (%day-1) 2 | 0.79±0.02 | 0.85±0.02 | 0.77±0.02 |
| Feed intake 3 | 0.57±0.02 | 0.55±0.01 | 0.54±0.01 |
| Feed conversion ratio 4 | 1.91±0.08 | 1.64±0.07 | 1.80±0.09 |
| Hepato-somatic index 5 | 1.64±0.05b | 1.82±0.04b | 2.05±0.06a |
| Viscera-somatic index 6 | 6.73±0.12b | 7.37±0.16b | 8.17±0.19a |
| Total cholesterol (mmol/L) | 2.38±0.28 | 2.88±0.36 | 3.08±0.47 |
| Triacylglycerol (mmol/L) | 2.77±0.11c | 4.55±0.03b | 5.85±0.36a |
| NEFA (μmol/L) 7 | 96.57±12.97b | 123.57±8.31ab | 154.72±5.49a |
| HDL-cholesterol (mmol/L) | 0.40±0.05 | 0.42±0.06 | 0.38±0.05 |
| LDL-cholesterol (mmol/L) | 0.13±0.02b | 0.21±0.03b | 0.39±0.03a |
*Values (means±S.E.M.) that share the same letter in the same row are not significantly different (P<0.05; Tukey’s test) among treatments (n = 3).
1Survival rate (%) = 100×final fish number/initial fish number.
2Specific growth rate (%day-1) = 100×[ln (final weight)-ln (initial weight)]/days.
3Feed intake (day-1) = feed consumption (g)/(days×(final body weight + initial body weight)/2).
4Feed conversion ratio = feed consumption (g)/wet weight gain (g).
5Hepato-somatic index = 100×(liver weight/body weight).
6Viscera-somatic index = 100×(viscera weight/body weight).
7NEFA: non-esterified fatty acids.
Moisture and lipid contents
The whole body and liver moisture contents decreased as dietary lipids increased, and these levels were significantly reduced in fish fed the high-lipid diet compared to fish fed the low-lipid diet (P<0.05) (Table 5). However, whole body and liver lipid contents were significantly elevated as dietary lipid content increased from 6 to 18% (P<0.05). In particular, the liver lipid content determined in fish fed the high-lipid diet (26.9%) was up to 77% increased compared with fish fed the low-lipid diet (15.1%) in terms of wet weight.
Table 5. Effects of dietary lipid levels on the moisture and lipid contents of the whole body, liver and muscle of large yellow croaker (Larimichthys crocea) (%, wet weight)*.
| Dietary lipid levels (%) | |||
|---|---|---|---|
| Low (6) | Moderate (12) | High (18) | |
| Whole body (g/100 g) | |||
| Moisture | 72.36±0.19a | 68.88±0.51b | 68.86±0.39b |
| Lipid | 9.56±0.12b | 11.45±0.2a | 11.83±0.25a |
| Liver (g/100 g) | |||
| Moisture | 61.41±0.40a | 56.90±0.44b | 55.12±0.31c |
| Lipid | 15.14±0.37c | 23.01±0.26b | 26.91±0.82a |
*Values (means±S.E.M.) that share the same letter in the same row are not significantly different (P<0.05; Tukey’s test) among treatments (n = 3).
Expression of proteins related to VLDL assembly in the liver
Among the three proteins (apoAI, apoB100 and MTP) involved in VLDL assembly in the liver, only apoB100 mRNA levels exhibited a significant difference among the three groups (Fig 1). The expression of apoB100 was significantly down-regulated in fish fed the high-lipid diet compared with those fed the low-lipid diet (P<0.05).
Fig 1. Expression of genes related to VLDL assembly in the liver of large yellow croaker.
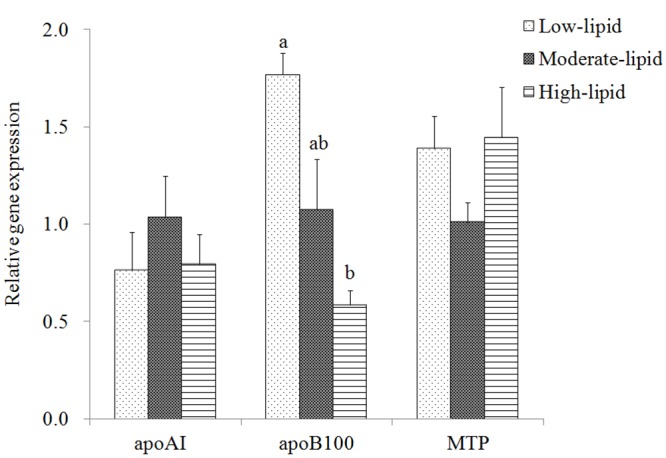
Values (means±S.E.M.) in bars that have the same letter are not significantly different (P>0.05; Tukey’s test) among treatments (n = 3). apo: apolipoprotein; MTP: microsomal triacylglycerol transfer protein.
Expression of lipoprotein lipase and lipoprotein receptors
LPL expression did not show any significant changes in the liver among the dietary treatments (P>0.05) (Fig 2). The lowest expression of LDLR and LRP-1 was noted in the control group, whereas the highest expression was observed in the low-lipid group, and there were significant differences between these two groups (P<0.05). SRBI expression was not significantly affected by dietary lipids (P>0.05).
Fig 2. Expression of genes encoding LPL and lipoprotein receptors in the liver of large yellow croaker.
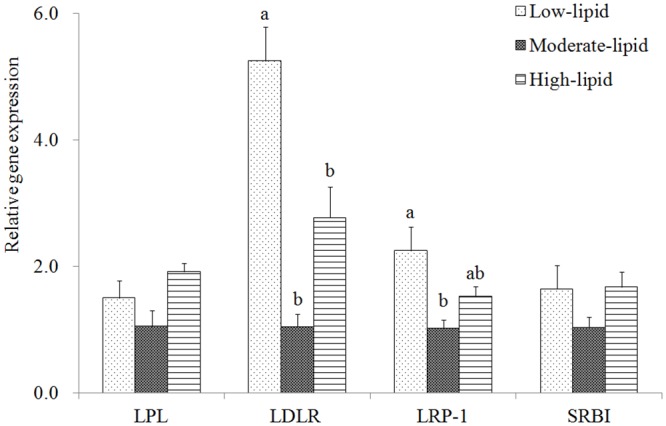
Values (means±S.E.M.) in bars that have the same letter are not significantly different (P>0.05; Tukey’s) among treatments (n = 3). LPL: lipoprotein lipase; LRP-1: LDL receptor-related protein-1; SRBI: scavenger receptor class BI.
Expression of genes related to fatty acid uptake
Compared with the control group, only FABP11 expression was significantly increased in the high-lipid group (P<0.05)(Fig 3). No significant changes in the expression of CD36, FATP1, FABP10 and FABP3 were observed between the high-lipid and control groups (P>0.05) though FATP1 was 1.4-fold up-regulated in the high-lipid group. In the low-lipid group, the expression of CD36, FATP1 and FABP3 was significantly up-regulated compared with the control group (P<0.05), whereas FABP10 and FABP11 mRNA levels did not significantly differ in these two groups. The expression of all of the investigated genes involved in FA uptake (CD36, FATP1, FABP3 and FABP10)was significantly increased in the low-lipid group compared with the high-lipid group with the exception of FABP11, which was expressed at a higher level in the high-lipid group (P<0.05).
Fig 3. Expression of genes related to fatty acid uptake in the liver of large yellow croaker.
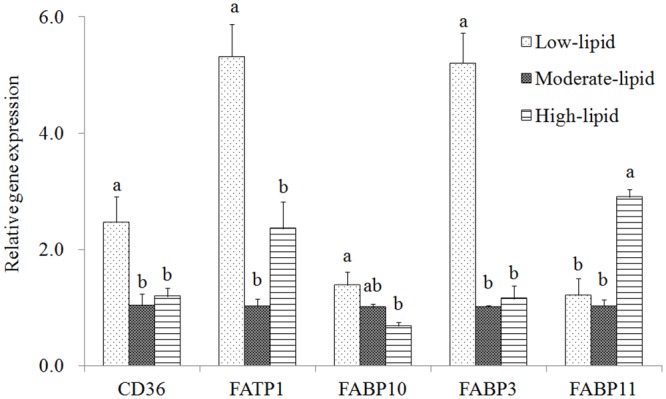
Values (means±S.E.M.) in bars that have the same letter are not significantly different (P>0.05; T test for FABP3 in the liver, and Tukey’s test for the other genes) among treatments (n = 3). CD36: cluster of differentiation; FATP1: fatty acid transport protein1; FABP: fatty acid binding protein.
Expression of genes involved in triacylglycerol synthesis and catabolism
In fish fed the high-lipid diet, the expression of DGAT2 and CPT I was significantly down-regulated compared with fish fed the control diet (P<0.05), whereas the expression of FAS, ATGL and ACO was not significantly altered between fish fed the high-lipid and control diets (P>0.05)(Fig 4). In fish fed the low-lipid diet, FAS and ACO were significantly up-regulated, whereas ATGL and CPT I were down-regulated compared with fish fed the control diet (P<0.05). TAG synthesis related genes (FAS and DGAT2) were expressed at higher levels in the low-lipid group compared with the high-lipid group, whereas genes related to TAG catabolism (ATGL and CPT I) were expressed at higher levels in the high-lipid group with the exception of ACO (P<0.05).
Fig 4. Expression of genes related totriacylglycerol synthesis and catabolism in the liver of large yellow croaker.
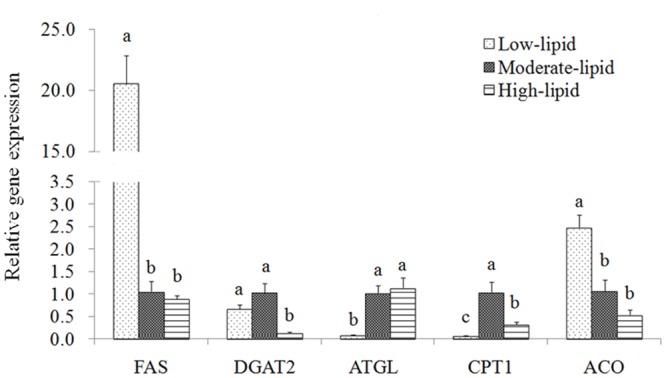
Values (means±S.E.M.) in bars that have the same letter are not significantly different (P>0.05; T test for FAS, DGAT2 and CPT I in the liver, and Tukey’s test for the other genes) among treatments (n = 3). FAS: fatty acid synthase; DGAT2: acyl-CoA:diacylglycerol acyltransferase 2; ATGL: adipose triglyceride lipase; CPT I: carnitine palmitoyltransferase I; ACO: acyl-CoA oxidase.
Discussion
In the present study, the final weight and specific growth rate of fish were not significantly affected by dietary lipid levels, which were consistent with some previous studies conducted in large yellow croaker [33], Atlantic cod [34], European sea bass [35] and rainbow trout [36]. However, several studies have shown that an increase in dietary lipid can promote growth[1,2,37] or suppress growth in fish[38–41]. The growth-promoting effect of high dietary lipids was general correlated with the protein-sparing effect of dietary lipids [1,37]. Although a protein-sparing effect was reported in juvenile large yellow croaker (10 g) [25], this effect was not observed in middle-sized fish of the same species in the study of Wang et al. (240 g) [33] or in the present study (150 g). In contrast, growth suppression by high dietary lipids was primarily due to high-lipid-induced reduction in feed intake [40,41]. However, this phenomenon was not observed in the study of Wang et al. or in the present study potentially due to the high capacity of lipid tolerance in large yellow croaker[33]. Therefore, the effects of dietary lipid levels on fish growth performance are complex, which may be related to fish life stages, fish species, feed composition and feeding strategies.
The lipid content was significantly increased in the whole body and liver when dietary lipids were increased from 6 to 18%, which was supported by the findings of Wang et al.[33] in the same fish species. The increase of dietary lipid levels lead to higher lipid deposition has also been observed in turbot [39], Atlantic cod [34]and juvenile cobia [40]. Also, increased dietary lipid levels resulted in increased plasma TAG, NEFA and LDL-c levels, which was consistent with results previously obtained in large yellow croaker [33], grass carp [3,42] and tiger puffer [41]. Adipose tissue plays an important role in storing excessive amounts of circulating FAs in the form of TAG and in mobilizing TAG via breakdown into NEFAs [43]. The increased plasma NEFAs level in fish fed the high-lipid diet may be an important source of lipid accumulation in the liver of large yellow croaker. In a study on gilthead sea bream, the authors also suggested that increased adipocyte lipolysis rates in adipose tissue could be an important factor resulting in excess lipid accumulation in the liver [44]. More precisely, Donnelly et al.[45] reported that 59% of the TAG that accumulated in the liver of the NAFLD patients originated from NEFAs, and greater than 60% of NEFAs were from adipose tissue. Therefore, further studies are needed to determine the mechanism by which adipose tissue influences lipid homeostasis and deposition.
In the present study, increased plasma TAG level potentially indicated a higher hepatic VLDL secretion in fish fed the high-lipid diet. However, apoB100 was down-regulated as the dietary lipid content increased up to 18%, which, to some extent, may be attributable to higher n-3 LC-polyunsaturated FA (LC-PUFA) level in the high-lipid diet. Nevertheless, hepatic lipids content is a predominant driver of VLDL assembly, and insufficient lipids supplies have evidenced to promote apoB100 degradation and reduce VLDL assembly and secretion[46,47]. Moreover, plasma NEFA could be directly incorporated in to VLDL-TAG and obviously stimulate VLDL secretion [48,49]. Therefore, the increase of plasma TAG and NEFA level and hepatic lipid content in the high-lipid group suggested that hepatic VLDL secretion to peripheral tissues increased in order to reduce lipid deposition.
LPL mRNA levels were not significantly affected by dietary lipid levels in the liver, which is consistent with results reported for red sea bream [50] and large yellow croaker [33]. However, Lu et al. [51]reported that a high-fat diet significantly up-regulated LPL in the liver of blunt snout bream. The different fish species used in these studies potentially accounted for this disparity. Abdominal (or perivisceral) adipose tissue is the primarily lipids storage tissue in red sea bream and large yellow croaker [52], whereas the liver is the main storage tissue and stored more lipids in blunt snout bream fed a high-fat diet [51]. Even though this tissue-specific regulation of LPL by dietary lipids exists in various fish species, further studies are needed to confirm, given that LPL plays a pivotal role in LP metabolism and lipid deposition and limited researches in nutritional regulation of LPL in fish.
LDLR and LRP-1 are closely associated with LP clearance in plasma, and increased expression of these two LP receptors has been reported to improve LP clearance in hamsters [53,54]. In the present study, based on the up-regulation of LDLR and LRP-1 expression in the low- and high-lipid groups, it was supposed that plasma LP clearance was enhanced in these two groups. Among the three receptors (LDLR, LRP-1 and SRBI), the highest elevation of expression was found for the LDLR, which indicated that this receptor is vulnerable to the regulation by dietary lipids and plays a critical role in LP clearance. In mammals, loss of the LDLR leads to decreased LDL catabolism and elevated LDL levels [55]. Additionally, LDLR gene mutations result in familial hypercholesterolemia [56,57]. Compared with the control mice, trans-10, cis-12-treated C57BL/6j mice exhibit increased LDLR expression with lower plasma TAG levels [58], and LDLR-/- apoB100/100 mice exhibit increased LRP-1 expression [11]. In the present study, SRBI expression was not significantly affected by dietary lipid content, possibly because SRBI is mainly associated with reverse cholesterol transport[59,60].
Regarding the FA transporters (CD36, FATP1 and FABPs), it was interesting that the expression of CD36, FATP1, FABP10 and FABP3 was the highest in the low-lipid group, whereas FABP11 expression was the highest in the high-lipid group. Increased expression of these genes was assumed to increased FA uptake. However, the importance and detail role of individual FA transporter may differ under various dietary lipid levels. In rats, greater transport efficiency of CD36 is found compared with FATP1 in skeletal muscle [61]. Regarding FABPs in mammals, FABP1 (homologous to fish FABP10 [62]) mainly acts as a LCFA transporter in the liver, particularly targeting ligand to β-oxidation. FABP3 is required for LCFA transport for mitochondria β-oxidation in muscle and FABP4 (homologous to fish FABP11 [62]) plays a role in TAG storage in the adipose tissue [63]. These transporters may have the similar roles in fish. Therefore, the highest expression of FABP11 in the high-lipid group could be mainly used to transport FAs to synthesis TAG, whereas FAs absorbed in the low-lipid group are potentially used for other metabolism pathways. However, these potential roles must be confirmed in further studies.
TAG synthesis and catabolism are also pivotal factors affecting lipid accumulation for a specific tissue. FAS is a key enzyme involved in de novo lipogenesis, and acyl-CoA:diacylglycerol acyltransferase (DGAT) catalyzes the final and only committed step in the biosynthesis of TAG [43]. ATGL has been proven to catalyze the initial step of TAG hydrolysis [64,65], and CTP1 and ACO both catalyze the rate-limiting step in FA β-oxidation. In the present study, hepatic gene expression related to TAG synthesis (DGAT2) and FA oxidation (CPT I and ACO) was attenuated in the high-lipid group, which was supported by results previously obtained in blunt snout bream [5] and large yellow croaker [33]. Reduced DGAT2 expression could be partially due to a feedback mechanism involving excessive lipid accumulation in the liver, which has been observed in a mouse model of high-fat diet-induced obesity [66–68]. Furthermore, the inhibitory effect of lipogenesis by increased n-3 LC-PUFA level in the high-lipid diet could be another factor involved in the reduction of DGAT2 expression [69,70]. However, n-3 LC-PUFA could lead to the unexpected down-regulation of CPT I and ACO expression in the high-lipid group. Lu et al. [5] reported that high levels of n-3 LC-PUFA significantly altered the hepatic mitochondrial membrane FA composition and CPT I kinetics in blunt snout bream, and the expression of hepatic CPT I and ACO was also down-regulated. Similar results have also been obtained in rats [71]. Furthermore, this decreased FA oxidation activity could result in increased lipid deposition in the liver of large yellow croaker. Increased hepatic FAS expression was observed in the low-lipid group compared with the control group, which was potentially due to the elevation of de novo lipogenesis in response to the excess carbohydrates in the low-lipid diet [72]. However, the de novo lipogenesis process is quite limited in fish [72]. Therefore, under the condition of reduced availability of FA sources, DGAT2 expression in the low-lipid group reduced compared with the control group and accompanied by a lower lipid content in the liver. In addition, the down-regulation of ATGL and CPT I expression potentially correlated with higher carbohydrate and lower lipid levels in the low-lipid group compared with the control group because that the provision of digestible carbohydrates in diets could spare the use of lipids as sources of energy [72,73].
Conclusions
Results of the present study demonstrated that different dietary lipid levels could regulate various metabolic pathways to affect hepatic lipid deposition in large yellow croaker at the transcriptional level. In fish fed the high-lipid diet, increased lipoprotein clearance and fatty acid uptake and decreased fatty acid β-oxidation were potentially involved in the increased hepatic lipid deposition. For fish fed the low-lipid diet, to some extent, lipoprotein clearance together with fatty acid uptake and de novo synthesis increased, whereas triacylglycerol catabolism decreased. However, lipid accumulation in the liver decreased, primarily due to low dietary lipid intake in fish fed the low-lipid diet.
Acknowledgments
We would like to thank Y. L. Liu, X. W. Yi, K. D. Wang, S. L. Li, K. Lu, Z. H. Wei and J. Ma for their help with the study.
Data Availability
All relevant data are within the paper.
Funding Statement
This research was supported by the National Natural Foundation of China (31372541 and 31172425), the Doctoral Scientific Fund Project of the Ministry of Education of China (20120132110007), and the National Basic Research Program of China (973 Program, 2014CB138600). The funder's websites are http://www.nsfc.gov.cn/, http://www.cutech.edu.cn/cn/index.htm and http://www.most.gov.cn/index.htm, respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Boujard T, Gélineau A, Covès D, Corraze G, Dutto G, Gasset E, et al. (2004) Regulation of feed intake, growth, nutrient and energy utilisation in European sea bass (Dicentrarchus labrax) fed high fat diets. Aquaculture 231: 529–545. [Google Scholar]
- 2. Li X, Jiang Y, Liu W, Ge X (2012) Protein-sparing effect of dietary lipid in practical diets for blunt snout bream (Megalobrama amblycephala) fingerlings: effects on digestive and metabolic responses. Fish Physiol Biochem 38: 529–541. 10.1007/s10695-011-9533-9 [DOI] [PubMed] [Google Scholar]
- 3. Du Z, Clouet P, Huang L, Degrace P, Zheng W, He J, et al. (2008) Utilization of different dietary lipid sources at high level in herbivorous grass carp (Ctenopharyngodon idella): mechanism related to hepatic fatty acid oxidation. Aquacult Nutr 14: 77–92. [Google Scholar]
- 4. Rueda-Jasso R, Conceiçao LE, Dias J, De Coen W, Gomes E, Rees J- F,et al. (2004) Effect of dietary non-protein energy levels on condition and oxidative status of Senegalese sole (Solea senegalensis) juveniles. Aquaculture 231: 417–433. [Google Scholar]
- 5. Lu K-L, Xu W-N, Wang L-N, Zhang D-D, Zhang C-N, Liu W-B (2014) Hepatic β-Oxidation and Regulation of Carnitine Palmitoyltransferase (CPT) I in Blunt Snout Bream Megalobrama amblycephala Fed a High Fat Diet. PLoS ONE 9: e93135 10.1371/journal.pone.0093135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shearer GC, Savinova OV, Harris WS (2012) Fish oil -- how does it reduce plasma triglycerides? Biochim Biophys Acta 1821: 843–851. 10.1016/j.bbalip.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheridan MA (1988) Lipid dynamics in fish: aspects of absorption, transportation, deposition and mobilization. Comp Biochem Physiol B Comp Biochem 90: 679–690. [DOI] [PubMed] [Google Scholar]
- 8. Weil C, Lefèvre F, Bugeon J (2012) Characteristics and metabolism of different adipose tissues in fish. Rev Fish Biol Fish 23: 157–173. [Google Scholar]
- 9. Magkos F (2009) Basal very low-density lipoprotein metabolism in response to exercise: mechanisms of hypotriacylglycerolemia. Prog Lipid Res 48: 171–190. 10.1016/j.plipres.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 10. Descamps O, Bilheimer D, Herz J (1993) Insulin stimulates receptor-mediated uptake of apoE-enriched lipoproteins and activated alpha 2-macroglobulin in adipocytes. J Biol Chem 268: 974–981. [PubMed] [Google Scholar]
- 11. Degrace P, Moindrot B, Mohamed I, Gresti J, Du Z-Y, Chardigny J-M, et al. (2006) Upregulation of liver VLDL receptor and FAT/CD36 expression in LDLR-/- apoB100/100 mice fed trans-10, cis-12 conjugated linoleic acid. J Lipid Res 47: 2647–2655. [DOI] [PubMed] [Google Scholar]
- 12. Ilmonen M. Genetic Variation of Apolipoprotein B in the Finns: Effects of Serum Lipid Levels. Helsinki: University of Helsinki; 2000. [Google Scholar]
- 13. Zhou J, Stubhaug I, Torstensen BE (2010) Trans-Membrane Uptake and Intracellular Metabolism of Fatty Acids in Atlantic Salmon (Salmo salar L.) Hepatocytes. Lipids 45: 301–311. 10.1007/s11745-010-3396-1 [DOI] [PubMed] [Google Scholar]
- 14. Sanchez-Gurmaches J, Cruz-Garcia L, Gutierrez J, Navarro I (2012) mRNA expression of fatty acid transporters in rainbow trout: in vivo and in vitro regulation by insulin, fasting and inflammation and infection mediators. Comp Biochem Physiol Part A Mol Integr Physiol 163: 177–188. [DOI] [PubMed] [Google Scholar]
- 15. Sanchez-Gurmaches J, Ostbye TK, Navarro I, Torgersen J, Hevroy EM, Ruyter B,et al. (2011) In vivo and in vitro insulin and fasting control of the transmembrane fatty acid transport proteins in Atlantic salmon (Salmo salar). Am J Physiol Regul Integr Comp Physiol 301: 947–957. 10.1152/ajpregu.00289.2011 [DOI] [PubMed] [Google Scholar]
- 16. Stahl A, Evans JG, Pattel S, Hirsch D, Lodish HF (2002) Insulin causes fatty acid transport protein translocation and enhanced fatty acid uptake in adipocytes. Dev Cell 2: 477–488. [DOI] [PubMed] [Google Scholar]
- 17. Haunerland NH, Spener F (2004) Fatty acid-binding proteins–insights from genetic manipulations. Prog Lipid Res 43: 328–349. [DOI] [PubMed] [Google Scholar]
- 18. Torstensen BE, Nanton DA, Olsvik PA, Sundvold H, Stubhaug I (2009) Gene expression of fatty acid-binding proteins, fatty acid transport proteins (cd36 and FATP) and β-oxidation-related genes in Atlantic salmon (Salmo salar L.) fed fish oil or vegetable oil. Aquacult Nutr 15: 440–451. [Google Scholar]
- 19. Agulleiro MJ, André M, Morais S, Cerdà J, Babin PJ (2007) High transcript level of fatty acid-binding protein 11 but not of very low-density lipoprotein receptor is correlated to ovarian follicle atresia in a teleost fish (Solea senegalensis). Biol Reprod 77: 504–516. [DOI] [PubMed] [Google Scholar]
- 20. Todorčević M, Vegusdal A, Gjøen T, Sundvold H, Torstensen BE, Kjær MA,et al. (2008) Changes in fatty acids metabolism during differentiation of Atlantic salmon preadipocytes; Effects of n-3 and n-9 fatty acids. Biochim Biophys Acta Mol Cell Biol L 1781: 326–335. 10.1016/j.bbalip.2008.04.014 [DOI] [PubMed] [Google Scholar]
- 21. Huang TS, Todorčević M, Ruyter B, Torstensen B (2010) Altered expression of CCAAT/enhancer binding protein and FABP11 genes during adipogenesis in vitro in Atlantic salmon (Salmo salar). Aquacult Nutr 16: 72–80. [Google Scholar]
- 22. Gu M, Kortner TM, Penn M, Hansen AK, Krogdahl Å (2014) Effects of dietary plant meal and soya-saponin supplementation on intestinal and hepatic lipid droplet accumulation and lipoprotein and sterol metabolism in Atlantic salmon (Salmo salar L.). Br J Nutr 111: 432–444. 10.1017/S0007114513002717 [DOI] [PubMed] [Google Scholar]
- 23. Leaver MJ, Bautista JM, Björnsson BT, Jönsson E, Krey G, Tocher DR,et al. (2008) Towards Fish Lipid Nutrigenomics: Current State and Prospects for Fin-Fish Aquaculture. Rev Fish Sci 16: 73–94. [Google Scholar]
- 24. Tocher DR (2003) Metabolism and functions of lipids and fatty acids in teleost fish. Rev Fish Sci 11: 107–184. [Google Scholar]
- 25. Yi X, Zhang F, Xu W, Li J, Zhang W, Mai K (2014) Effects of dietary lipid content on growth, body composition and pigmentation of large yellow croaker Larimichthys croceus . Aquaculture 434: 355–361. [Google Scholar]
- 26. Scheletter G, Nussel E (1975) Quantitative enzymatic Colorimetric determination of triglycerides in serum or plasma. Arbeitsmed Sozialmed Pracentimed 10: 25. [Google Scholar]
- 27. Richmond W (1973) Preparation and properties of a cholesterol oxidase from Nocardia sp. and its application to the enzymatic assay of total cholesterol in serum. Clin Chem 19: 1350–1356. [PubMed] [Google Scholar]
- 28. Okada M, Matsui H, Ito Y, Fujiwara A, Inano K (1998) Low-density lipoprotein cholesterol can be chemically measured: a new superior method. J Lab Clin Med 132: 195–201. [DOI] [PubMed] [Google Scholar]
- 29. Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR (1977) High density lipoprotein as a protective factor against coronary heart disease: the Framingham Study. Am J Med 62: 707–714. [DOI] [PubMed] [Google Scholar]
- 30. Falholt K, Lund B, Falholt W (1973) An easy colorimetric micromethod for routine determination of free fatty acids in plasma. Clinica Chimica Acta 46: 105–111. [DOI] [PubMed] [Google Scholar]
- 31. Folch J, Lees M, Sloane-Stanley G (1957) A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226: 497–509. [PubMed] [Google Scholar]
- 32. Livak KJ, Schmittgen TD (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2− ΔΔCT Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 33. Wang X, Li Y, Hou C, Gao Y, Wang Y (2013) Physiological and molecular changes in large yellow croaker (Pseudosciaena crocea R.) with high-fat diet-induced fatty liver disease. Aquacult Res: 1–11. [Google Scholar]
- 34. Hansen JØ, Berge GM, Hillestad M, Krogdahl Å, Galloway TF, Holm H, et al. (2008) Apparent digestion and apparent retention of lipid and fatty acids in Atlantic cod (Gadus morhua) fed increasing dietary lipid levels. Aquaculture 284: 159–166. [Google Scholar]
- 35. Peres H, Oliva-Teles A (1999) Effect of dietary lipid level on growth performance and feed utilization by European sea bass juveniles (Dicentrarchus labrax). Aquaculture 179: 325–334. 10228951 [Google Scholar]
- 36. Jobling M, Koskela J, Savolainen R (1998) Influence of dietary fat level and increased adiposity on growth and fat deposition in rainbow trout, Oncorhynchus mykiss (Walbaum). Aquacult Res 29: 601–607. [Google Scholar]
- 37. López LM, Durazo E, Viana MT, Drawbridge M, Bureau DP (2009) Effect of dietary lipid levels on performance, body composition and fatty acid profile of juvenile white seabass, Atractoscion nobilis . Aquaculture 289: 101–105. [Google Scholar]
- 38. Borges P, Oliveira B, Casal S, Dias J, Conceiçao L, Valente LM (2009) Dietary lipid level affects growth performance and nutrient utilisation of Senegalese sole (Solea senegalensis) juveniles. Br J Nutr 102: 1007–1014. 10.1017/S0007114509345262 [DOI] [PubMed] [Google Scholar]
- 39. Regost C, Arzel J, Cardinal M, Robin J, Laroche M, Kaushik S (2001) Dietary lipid level, hepatic lipogenesis and flesh quality in turbot (Psetta maxima). Aquaculture 193: 291–309. [Google Scholar]
- 40. Wang J-T, Liu Y-J, Tian L-X, Mai K-S, Du Z-Y, Wang Y,et al. (2005) Effect of dietary lipid level on growth performance, lipid deposition, hepatic lipogenesis in juvenile cobia (Rachycentron canadum). Aquaculture 249: 439–447. [Google Scholar]
- 41. Kikuchi K, Furuta T, Iwata N, Onuki K, Noguchi T (2009) Effect of dietary lipid levels on the growth, feed utilization, body composition and blood characteristics of tiger puffer Takifugu rubripes Aquaculture 298: 111–117. [Google Scholar]
- 42. Du Z-Y, Clouet P, Zheng W-H, Degrace P, Tian L-X, Liu Y-J (2006) Biochemical hepatic alterations and body lipid composition in the herbivorous grass carp (Ctenopharyngodon idella) fed high-fat diets. Br J Nutr 95: 905–915. [DOI] [PubMed] [Google Scholar]
- 43. Yen C-LE, Stone SJ, Koliwad S, Harris C, Farese RV (2008) Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res 49: 2283–2301. 10.1194/jlr.R800018-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cruz-Garcia L, Sanchez-Gurmaches J, Bouraoui L, Saera-Vila A, Perez-Sanchez J, Gutierrez J,et al. (2011) Changes in adipocyte cell size, gene expression of lipid metabolism markers, and lipolytic responses induced by dietary fish oil replacement in gilthead sea bream (Sparus aurata L.). Comp Biochem Physiol Part A Mol Integr Physiol 158: 391–399. [DOI] [PubMed] [Google Scholar]
- 45. Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, Parks EJ (2005) Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest 5: 1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adiels M, Taskinen M-R, Packard C, Caslake M, Soro-Paavonen A, Westerbacka J,et al. (2006) Overproduction of large VLDL particles is driven by increased liver fat content in man. Diabetologia 49: 755–765. [DOI] [PubMed] [Google Scholar]
- 47. Adiels M, Westerbacka J, Soro-Paavonen A, Häkkinen A, Vehkavaara S, Caslake M,et al. (2007) Acute suppression of VLDL1 secretion rate by insulin is associated with hepatic fat content and insulin resistance. Diabetologia 50: 2356–2365. [DOI] [PubMed] [Google Scholar]
- 48. Hellerstein M, Christiansen M, Kaempfer S, Kletke C, Wu K, Reid J,et al. (1991) Measurement of de novo hepatic lipogenesis in humans using stable isotopes. J Clin Invest 87: 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vedala A, Wang W, Neese RA, Christiansen MP, Hellerstein MK (2006) Delayed secretory pathway contributions to VLDL-triglycerides from plasma NEFA, diet, and de novo lipogenesis in humans. J Lipid Res 47: 2562–2574. [DOI] [PubMed] [Google Scholar]
- 50. Liang X-F, Oku H, Ogata HY (2002) The effects of feeding condition and dietary lipid level on lipoprotein lipase gene expression in liver and visceral adipose tissue of red sea bream Pagrus major Comp Biochem Physiol Part A Mol Integr Physiol 131: 335–342. [DOI] [PubMed] [Google Scholar]
- 51. Lu K-L, Xu W-N, Li X-F, Liu W-B, Wang L-N, Zhang C-N (2013) Hepatic triacylglycerol secretion, lipid transport and tissue lipid uptake in blunt snout bream (Megalobrama amblycephala) fed high-fat diet. Aquaculture 408–409: 160–168. [Google Scholar]
- 52. Oku H,Ogata HY (2000) Body lipid deposition in juveniles of red sea bream Pagrus major, yellowtail Seriola quinqueradiata, and Japanese flounder Paralichthys olivaceus . Fish Sci 66: 25–31. [Google Scholar]
- 53. Bennett A, Billett M, Salter A, Mangiapane E, Bruce J, Anderton K,et al. (1995) Modulation of hepatic apolipoprotein B, 3-hydroxy-3-methylglutaryl-CoA reductase and low-density lipoprotein receptor mRNA and plasma lipoprotein concentrations by defined dietary fats. Comparison of trimyristin, tripalmitin, tristearin and triolein. Biochem J 311: 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dorfman SE, Lichtenstein AH (2006) Dietary fatty acids differentially modulate messenger RNA abundance of low-density lipoprotein receptor, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and microsomal triglyceride transfer protein in Golden-Syrian hamsters. Metabolism 55: 635–641. [DOI] [PubMed] [Google Scholar]
- 55. Murayama T, Yokode M, Horiuchi H, Yoshida H, Sano H, Kita T (2000) Overexpression of low density lipoprotein receptor eliminates apolipoprotein B100-containing lipoproteins from circulation and markedly prevents early atherogenesis in apolipoprotein E-deficient mice. Atherosclerosis 153: 295–302. [DOI] [PubMed] [Google Scholar]
- 56. Brown MS, Goldstein JL (1986) A receptor-mediated pathway for cholesterol homeostasis. Science 232: 34–47. [DOI] [PubMed] [Google Scholar]
- 57. Twisk J, Gillian-Daniel DL, Tebon A, Wang L, Barrett PHR, Attie AD (2000) The role of the LDL receptor in apolipoprotein B secretion. J Clin Invest 105: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Degrace P, Demizieux L, Gresti J, Chardigny J-M, Sébédio J-L, Clouet P (2003) Association of liver steatosis with lipid oversecretion and hypotriglyceridaemia in C57BL/6j mice fed trans-10, cis-12-linoleic acid. FEBS Lett 546: 335–339. [DOI] [PubMed] [Google Scholar]
- 59. Acton S, Rigotti A, Landschulz KT, Xu SZ, Hobbs HH, et al. (1996) Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271: 518–520. [DOI] [PubMed] [Google Scholar]
- 60. Rader DJ, Hobbs HH. Disorders of lipoprotein metabolism In: Longo DL, Fauci AS, Kasper DL, Hauser SL, Jameson JL, Loscalzo J, editors.Harrisons Principles of Internal Medicine. New York: McGraw-Hill Companies; 2008. pp. 334–337. [Google Scholar]
- 61. Nickerson JG, Alkhateeb H, Benton CR, Lally J, Nickerson J, Han XX, et al. (2009) Greater transport efficiencies of the membrane fatty acid transporters FAT/CD36 and FATP4 compared with FABPpm and FATP1 and differential effects on fatty acid esterification and oxidation in rat skeletal muscle. J Biol Chem 284: 16522–16530. 10.1074/jbc.M109.004788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thirumaran A, Wright JM, Bell J (2014) Fatty acid-binding protein (fabp) genes of spotted green pufferfish (Tetraodon nigroviridis): comparative genomics and spatial transcriptional regulation. Genome 57: 289–301. 10.1139/gen-2014-0059 [DOI] [PubMed] [Google Scholar]
- 63. Storch J, Thumser AE (2010) Tissue-specific functions in the fatty acid-binding protein family. J Biol Chem 285: 32679–32683. 10.1074/jbc.R110.135210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M,et al. (2004) Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306: 1383–1386. [DOI] [PubMed] [Google Scholar]
- 65. Smirnova E, Goldberg EB, Makarova KS, Lin L, Brown WJ, Jackson CL (2006) ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep 7: 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kim S, Sohn I, Ahn J-I, Lee K-H, Lee YS, Lee YS (2004) Hepatic gene expression profiles in a long-term high-fat diet-induced obesity mouse model. Gene 340: 99–109. [DOI] [PubMed] [Google Scholar]
- 67. Kreeft AJ, Moen CJ, Porter G, Kasanmoentalib S, Sverdlov R, van Gorp PJ,et al. (2005) Genomic analysis of the response of mouse models to high-fat feeding shows a major role of nuclear receptors in the simultaneous regulation of lipid and inflammatory genes. Atherosclerosis 182: 249–257. [DOI] [PubMed] [Google Scholar]
- 68. Inoue M, Ohtake T, Motomura W, Takahashi N, Hosoki Y, Miyoshi S,et al. (2005) Increased expression of PPARγ in high fat diet-induced liver steatosis in mice. Biochem Biophys Res Commun 336: 215–222. [DOI] [PubMed] [Google Scholar]
- 69. Alvarez M, Diez A, Lopez-Bote C, Gallego M, Bautista J (2000) Short-term modulation of lipogenesis by macronutrients in rainbow trout (Oncorhynchus mykiss) hepatocytes. Br J Nutr 84: 619–628. [PubMed] [Google Scholar]
- 70. Kjaer MA, Vegusdal A, Gjoen T, Rustan AC, Todorcevic M, Ruyter B (2008) Effect of rapeseed oil and dietary n-3 fatty acids on triacylglycerol synthesis and secretion in Atlantic salmon hepatocytes. Biochim Biophys Acta 1781: 112–122. 10.1016/j.bbalip.2007.12.004 [DOI] [PubMed] [Google Scholar]
- 71. Colquhoun A (2002) Gamma-linolenic acid alters the composition of mitochondrial membrane subfractions, decreases outer mitochondrial membrane binding of hexokinase and alters carnitine palmitoyltransferase I properties in the Walker 256 rat tumour. Biochim Biophys Acta Mol Cell Biol L 1583: 74–84. [DOI] [PubMed] [Google Scholar]
- 72. National Research Council. Committee on the Nutrient Requirements of Fish and Shrimp Nutrient requirements of fish and shrimp. Washington, DC: National Academies Press; 2011. [Google Scholar]
- 73. García-Meilán I, Ordóñez-Grande B, Gallardo M (2014) Meal timing affects protein-sparing effect by carbohydrates in sea bream: Effects on digestive and absorptive processes. Aquaculture 434: 121–128. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


