Abstract
Purpose of review
Recent studies have enhanced our understanding the role of the SIRT1 deacetylase in regulation of normal hematopoietic stem cells (HSC) and leukemia stem cells (LSC), and its importance in regulating autophagy and epigenetic reprogramming in response to metabolic alterations.
Recent findings
Studies employing conditional deletion mouse models indicate an important role for SIRT1 in maintenance of adult HSC under conditions of stress. SIRT1 is significantly overexpressed in LSC populations from acute myeloid leukemia (AML) patients with the FLT3-ITD mutation, and maintains their survival, growth and drug resistance, as previously described for chronic myelogenous leukemia (CML). SIRT1 can also enhance leukemia evolution and drug resistance by promoting genetic instability. Recent studies indicate an important role for SIRT1 in regulating autophagy in response to oxidative stress and nutrient requirements, and have elucidated complex mechanisms by which SIRT1 regulates epigenetic reprogramming of stem cells.
Summary
SIRT1 inhibition holds promise as a novel approach for ablation of leukemia stem cells in chronic phase CML or FLT3-ITD associated AML. Additional studies to understand the role of SIRT1 in linking metabolic alterations to genomic stability, autophagy and epigenetic reprogramming of stem cells are warranted.
Keywords: Sirtuins, drug resistance, metabolism, chromatin modification, autophagy
Introduction
Silent information regulator-2 (Sir-2) proteins, or sirtuins, are a highly conserved protein family of NAD-dependent HDACs (class III HDACs, SIRT1-7) that promote longevity and are conserved from lower organisms to mammalian cells.(1*) Mammalian sirtuins are recognized as critical regulators of cellular stress resistance, energy metabolism, and tumorigenesis. There are seven mammalian sirtuins that exhibit distinct expression patterns, catalytic activities, and biological functions. SIRT1 shares the highest homology with yeast Sir2 and is the most extensively studied of the sirtuins. In addition to its roles in gene silencing and heterochromatin formation, related to histone H4K16 and H1K26 deacetylation, SIRT1 also deacetylates several non-histone proteins to regulate a variety of biological processes including cell growth, apoptosis, and adaptation to calorie restriction, metabolism and cell senescence.(2) Interestingly both tumor suppressors and oncogenes can be modulated by SIRT1 deacetylation, and SIRT1 can function as a tumor suppressor or oncogene depending on the specific cancer type.(3)
Previous studies have indicated a potential role for SIRT1 in embryonic hematopoiesis, in adult hematopoiesis under hypoxia, and in regulation of leukemic hematopoiesis through regulation of p53 activity.(4, 5) The current review summarizes recent studies that enhance our understanding the role of SIRT1 in regulation of normal hematopoietic stem cells (HSC) under conditions of stress, in maintenance and drug resistance of leukemia stem cells (LSC), and in regulating autophagy and epigenetic reprogramming in response to metabolic alterations.
The role of SIRT1 in regulation of normal HSC
Hematopoietic stem cells (HSC) are characterized by capacity for both extensive self-renewal as well as generation of hematopoietic cells of different lineages. Several studies have evaluated the role of SIRT1 in normal hematopoietic stem cell regulation. SIRT1 inhibition by RNA interference (RNAi) or a pharmacological inhibitor had only a minor impact on normal human CD34+ hematopoietic cells or CD34+ CD38− primitive progenitors.(4) SIRT1 knockout mouse models have been established, and although significant embryonic or perinatal mortality is seen, a fraction of mice survive to adulthood. Previous studies showed that SIRT1 regulates apoptosis expression in mouse embryonic stem cells (ESC) by controlling p53 subcellular localization and that SIRT1−/− ESCs formed fewer mature blast cell colonies, and SIRT1−/− yolk sacs manifested fewer primitive erythroid precursors. (5, 6) These results support an important role for SIRT1 during embryonic hematopoietic development. Adult SIRT1−/− mice demonstrated decreased numbers of bone marrow hematopoietic progenitors. Hematopoietic defects were more apparent under hypoxic rather than normoxic condition. Matsui et al. observed that SIRT1 was widely expressed in human and murine hematopoietic cells of all lineages and stages of maturation.(7) HSC from SIRT1−/− mice showed enhanced differentiation and loss of stem cell characteristics, suggesting that SIRT1 suppresses HSC differentiation and contributes to the maintenance of the HSC pool. HSC maintenance was related to ROS elimination, FOXO activation, and p53 inhibition. On the other hand, Leko et al reported that SIRT1 exon 4 deleted C57BL/6 mice, which exhibit all of the stigmata of SIRT1 deletion, did not exhibit any phenotypic or functional abnormalities in their HSC compartment. (8) HSC from young SIRT1-deficient mice were capable of stable long-term reconstitution in competitive repopulation and serial transplantation experiments, arguing against an essential role for SIRT1 in HSC maintenance in adult mice, at least in steady state.
A conditional deletion approach has recently been used to further evaluate the role of SIRT1 in HSC homeostasis. Rimmele et al. using a tamoxifen-inducible SIRT1 knockout mouse model showed that SIRT1 deletion was associated with anemia, expansion of myeloid cells and depletion of lymphoid cells.(9**) These phenotypic changes were combined with DNA damage accumulation and gene expression changes associated with ageing, suggesting that SIRT1-deleted HSCs demonstrated features associated with ageing. Their studies suggested an important role for transcription factor FOXO3 in mediating the homeostatic effects of SIRT1 in HSCs. Singh et al. showed that SIRT1 deletion in the tamoxifen-inducible model resulted in increased cycling and aberrant expansion of hematopoietic stem/progenitor cells (HSPCs) in vivo. (10**) On the other hand constitutive SIRT1 ablation using Vav-Cre specific to hematopoietic cells resulted with progressive loss of HSC but only in the context of hematopoietic stress. SIRT1 loss was associated with the accumulation of DNA damage and genomic aberrations. SIRT1 physically associated with and negatively regulated the Hoxa9 gene, a key HSC regulator. Together, these findings indicate that SIRT1 contributes to HSC maintenance under conditions of stress.
SIRT1 dysregulation in leukemia stem cells
Several leukemias including chronic myelogenous leukemia (CML) and acute myeloid leukemia (AML) are propagated by small populations of leukemia stem cells (LSC) that resist elimination by conventional and targeted therapies. It was previously shown that SIRT1 protein and RNA levels are significantly increased in CML compared with normal HSPC.(4) Li et al. showed that the majority of AML CD34+CD38− cells showed increased SIRT1 expression compared to normal counterparts from healthy donors.(11**) Unlike CML, AML is a heterogeneous disease, with a number of distinct genetic abnormalities described. SIRT1 expression was higher in cells from patients with poor and intermediate compared with better risk genetic lesions. An internal tandem duplication (ITD) in the Fms-Like Tyrosine kinase (FLT3) mutation is the most commonly observed mutation in AML, seen in 25–30% of AML patients.(12) SIRT1 protein but not mRNA expression is increased in CD34+ cells from FLT3-ITD positive AML patients compared to FLT3 wild-type AML patients. Enhanced SIRT1 protein expression was seen after enforced expression of FLT3-ITD in normal human CD34+ cells compared to FLT3 wild type transduced cells. Sasca et al. suggested that SIRT1 modulation by FLT3-ITD was mediated by the ATM-DBC1-SIRT1 axis.(13**) However, recent studies suggest that DBC1 may function independent of SIRT1 by directly binding and stabilizing p53.(14) Li et al. showed that SIRT1 overexpression in FLT3-ITD positive AML cells resulted from reduced protein degradation, related to a novel c-MYC associated post-transcriptional regulatory network.(11**) Significant enrichment of c-MYC-related genes was seen in FLT3-ITD compared to FLT3-WT AML blasts. Overexpression of c-MYC resulted in SIRT1 deubiquitination, whereas c-MYC knockdown led to decrease in SIRT1 protein stability and expression. USP22 deubiquitinase was overexpressed in FLT3-ITD positive AML CD34+ cells. USP22 expression was regulated by c-MYC and contributed to c-MYC mediated reduction in SIRT1 polyubiquitination and degradation. USP22 directly interacted with and removing polyubiquitin chains from SIRT1 to increase SIRT1 protein stabilization and expression. These results support a role for USP22 in MYC-mediated increase in SIRT1 protein stabilization, and indicate that FLT3-ITD, c-MYC and USP22 form an oncogenic network that enhances SIRT1 expression and activity in leukemic cells.
Role of SIRT1 mediated regulation of p53 in leukemia stem cell maintenance
The p53 protein plays a critical role in the cellular response to stress, by inducing cell cycle arrest or apoptosis. Loss of p53 function is a common feature of human malignancies. p53 inactivation is also required for maintenance of established malignancies, in the setting of continued oncogenic stress. In addition to mutations, p53 transcriptional activity can be modulated via post-translational modifications. Acetylation of p53 modulates protein stability and transcriptional activity independent of phosphorylation status. (15) Deacetylation of p53 at lysine sites by SIRT1 negatively regulates p53 transcriptional activity. (2)
CML results from BCR-ABL oncogene transformed normal HSC. Although BCR-ABL tyrosine kinase inhibitors (TKI) are effective in inducing remission in CML, they do not eliminate CML LSC even in responsive patients, and leukemia usually recurs when treatment is stopped.(16) It was previously shown that pharmacological inhibition of SIRT1, or RNAi of SIRT1 in CML CD34+ cells resulted in increased p53 acetylation and transcriptional activation, and increased CML CD34+ cells apoptosis and reduced CML CD34+ cells growth in vitro and in vivo.(4, 17) SIRT1 inhibition further enhanced targeting of CML CD34+ cells in combination with the BCR-ABL TKI. These inhibitory effects of SIRT1 targeting on CML cells were dependent on p53 expression and acetylation.
Similar to CML, AML is hierarchically organized with small populations of self-renewing LSC that generate the bulk of leukemic cells. Small molecule FLT3 TKIs inhibit growth but do not effectively induce apoptosis of human FLT3-ITD positive AML LSC. In contrast to CML, TKI demonstrate limited clinical activity in FLT3-ITD positive AML patients. (12) Li et al reported that SIRT1 overexpression in FLT3-ITD AML LSC is associated with down-modulation of p53 activity.(11**) AML cells bearing the FLT3-ITD mutation demonstrated increased sensitivity to SIRT1 inhibition.(11**, 13**) SIRT1 inhibition using RNAi or the small molecule inhibitor Tenovin-6 (TV-6) enhanced acetylated p53 levels and p53 target gene expression. The inhibitory effects of TV-6 on FLT3-ITD positive CD34+ cells are to a greater extent than FLT3-WT AML and normal CD34+ cells. The combination of SIRT1 inhibition and FLT3 TKI treatment resulted in significantly increased inhibition of survival and in vivo growth of FLT3-ITD positive AML CD34+ cells compared to either drug alone. These results indicate a role for SIRT1 mediated inhibition of p53 activity in maintenance of AML LSC in the setting of FLT3-ITD induced oncogenic stress. (11**) Interestingly, SIRT1 activation was not a feature of AML cells with FLT3-tyrosine kinase domain mutation (TKD) mutations, which have different signaling and transformative properties than FLT3-ITD mutations. Sasca et al. also showed that leukemic blasts in murine MLL-AF9 or AML1-ETO leukemia models coexpressing FLT3-ITD become dependent on SIRT1 activity to counteract oncogene-induced stress. (13**) Pharmacologic or RNAi-mediated SIRT1 inhibition reduced cell growth and sensitized AML cells to TKI treatment and chemotherapy through restoration of p53 activity. These results further support targeting of SIRT1 as a therapeutic strategy in defined subsets of patients with AML, like FLT3-ITD positive patients.
In addition to p53, SIRT1 can also deacetylate several other proteins that regulate cell growth and survival. The role of additional SIRT1 targets in LSC transformation and drug resistance requires further investigation. SIRT1 appears to be an attractive molecular target against FLT3-ITD positive AML LSC, small molecule inhibitors of SIRT1 are being investigated as potential anti-cancer treatments. (18) Further studies to determine the spectrum and nature of oncogenic stimuli that induce SIRT1 activation and sensitivity to SIRT1 inhibition will be helpful to identify other malignancies that may benefit from SIRT1 inhibitors treatment.
The role of SIRT1 in mutagenesis
SIRT1 may also play a role in leukemia evolution and drug resistance by promoting genetic instability. Wang et al. showed that inhibition of SIRT1 by small molecule inhibitors or gene knockdown blocked acquisition of BCR-ABL secondary mutations in CML cells treated with TKI.(19) SIRT1 knockdown also suppressed de novo mutations of the hypoxanthine phosphoribosyl transferase gene in CML and non-CML cells following camptothecin treatment. Acquisition of genetic mutations was associated with SIRT1 stimulation of error-prone DNA damage repair activity. The same group subsequently showed that ATRA induced expression of CD38, a cell surface marker and cellular NADase, inhibited the activity of SIRT1 by reducing intracellular NAD+ levels.(20*) SIRT1 inhibition following ATRA treatment decreased DNA damage repair and suppressed acquisition of BCR-ABL secondary mutations. This study suggested a potential benefit of combining ATRA with TKIs in treating CML, particularly in advanced phases.
Role of SIRT1 in regulating autophagy in stem cells
Previous studies have shown that SIRT1 plays a critical role in ROS-triggered apoptosis in mouse ES cells, and in modulating the PTEN/JNK/FOXO1 response to cellular ROS. Ou et al. evaluated connections between SIRT1 activity and induction of autophagy upon ROS challenge in murine and human ESCs. (21*) SIRT1−/− ESC demonstrated increased apoptosis with decreased induction of autophagy in response to oxidative stress. Knockout of SIRT1 also decreased ROS-induced autophagy in human ESC. SIRT1 effects were mediated at least in part by PI3K/Beclin 1 and mTOR pathways. Huang et al. showed that LC3, a key initiator of autophagy, is selectively activated in the nucleus during starvation through deacetylation by SIRT1.(22*) Deacetylation of LC3 allows it to return to the cytoplasm, binds Atg7 and other autophagy factors, and promote autophagy to help cope with the lack of external nutrients. Tang et al. found that SIRT1 regulates autophagic flux in muscle stem cell progeny. (23*) Autophagy was induced during muscle stem cell activation. SIRT1 deficiency leads to delayed muscle stem cell activation that was partially rescued by exogenous pyruvate, supporting the importance for SIRT1 regulation of autophagy in meeting bioenergetic demands during muscle stem cell activation.
Role of SIRT1 in epigenetic reprogramming of stem cells
SIRT1 deacetylase activity is known to contribute to gene silencing and heterochromatin formation via histone H4K16 and H1K26 deacetylation. Recent studies reveal additional mechanisms and roles for SIRT1 in chromatin regulation in stem cells. The mixed-lineage leukemia 1 (MLL1) gene is responsible for H3K4 trimethylation at circadian promoters. Aguiar-Amal et al showed that SIRT1 controlled MLL1 mediated H3K4 trimethylation through circadian deacetylation of the protein.(24*) MLL1 is deacetylated by SIRT1 at K1130 and K1133, leading to modulation of its methyltransferase activity. MLL-mediated H3K4 methylation at clock-controlled-gene promoters is affected by inactivation of SIRT1, which also depends on circadian levels of NAD+. These findings indicate that SIRT1 in addition to effects on histone acetylation could also impact alterations in histone methylation in response to energy metabolism.
Mishra et al. found that deletion of MLL1 selectively depletes histone H4K16 acetylation at MLL1 target genes, together with reduced gene transcription. (25*) Endogenous MLL1 histone methyltransferase activity was not required for HSC gene expression and MLL-AF9 transformation. Inhibition of SIRT1 prevents loss of H4K16 acetylation and the reduction in MLL1 target gene expression. This result indicated MLL1-recruited H4K16Ac activity as the major mechanism by which gene expression is maintained in hematopoietic stem/progenitors, and SIRT1 as an antagonist for MLL1 dependent gene activation.
The studies of Ryall et al. showed a role for SIRT1 in integrating muscle stem cell metabolism with epigenetic changes required for myogenic commitment. (26*) During transition from quiescence to proliferation, skeletal muscle stem cells undergo a metabolic switch from fatty acid oxidation to glycolysis. This metabolic reprogramming leads to decrease in NAD+ levels and SIRT1 activity, resulting in elevated H4K16 acetylation and activation of muscle gene transcription. These observations support a role for SIRT1 in translating metabolic cues into epigenetic modifications that regulate stem cell fate. A role for SIRT1 in linking metabolism to hematopoietic stem cell fate has not as yet been described.
Conclusions
Current understanding of SIRT1 in normal hematopoiesis indicates an important role in maintaining HSC homeostasis under the hematopoietic stress (Figure 1). There is also an increasing appreciation of the role of SIRT1 in regulating stem cell fate determination, and in coordinating cellular responses to metabolic cues. Previous studies demonstrating an important role for SIRT1 overexpression in maintaining CML LSC have been extended to AML, and have revealed aberrant activation of SIRT1 in LSC from AML patients with the common FLT3-ITD mutation, through post-transcriptional mechanism mediated by a MYC-associated oncogenic network (Figure 1). SIRT1 activation promotes maintenance and TKI resistance of FLT3-ITD AML LSC These observations support further investigation of SIRT1 inhibition as a strategy to target CML and FLT3-ITD leukemia. Further evaluation of the spectrum and nature of oncogenic stimuli that induce SIRT1 activation and sensitivity to SIRT1 inhibition and identification of other malignancies that may benefit from SIRT1 inhibitor treatment are warranted.
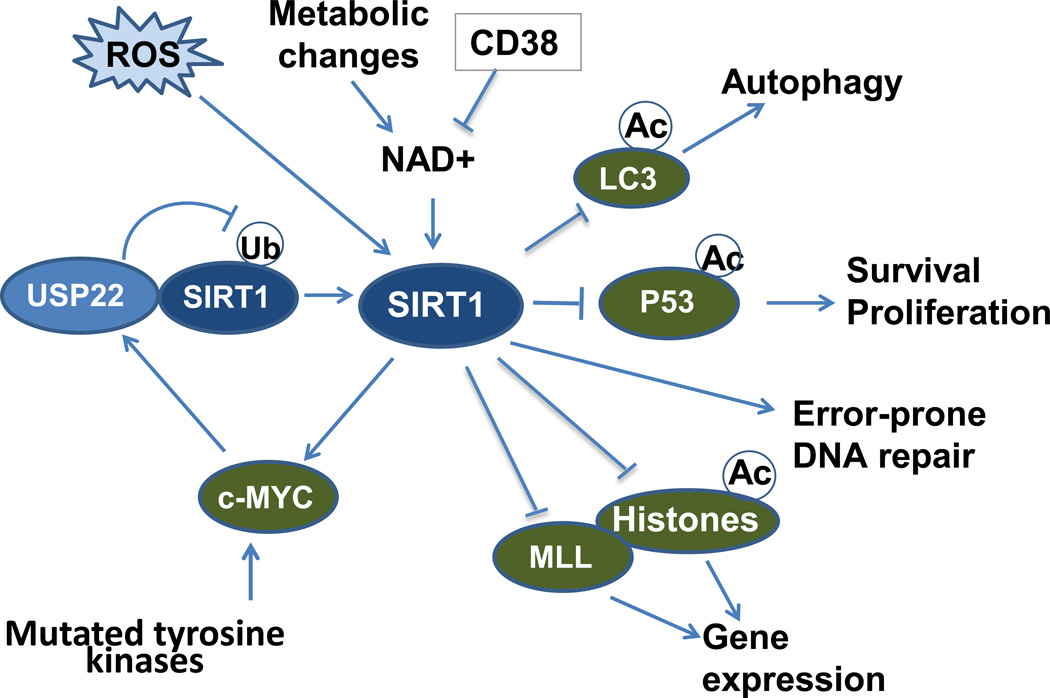
Figure 1. Regulation and downstream targets of SIRT1 in stem cells.
SIRT1 plays an important role in maintaining HSC homeostasis under conditions of stress. SIRT1 activity is regulated by ROS. Nutritional deprivation, metabolic alterations or CD38 activity can alter levels of NAD+ required for SIRT1 activity. SIRT1 can regulate stem cell fate determination by direct effects on histone acetylation and indirectly by altering other chromatin modulators such as MLL1. SIRT1 also regulates cellular function by deacetylating non-histone proteins affecting cell survival and proliferation (p53), autophagy (LC3), and DNA repair. In leukemia stem cells SIRT1 can be upregulated by mutant tyrosine kinases through activation of c-MYC leading to increased expression of the USP22 deubiquitinase with reduces SIRT1 degradation. SIRT1 activation also stabilizes c-MYC leading to a feed-forward loop enhancing SIRT1 expression. Increased SIRT1 expression and activity results in deacetylation and deactivation of p53, contributing to leukemia evolution, maintenance and drug resistance.
Key points.
Sirtuins are a highly conserved family of NAD-dependent HDACs that are critical regulators of cellular stress resistance, metabolism, and tumorigenesis.
SIRT1 is required normal murine HSC maintenance under conditions of hematopoietic stress.
There is increasing appreciation of the role of SIRT1 in regulating stem cell fate determination, and in coordinating cellular responses to metabolic cues.
SIRT1 is aberrantly activated in FLT3-ITD+ AML LSC through a MYC-associated oncogenic network
SIRT1 activation promotes maintenance and TKI resistance of CML and FLT3-ITD AML LSC supporting further investigation of SIRT1 inhibition as a therapeutic strategy in these leukemias.
Acknowledgements
Financial support and sponsorship
This work was supported by NIH NCI grants R01 CA95684 and the Leukemia and Lymphoma Society, and NIH NCI grant K99CA184411
Footnotes
Conflicts of interest
None
References
- 1. Anderson KA, Green MF, Huynh FK, et al. SnapShot: Mammalian Sirtuins. Cell. 2014;159(4):956- e1. doi: 10.1016/j.cell.2014.10.045.. A useful, brief, visual overview of mammalian sirtuins.
- 2.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nature reviews Cancer. 2009;9(2):123–128. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer research. 2009;69(5):1702–1705. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Wang L, Li L, et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer cell. 2012;21(2):266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ou X, Chae HD, Wang RH, et al. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood. 2011;117(2):440–450. doi: 10.1182/blood-2010-03-273011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han MK, Song EK, Guo Y, et al. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell stem cell. 2008;2(3):241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsui K, Ezoe S, Oritani K, et al. NAD-dependent histone deacetylase, SIRT1, plays essential roles in the maintenance of hematopoietic stem cells. Biochemical and biophysical research communications. 2012;418(4):811–817. doi: 10.1016/j.bbrc.2012.01.109. [DOI] [PubMed] [Google Scholar]
- 8.Leko V, Varnum-Finney B, Li H, et al. SIRT1 is dispensable for function of hematopoietic stem cells in adult mice. Blood. 2012;119(8):1856–1860. doi: 10.1182/blood-2011-09-377077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimmele P, Bigarella CL, Liang R, et al. Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem cell reports. 2014;3(1):44–59. doi: 10.1016/j.stemcr.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Singh SK, Williams CA, Klarmann K, et al. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. The Journal of experimental medicine. 2013;210(5):987–1001. doi: 10.1084/jem.20121608.. These two papers describe results of studies with conditional knockout mouse models showing an important role for SIRT1 in protection of adult HSC from stress
- 11. Li L, Osdal T, Ho Y, et al. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute Myeloid Leukemia stem cells. Cell stem cell. 2014;15(4):431–446. doi: 10.1016/j.stem.2014.08.001.. This paper describes the role of a MYC-associated oncogenic network in SIRT1 activation in FLT3-ITD AML stem cells, and the importance of SIRT1 in AML LSC maintenance and drug resistance by p53 suppression.
- 12.Kindler T, Lipka DB, Fischer T. FLT3 as a therapeutic target in AML: still challenging after all these years. Blood. 2010;116(24):5089–5102. doi: 10.1182/blood-2010-04-261867. [DOI] [PubMed] [Google Scholar]
- 13. Sasca D, Hahnel PS, Szybinski J, et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood. 2014;124(1):121–133. doi: 10.1182/blood-2013-11-538819.. As with 11 above, this paper also describes that SIRT1 is activated in FLT3-ITD AML stem cells, and plays an important role in AML LSC maintenance by p53 suppression.
- 14.Qin B, Minter-Dykhouse K, Yu J, et al. DBC1 Functions as a Tumor Suppressor by Regulating p53 Stability. Cell reports. 2015;10(8):1324–1334. doi: 10.1016/j.celrep.2015.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Y, Zhao W, Chen Y, et al. Acetylation is indispensable for p53 activation. Cell. 2008;133(4):612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross DM, Branford S, Seymour JF, et al. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER study. Blood. 2013;122(4):515–522. doi: 10.1182/blood-2013-02-483750. [DOI] [PubMed] [Google Scholar]
- 17.Yuan H, Wang Z, Li L, et al. Activation of stress response gene SIRT1 by BCR-ABL promotes leukemogenesis. Blood. 2012;119(8):1904–1914. doi: 10.1182/blood-2011-06-361691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kokkonen P, Mellini P, Nyrhila O, et al. Quantitative insights for the design of substrate-based SIRT1 inhibitors. European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences. 2014;59:12–19. doi: 10.1016/j.ejps.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Yuan H, Roth M, et al. SIRT1 deacetylase promotes acquisition of genetic mutations for drug resistance in CML cells. Oncogene. 2013;32(5):589–598. doi: 10.1038/onc.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Z, Liu Z, Wu X, et al. ATRA-induced cellular differentiation and CD38 expression inhibits acquisition of BCR-ABL mutations for CML acquired resistance. PLoS genetics. 2014;10(6):e1004414. doi: 10.1371/journal.pgen.1004414.. This study describes how CD38 NADase activity can modulate SIRt1 activity.
- 21.Ou X, Lee MR, Huang X, et al. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem cells. 2014;32(5):1183–1194. doi: 10.1002/stem.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang R, Xu Y, Wan W, et al. Deacetylation of Nuclear LC3 Drives Autophagy Initiation under Starvation. Molecular cell. 2015;57(3):456–466. doi: 10.1016/j.molcel.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 23. Tang AH, Rando TA. Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. The EMBO journal. 2014;33(23):2782–2797. doi: 10.15252/embj.201488278.. These three papers describe the importance of SIRT1 in regulating autophagy in stem cells under conditions of oxidative stress, starvation and stem cell activaation.
- 24.Aguilar-Arnal L, Katada S, Orozco-Solis R, Sassone-Corsi P. NAD-SIRT1 control of H3K4 trimethylation through circadian deacetylation of MLL1. Nature structural & molecular biology. 2015 doi: 10.1038/nsmb.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishra BP, Zaffuto KM, Artinger EL, et al. The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell reports. 2014;7(4):1239–1247. doi: 10.1016/j.celrep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ryall JG, Dell'Orso S, Derfoul A, et al. The NAD(+)-Dependent SIRT1 Deacetylase Translates a Metabolic Switch into Regulatory Epigenetics in Skeletal Muscle Stem Cells. Cell stem cell. 2015;16(2):171–183. doi: 10.1016/j.stem.2014.12.004.. These three articles provide new insights into direct and indirect mechanisms by which SIRT1 responds to extrinsic signals to regulate chromatin modifications affecting stem cell state