Abstract
Background
Cancer stemness, observed in several types of glioma stem cells (GSCs), has been demonstrated to be an important barrier for efficient cancer therapy. We have previously reported that cancerous neural stem cells (F3.Ras.CNSCs), derived from immortalized human neural stem cells by a single oncogenic stimulation, form glial tumors in vivo.
Method
We searched for a commonly expressed stress modulator in both F3.Ras.CNSCs and GSCs and identified silent mating type information regulation 2, homolog (SIRT1) as a key factor in maintaining cancer stemness.
Result
We demonstrate that the expression of SIRT1, expressed in “cancer cells with neural stemness,” is critical not only for the maintenance of stem cells, but also for oncogenic transformation. Interestingly, SIRT1 is essential for the survival and tumorigenicity of F3.Ras.CNSCs and GSCs but not for the U87 glioma cell line.
Conclusion
These results indicate that expression of SIRT1 in cancer cells with neural stemness plays an important role in suppressing p53-dependent tumor surveillance, the abrogation of which may be responsible not only for inducing oncogenic transformation but also for retaining the neural cancer stemness of the cells, suggesting that SIRT1 may be a putative therapeutic target in GSCs.
Keywords: glioma stem cells, human neural stem cell, H-Ras, p53, SIRT1
Uncontrolled proliferation by the mutation of proto-oncogenes confers independence from growth factors and has been considered one of the main causes of tumorigenic transformation.1 However, the aberrant mitogenic stimulation by an oncogene results in a permanent cell cycle arrest, such as cellular senescence,2 or apoptosis due to the protective action of a number of mediators of stress signaling such as p53 and p16Ink4a.3 Induction of cyclin dependent kinase inhibitors or pro-apoptotic factors through a tumor surveillance network, following p53 activation, reactivates retinoblastoma (Rb) to cause cell cycle arrest or induces apoptosis to avoid oncogenic transformation.4,5 Therefore, abrogation of the innate cellular defensive mechanism is critical for tumorigenic transformation.1 Thus, the suppression or mutation of key tumor suppressors or gene amplification of the inhibitory protein to target a number of tumor suppressors frequently occurs in many types of cancers. For example, mutation of TP53 or gene silencing of CDKN2A is frequently observed. Alternatively, gene amplification of wild type p53 induced phosphatase (Wip1), of which the ectopic expression is sufficient to deactivate tumor surveillance networks or B lymphoma Moloney murine leukemia virus insertion region 1 homolog (Bmi-1), suppressing p16Ink4a expression,6 also occurs in many types of cancers.7
Cancers originating from stem/progenitor cells but not from differentiated cells under the same level of oncogenic challenges in animal models are well documented.8,9 In particular, the deletion of key tumor suppressors in stem cells induces tumorigenesis of neural stem cells (NSCs) but does not affect their differentiated counterpart (eg, astrocytes in the brain), implying that stem cells somehow may have higher oncogenic susceptibility than their differentiated counterpart. This result is in agreement with a previous study demonstrating that the combination of 3 oncogenes (H-Ras, human telomerase reverse transcriptase, and Simian virus 40 T/t-antigens) is required for oncogenic transformation of human astrocytes to glioma-like cells,10 whereas only 2 oncogenes (v-myc and H-Ras) are sufficient for oncogenic transformation of human NSCs.11
The role of silent mating type information regulation 2, homolog (SIRT1), a nicotinamide adenine dinucleotide–dependent histone deacetylase in tumorigenesis, is controversial, as SIRT1 regulates both tumor suppressors such as p53 and fork-head class O transcription factor and proto-oncogenes such as β-catenin, survivin, and nuclear factor–kappaB, deacetylation by which affects their function.12 The neurodevelopmental defect found in SIRT1-null mice is consistent with the role of SIRT1 in neurogenesis13 and neural differentiation14 of neural precursors. Of interest, recent studies demonstrated that CD133-positive glioma cells (representing glioma stem cells [GSCs], which are characterized by higher tumorigenic potential and higher drug resistance15) but not CD133-negative glioma cells are more susceptible to apoptosis by depletion of SIRT1, which means that SIRT1 may be critical to the survival of “cancer cells with stemness.”
Previously, we demonstrated that human NSCs immortalized by v-myc (F3.NSCs)16 underwent oncogenic transformation by a single oncogenic challenge with H-Ras, forming heterogeneous glial tumors consisting of a mixture of nestin-positive or glial fibrillary acidic protein (GFAP)–positive cell population.11 In the current studies, we provide evidence that SIRT1 in F3.NSCs is responsible not only for maintenance of the growth potential but also for oncogenic transformation by H-Ras. As a result, SIRT1 is overexpressed in cancerous neural stem cells (CNSCs) and has a critical role in the maintenance of neural stemness in cancer cells with stemness (cancer cells showing stemness properties), including F3.Ras.CNSCs and GSCs isolated from glioma patients,17 rather than in the U87 glioma cell line. Therefore, the loss of SIRT1 in cancer cells with stemness, but not in the U87 glioma cell line, results in cell death in a p53-dependent manner. These results suggest that SIRT1 would be a promising molecular target in cancer cells with neural stemness (cancer cells showing neural stemness properties), including F3.Ras.CNSCs and GSCs.
Materials and Methods
Details of the methods are available in the online supplement.
Cell Culture and Animal Study
F3.Ras.CNSCs, human dermal fibroblasts, and U87 cells were maintained as previously described.11 Nude male mice at 6 weeks of age were subcutaneously injected with 5 × 105 short hairpin (sh) control (shCont)- or shSIRT1- F3.Ras.CNSCs in the thigh muscle, and tumor appearance was monitored after 6 weeks. The experiments with animals were reviewed and approved by the Institutional Animal Care and Use Committee of Chung-Ang University. All procedures were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals published by the US National Institutes of Health (publication 85-23, revised 1996).
Sphere Culture and Differentiation
As described previously,17–19 GSC 528 spheres (528NS) were maintained in a defined medium (Dulbecco's modified Eagle's medium [DMEM]/F12 supplemented with 0.1% B27, 0.1% gentamicin, 20 ng/mL basic fibroblast growth factor, and 20 ng/mL epidermal growth factor [Invitrogen]). Differentiation of 528NS was done with the defined medium (DMEM/F12 supplemented with 10% fetal bovine serum, 0.1% B27, and 0.1% gentamicin) for 12 days.
Lentiviral Gene Transfer
The shRNA SIRT1 plasmid (SHCLNG-NM_012238) was purchased from Sigma. All procedures were according to the manufacturer's instructions (ViraPower Lentiviral Expression Systems, Invitrogen).
Senescence-associated β-Galactosidase Staining
Senescence-associated β-galactosidase (SA-β-gal) activity at pH 6.0 was detected with an SA-β-gal staining kit (#9860, Cell Signaling Technology) according to the manufacturer's protocol.
Apoptosis Assay Using Annexin V and 7-Aminoactinomycin
The apoptotic cell population was analyzed using the Phycoerythrin Annexin V Apoptosis Detection Kit I (BD Biosciences) according to the manufacturer's instructions.
Dox-inducible Ras Plasmid Construct
This plasmid was constructed in the CSIV-TRE-RfA-UbC-KT vector using Gateway Technology with clonase II (Invitrogen) as described previously.20
Cell Proliferation Assay
The above procedure was followed based on the protocol for the Cell Proliferation Kit II (XTT) (Roche #11465015001).
Results
Increased Expression of SIRT1 in F3.Ras.CNSCs
As we have previously demonstrated, a defective p53 response in F3.NSCs is responsible for the tumorigenic transformation by H-Ras even in the presence of activating phosphorylation of p53.11 Thus, we speculated that a posttranslational modification other than phosphorylation might be responsible for the impaired p53 response. Recently, acetylation of p53 has been reported to be critical for transactivating the p53-dependent gene response,21 especially apoptotic gene induction.22 Thereby, we examined the expression level of SIRT1, a well-known deacetylase targeting p53 in F3.NSCs and F3.Ras.CNSCs. As shown in Fig. 1, the mRNA and subsequently the protein levels of SIRT1 were higher in the F3.NSCs compared with primary human NSCs, and even more in F3.Ras.CNSCs (Fig. 1A and B). To examine the SIRT1 induction in CNSCs, we attempted to generate CNSCs with other well-characterized oncogenes in gliomagenesis such as K-Ras and examined the subsequent expression of SIRT1. We found that ectopic expression of K-Ras, but not constitutively active Akt (v-Akt), could transform F3.NSCs into CNSCs, similar to that by F3.Ras, where SIRT1 expression was concurrently increased in F3 expressing H-Ras or K-Ras but not v-Akt (Supplementary Fig. S1A and B).
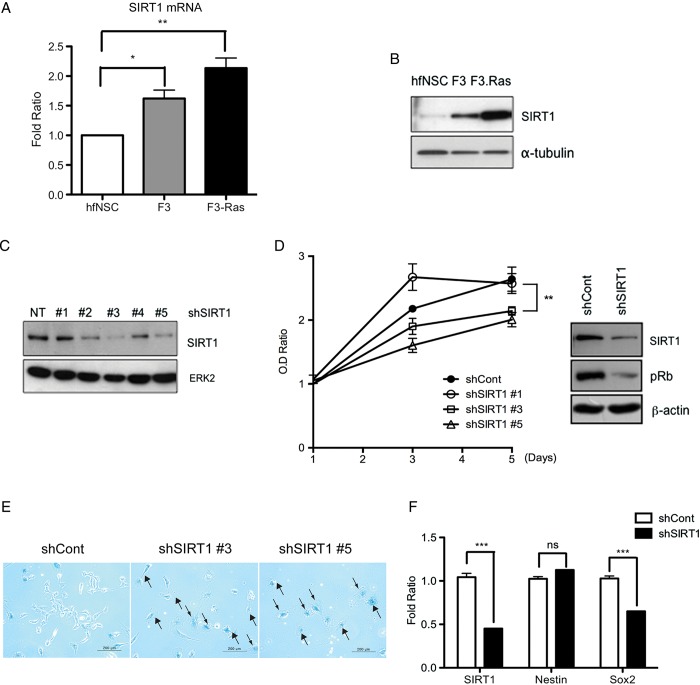
Fig. 1.
SIRT1 expression level was increased in F3.Ras.CNSCs (F3.Ras), and SIRT1 knockdown caused growth retardation in F3.NSCs. (A) The mRNA level of SIRT1 in human fetal NSC (hfNSC), F3.NSCs (F3), and F3.Ras.CNSCs (F3-Ras) was determined via real-time PCR (n = 3). (B) SIRT1 levels were determined via immunoblotting. (C) The lentiviral shRNA SIRT1 infection in F3.NSCs was confirmed by immunoblotting for SIRT1. ERK2, extracellular signal-regulated kinase 2. (D) ShCont (closed circle) and shSIRT1 #1, #3, and #5 (open circle, square, and triangle, respectively)–infected F3.NSCs were plated at 2 × 103 cells. Optical density (OD) ratio was measured using XTT every 2 days (left panel) (n = 3). The phospho-Rb levels were determined via immunoblotting in F3.NSCs infected with shCont and shSIRT1. (E) ShCont and shSIRT #3 or #5–infected F3.NSCs were analyzed by SA-β-gal staining assay (scale bars, 200 m). Black arrow for β-gal–positive senescent cell. (F) The mRNA level of SIRT1, Sox2, and nestin in shCont and shSIRT1 F3.NSCs was determined via real-time PCR (n = 3). Alpha-tubulin, β-actin, or ERK2 was a loading control.
Growth Retardation and Induction of Cellular Senescence Following Loss of SIRT1
Since SIRT1 expression was significantly higher in the F3.NSCs and F3.Ras.CNSCs, we next hypothesized that the induction of SIRT1 in F3.NSCs would be responsible for the rapid oncogenic transformation that occurs following H-Ras oncogenic stimulation, as previously described.11 To test this idea, we first generated SIRT1 knockdown F3.NSCs. Five different shRNA sequences of SIRT1 were introduced and the depletion of SIRT1 was monitored. We found that 2 out of the 5 shRNA sequences (#3 and #5) were most efficient in depleting SIRT1 in the F3.NSCs (Fig. 1C). The F3.NSCs whose SIRT1 expression was mostly depleted were then selected (eg, shRNA #3 and #5; Fig. 1C) and their growth rate was compared with control F3.NSCs (shCont) as well as F3.NSCs that lacked SIRT1 depletion (eg, shRNA #1). The SIRT1-depleted F3.NSCs were significantly growth retarded compared with the shRNA control F3.NSCs (Fig. 1D, left panel). The phosphorylation of Rb, a well-known readout for proliferation, was also significantly reduced following SIRT1 depletion (Fig. 1D, right panel). The apparent growth retardation in SIRT1-depleted F3.NSCs was concurrent with the onset of cellular senescence, as determined by senescence-associated β-galactosidase assay (Fig. 1E, black arrows) using another shRNA sequence of equivalent knockdown efficiency (shRNA #3 and #5; Fig. 1E). We next examined whether the depletion of SIRT1 influenced neural stemness and observed that the expression of human SRY (sex determining region Y)-box 2 (Sox2), a persistent marker for NSCs,23 but not human nestin, was suppressed in the SIRT1-depleted F3.NSCs (Fig. 1F and Supplementary Fig. S1C). These results clearly demonstrate the critical role of SIRT1 in maintaining the cellular growth potential as well as neural stemness in F3.NSCs.
SIRT1 Is Essential for the Oncogenic Transformation of F3.NSCs by H-Ras
Considering that SIRT1 modulates the p53 response and that inhibition of SIRT1 elicits a p53-dependent stress response,24,25 despite a contradictory report,26 we next hypothesized that the loss of SIRT1 may promote resistance of H-Ras–mediated oncogenic stimulation in F3.NSCs. To prove this hypothesis, H-Ras oncogenic stimulation was conducted in the presence of known SIRT1 inhibitors (nicotinamide [NIC] and sirtinol [SRT]). Distinct morphological changes and colony formation, both indicating oncogenic transformation, were markedly decreased in the presence of NIC under H-Ras oncogenic challenge. However, when NIC was removed, colony numbers of F3.NSCs were comparable to the control (Fig. 2A, right panels). The robust reduction in oncogenic colony formation by H-Ras was reproduced in the presence of SRT in a similar manner (Supplementary Fig. S1D). The F3.NSCs, stimulated by H-Ras in the presence or absence of NIC, were further grown in soft agar to determine the rate of oncogenic transformation. Consistently, the NIC-treated F3.NSCs exhibited a significantly lower number of colonies on soft agar compared with the control cells under H-Ras oncogenic challenge (Fig. 2B). The defective oncogenic transformation by SIRT1 inhibitor treatment was not a result of lower oncogenic stimulation by H-Ras, because the expression level of H-Ras, and the subsequent activation of extracellular signal-regulated kinase 1/2, was equivalent to that of mock F3.NSCs with H-Ras expression. However, apoptosis determined by the formation of active caspase-3 was moderately higher in the presence of NIC following oncogenic stimulation (Fig. 2C). In order to exclude any unexpected side effects of NIC or SRT, we depleted SIRT1 from F3.NSCs and then challenged them to oncogenic stimulation. Similar to the effect of NIC or SRT, F3.NSCs devoid of SIRT1 failed to undergo the tumorigenic morphological changes induced by H-Ras (Fig. 2D and Supplementary Fig. S1E) and also presented with reduced colony formation on soft agar (Fig. 2E). Next, we generated an inducible H-Ras F3 cell line, markedly showing induction of H-Ras by doxycycline (Dox) treatment (Supplementary Fig. S2A). Consistent with the stable expression of H-Ras, a continuous exposure of Dox for 4 days was sufficient to achieve not only tumorigenic morphological changes, but also soft-agar colonies in F3.NSCs (Supplementary Fig. S2B). Consistent with the observation of the stable expression of H-Ras in F3.NSCs, the inhibition of SIRT1 enzymatic activity by SRT or EX527, a more potent and selective SIRT1 chemical inhibitor,27 markedly lowered the number of cells undergoing tumorigenic morphological changes in the inducible F3.Ras cell line (dotted line, Supplementary Fig. S2C and D). The impaired tumorigenic transformation was concurrent with the induction of apoptosis and a marked decrease of H-Ras (Fig. 2F and Supplementary Fig. S2E), suggesting that apoptotic induction with p53 stabilization in the absence of SIRT1 may be associated with the elimination of H-Ras–expressing F3.NSCs that are responsible for tumorigenic transformation (Fig. 2F). As the initial tumorigenic transformation occurs at least 3 days after H-Ras induction (Supplementary Fig. S2C), we next collected F3.NSCs exposed to Dox for 3 days to induce tumorigenic transformation with the indicated treatments; the cells were then applied to an in vitro transformation (soft-agar) assay. As predicted, SRT treatment or SIRT1 depletion significantly inhibited anchorage-independent growth (Fig. 2G and H).
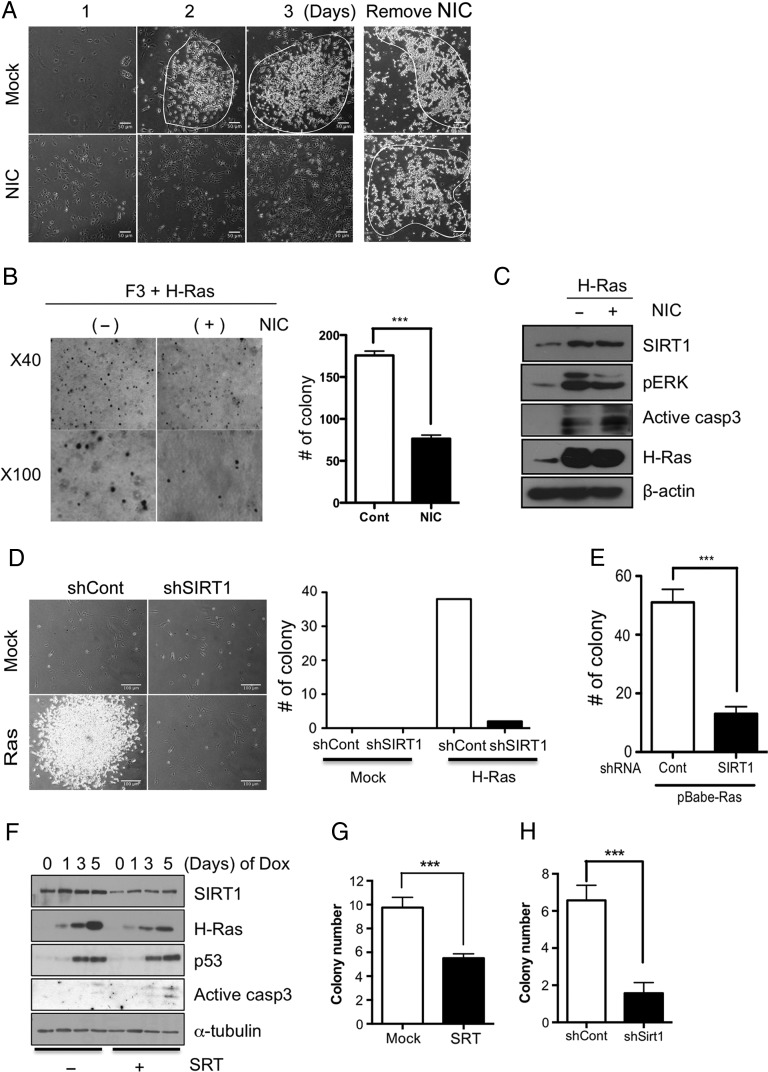
Fig. 2.
Knockdown of SIRT1 failed to transform cells following H-Ras infection. (A) The morphological image of Mock- or NIC (10 mM)-treated F3.NSCs after H-Ras infection for 3 days (scale bars, 50 m). (B) Colony formation by each indicated cell in soft agar (left panel) was determined by the number of colonies and then graphically presented (right panel) (n = 6). (C) The level of SIRT1, phosphorylated extracellular signal-regulated kinase (pERK), active caspase-3, H-Ras for F3, H-Ras–infected Mock- or NIC-treated F3 was determined by immunoblotting. (D) Morphological image of shCont- or shSIRT1-infected F3.NSCs after H-Ras infection (left panel) (scale bars, 100 m) and the number of transformed cells was graphically presented (right panel). (E) Colony formation by each indicated cell in soft agar was determined by the number of colonies and then graphically presented. (F) Dox-inducible F3.NSCs were pretreated with the SIRT1 inhibitor SRT (50 μM) for 1 h and then the cells were harvested at the indicated time after Dox treatment. The expression of SIRT1, H-Ras, p53, and active caspase-3 was determined by immunoblotting. (G) Mock- or SRT- (H) shCont- or shSIRT1-treated Ras-inducible F3.NSCs were treated with Dox every other day for 21 days. Colony formation of each indicated cell in soft agar was determined by the number of colonies and then graphically presented (n = 4). Alpha-tubulin or β-actin was loading control.
SIRT1 for Tumorigenic Potential in F3.Ras.CNSCs
As SIRT1 expression was obviously critical for the oncogenic transformation of F3.NSCs (Fig. 2), we next examined whether SIRT1, whose expression was further upregulated during the oncogenic transformation (Fig. 1), is important for the tumorigenic properties of F3.Ras.CNSCs. While SIRT1 depletion caused significant growth retardation in F3.NSCs (Fig. 1D), SIRT1 depletion had a higher impact on proliferation in F3.Ras.CNSCs (Fig. 3A). Unlike the F3.NSCs that underwent growth retardation and cellular senescence following SIRT1 depletion (Fig. 1D and E), the loss of SIRT1 triggers apoptosis in the F3.Ras.CNSCs (Fig. 3B). Apoptosis occurred only in the F3.Ras.CNSCs but not in human dermal fibroblasts in which SIRT1 expression appeared to be relatively lower than F3.Ras.CNSCs (Fig. 3C, inserted panel). On the basis of this result, we speculated that oncogenic stress by H-Ras would be responsible for distinct growth arrest as well as apoptosis in SIRT1-depleted F3.Ras.CNSCs. Considering the dramatic growth retardation and apoptosis (Fig. 3A–C), we surmised that the F3.Ras.CNSCs, in the absence of SIRT1, might fail to form heterogeneous primitive neuroectodermal–like tumor.11 As expected, none of the 3 mice injected with the SIRT1-depleted F3.Ras.CNSCs formed tumors, whereas all 3 mice that were injected with F3.Ras.CNSCs formed a tumor mass (Fig. 3D). Following detailed analysis, we discovered that the tumor generated by the F3.Ras.CNSCs was consistently heterogeneous, consisting of an hNestin- and GFAP-positive and a microtubule-associated protein 2–negative population, with an active proliferating potential as determined by Ki-67–positive staining (Fig. 3E). Thus, SIRT1 induction during tumorigenesis (Fig. 1) may be strongly associated with the tumorigenic potential in F3.Ras.CNSCs. It is noteworthy that higher SIRT1 expression was observed in the CD133-positive cancer stem cells isolated from gliomas.28
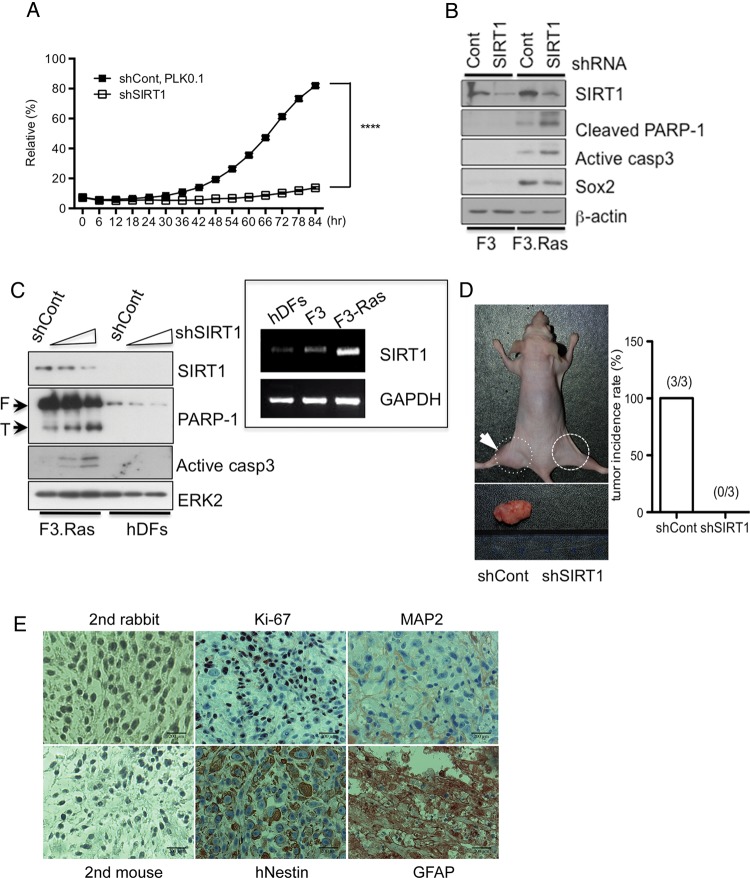
Fig. 3.
Knockdown of SIRT1 failed to form tumors in F3.Ras.CNSCs. (A) Cell proliferation in shCont- or shSIRT1-infected F3.Ras.CNSCs cells was measured every 6 h using Incucyte (ESSEN Bioscience) (n = 3). (B) The level of SIRT1, cleaved poly(ADP-ribose) polymerase 1 (PARP-1), active caspase-3, and Sox2 was determined by immunoblotting in shCont- or shSIRT1-infected F3.NSCs and F3.Ras.CNSCs. (C) The level of SIRT1, PARP-1, and active caspase-3 was determined by immunoblotting in shCont - or shSIRT1-gradually infected F3.Ras.CNSCs and human dermal fibroblasts (hDFs). The mRNA level of SIRT1 in hDFs, F3, and F3.Ras.CNSCs was determined by real-time PCR. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was loading control. ERK2, extracellular signal-regulated kinase 2. (D) Male nude mice (6 wk old) were injected intramuscularly with 5 × 105 cells of shCont (left thigh: dotted line) or shSIRT1 (right thigh: solid line) F3.Ras.CNSCs, and tumor appearance was assessed after 6 weeks. A total of 3 mice were isolated and processed for histological analysis (right panel). (E) Tumor tissues were immunostained with a proliferation marker (Ki-67), NSC marker (hNestin), astrocyte marker (GFAP), and neuron marker (microtubule-associated protein 2 [MAP2]). Each tissue was counterstained with hematoxylin for nuclear staining. Immunohistochemical images without the primary antibody (mouse or rabbit) were used as negative control (scale bars, 200 m).
SIRT1 for Survival in Cancer Cells With Neural Stemness
Based on the results that SIRT1 expression in F3.NSCs as well as in F3.Ras.CNSCs, both of which retain their “stemness,” may play an important role in maintaining both survival and tumorigenic potential, we speculated that SIRT1 may play a key role in cancer cells with neural stemness. To test this hypothesis, we initially compared gene expression profiles such as for SIRT1, Sox2, GFAP, CD133, and nestin among the 2 types of glioma cells isolated from the glioma patients, 1 human astrocyte, 4 glioma cell lines (U138, T98G, A172, and U87MG), 3 types of cells that originated from human fetal NSCs (hNSCs, F3.NSCs, and F3.Ras.CNSCs), and 3 GSCs derived from glioma patients (83NS, 528NS, 1123NS) (Supplementary Fig. S3A)17,18,29 and applied their relative mRNA expression levels to a 2D scatter plot. Of note, each group (glioma = glioma cell lines and glioma cells isolated from the patients; GSCs = 83NS, 528NS, 1123NS; astrocyte; and NSCs = fetal NSCs, F3.NSCs, and F3.Ras.CNSCs) appeared to be separated when plotting the expression level of Sox2 and SIRT1. Cells derived from NSCs (closed circle) appeared to be closely located to GSCs (open circle), whereas gliomas were placed separately (Fig. 4A), which is in parallel with previous reports stating that high SIRT1 expression was observed in CD133-positive GSCs.28 Thus, it could be readily surmised that the inhibition of SIRT1 would have varied effects on cancer cells with neural stemness (eg, F3.Ras.CNSCs, GSCs) and glioma cells. To test this belief, we compared cellular apoptosis in the absence of SIRT1 between F3.Ras.CNSCs and the U87 glioma cell line (open square in Fig. 4A). Inhibition of the deacetylase activity of SIRT1 by SRT was sufficient to induce apoptosis only in F3.Ras.CNSCs but not in U87 cells, which express very low levels of SIRT1 compared with F3.Ras.CNSCs (Fig. 4B and C). Consistently, the dose-dependent depletion of SIRT1 by the gradual increase of shRNA also validated the specific apoptotic event in F3.Ras.CNSCs but not in U87 (Fig. 4D). These results clearly demonstrated that SIRT1 might be a putative molecular target of cancer cells with neural stemness. This hypothesis may be consistent with the previous finding that SIRT1 expression, which is induced by genes specific to neural precursor cells,30 is important for neuronal cell survival during neural differentiation.31
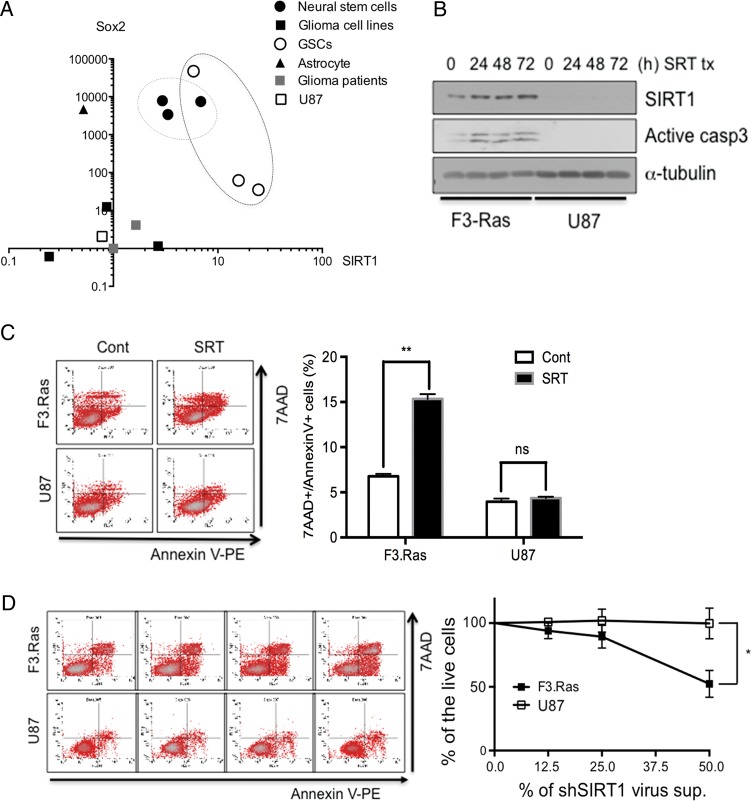
Fig. 4.
SIRT1 is important for the survival of cancer cells with neural stemness. (A) The relationship between the mRNA levels of SIRT1 and Sox2 was determined in a variety of cells via real-time PCR and analyzed as a 2D scatter plot. (B) F3.Ras.CNSCs and U87 cells were treated with SRT and harvested at the indicated times. The level of SIRT1 and active caspase-3 was determined by immunoblotting. Alpha-tubulin was loading control. (C) For apoptosis validation, F3.Ras.CNSCs and U87 cells were treated with SRT for 72 h, stained with 7-aminoactinomycin (7-AAD) and annexin V–phycoerythrin (PE), and then analyzed on a conventional flow cytometer (left panel). A percentage of annexin V–positive cells is graphically presented (right panel) (n = 3). (D) F3.Ras.CNSCs and U87 cells were infected with shCont or shSIRT1 virus supernatant, stained with 7-AAD and annexin V-PE, and then analyzed on a conventional flow cytometer (left panel). A percentage of annexin V–positive cells is graphically presented (right panel) (n = 3).
P53-dependent Cell Death by SIRT1 Depletion in Cancer Cells With Neural Stemness
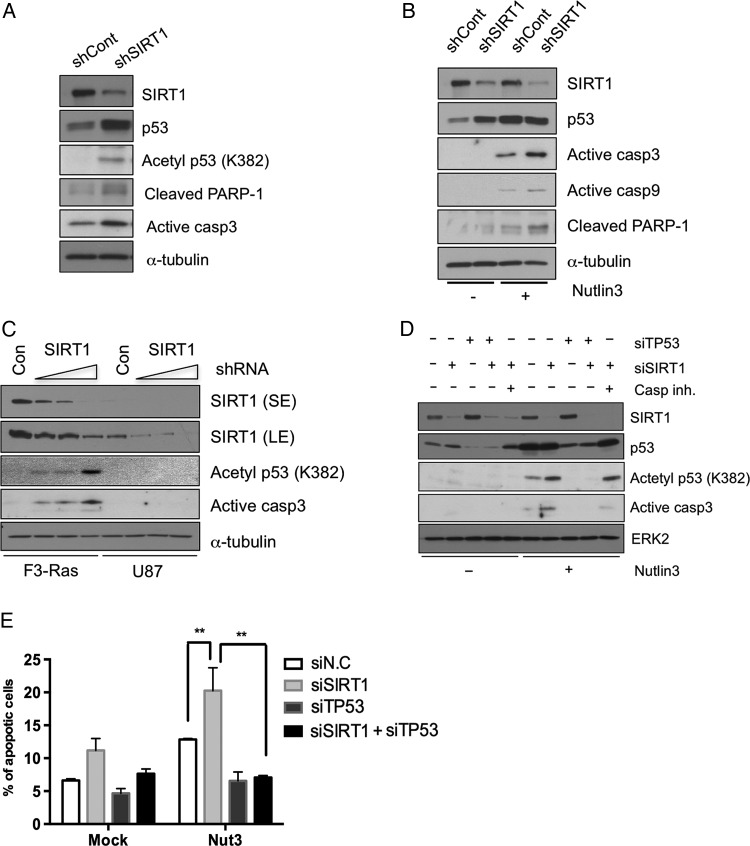
Since tumorigenicity of F3.NSCs upon oncogenic stimulation occurred in parallel with a defective p53 response even at the high level of phosphorylation11 and high expression of SIRT1 in F3.Ras.CNSCs, p53 expression and p53-dependent response in the absence of SIRT1 expression were carefully investigated. Following the loss of SIRT1 expression in F3.Ras.CNSCs by shRNA (Fig. 5A) or siRNA (Supplementary Fig. S3B), p53 expression and the acetylation of p53 were significantly increased. The increased level of p53 protein as well as acetylation (K382, a deacetylation site of SIRT1) by the loss of SIRT1 in F3.Ras.CNSCs were concurrent with the induction of apoptosis (Fig. 5A). Similarly, p53 acetylation in F3.CNSCs was observed by SIRT1 inhibitor treatment (Supplementary Fig. S3C). Apoptosis in the absence of SIRT1 seemed augmented when p53 protein level was induced following treatment with the murine double minute 2 (Mdm2) chemical inhibitor Nutlin3. Thus, the p53-dependent apoptotic event may be related to poor survival of SIRT1-depleted F3.Ras.CNSCs and the loss of tumorigenicity (Fig. 5B). Examination of the relative expression of representative p53 target genes such as Mdm2, Bax, Fas, and Apaf-1 further validated the recovery of the p53 response following SIRT1 depletion in F3.Ras.CNSCs. As the SIRT1-depleted cells gradually regained SIRT1 expression over time (Supplementary Fig. S3D), the expression level of the representative p53 target genes subsequently affected was altered accordingly (Supplementary Fig. S3E), indicating that SIRT1 depletion is closely associated with p53-dependent transcriptional activity. Apoptosis following SIRT1 depletion (Fig. 4), in cancer cells with neural stemness but not in U87, may be regulated by different p53 responses, since SIRT1 deacetylates p53 and can affect its transcriptional function.27,32 To test this hypothesis, the p53 response was determined following SIRT1 depletion in either F3.Ras.CNSCs or U87 cells. The stable knockdown of SIRT1 markedly increased the formation of active caspase-3 and acetylated p53 in F3.Ras.CNSCs but not in the U87 cells (Fig. 5C). To confirm the dependence of p53 on SIRT1 depletion for its apoptosis in F3.Ras.CNSCs, we simultaneously depleted p53 with SIRT1, with or without Nutlin3 treatment. Apoptosis in F3.Ras.CNSCs following SIRT1 depletion was significantly reduced by codepletion of p53 after Nutlin3 treatment, indicating that alteration of p53 by SIRT1 depletion may be responsible for the apoptosis in F3.Ras.CNSCs (Fig. 5D and E).
Fig. 5.
P53 is dependent on SIRT1 depletion for cell death in cancer cells with neural stemness. (A) The expressions of SIRT1, p53, acetyl-p53 (K382), cleaved poly(ADP-ribose) polymerase 1 (PARP-1), and active caspase-3 were determined by immunoblotting. (B) F3.Ras.CNSCs (shCont or shSIRT1) treated with Nutlin3 (5 μM) and then harvested after 20 h. The levels of SIRT1, p53, active caspase-3, 9, and cleaved PARP-1 were determined by immunoblotting. (C) F3.Ras.CNSCs and U87 cells underwent a dose-dependent infection. The levels of SIRT1, acetyl-p53 (K382), and active caspase-3 were determined by immunoblotting. (D) F3.Ras.CNSCs were transfected with siRNA negative control (N.C; labeled as ‘-’), SIRT1 (siSIRT1), p53 (siTP53), and SIRT1/p53. The cells were pretreated with a caspase-9 inhibitor when SIRT1 was depleted using siSIRT1, prior to Nutlin3 treatment. The levels of SIRT1, p53, acetyl-p53 (K382), and active caspase-3 were determined by immunoblotting. ERK2, extracellular signal-regulated kinase 2. (E) F3.Ras.CNSCs were transfected with siRNA N.C, SIRT1, p53, and both SIRT1 and p53 and then treated with Nutlin3. These cells were stained with 7-aminoactinomycin and annexin V–phycoerythrin and then analyzed on a conventional flow cytometer. The percentage of apoptotic cell population is presented graphically (n = 4). Alpha-tubulin or ERK2 was loading control.
Induction of Cell Death by SIRT1 Depletion in Cancer Cells With Neural Stemness
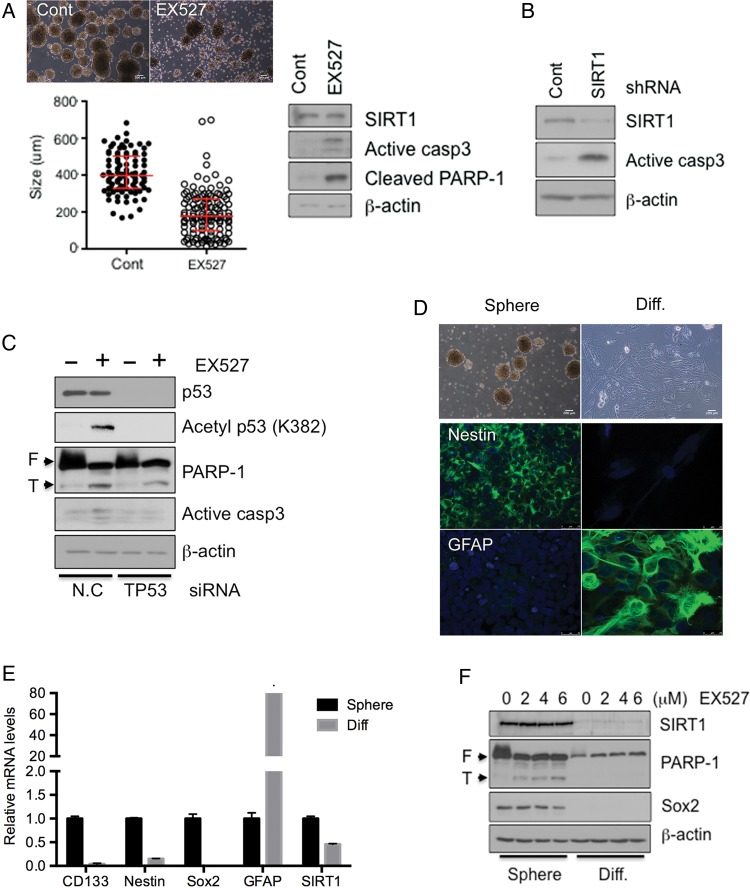
Considering both the requirement of SIRT1 expression in the process of oncogenic transformation of F3.NSCs (Fig. 2) and the survival/tumorigenic potential of F3.Ras.CNSCs (Figs. 3 and 4), we next surmised that SIRT1 would be a possible common molecular target of cancer cells with neural stemness. Thereby, using GSCs as a model for cancer cells with neural stemness, we determined the role of SIRT1 in cell survival and tumorigenic potential. To examine the role of SIRT1 in GSCs, we initially simply treated the GSC cell culture with EX527. As shown in Fig. 6A, the size of the GSC spheres was markedly reduced in the presence of EX527 in culture, and apoptosis was also observed. Consistently, the dependence of GSCs on SIRT1 for survival was confirmed by the apoptosis in GSCs following SIRT1 depletion by shRNA (Fig. 6B), small interfering (si)RNA (Supplementary Fig. S4A), or SIRT1 inhibition by EX527 treatment (Supplementary Fig. S4B). Furthermore, Sox2 expression in GSCs was also markedly reduced by SIRT1 depletion in parallel with apoptosis (Supplementary Fig. S4C). Similar to the F3.Ras.CNSCs, apoptosis of GSCs was attenuated following codepletion of p53, indicating that the induction of cell death by SIRT1 depletion in GSCs is also p53 dependent (Fig. 6C). In order to examine SIRT1 depletion-dependent cell death as an event specific to cancer stemness, GSCs were forced to differentiate and lose their cancer stemness as described previously.18 After spontaneous differentiation of GSCs, expressions of CD133, Sox2, and nestin were markedly attenuated, whereas GFAP expression was increased (Fig. 6D and E).15 As expected, SIRT1 expression was concurrently suppressed in the glial type of cells after differentiation from GSCs (Fig. 6E and F). Additionally, the GSCs and their differentiated counterparts displayed varying apoptotic susceptibility following EX527 treatment. The differentiated glial type cells from GSCs were more resistant to apoptosis by EX527 treatment, indicating that dependence on SIRT1 for survival is specific to cancer cells with neural stemness.
Fig. 6.
Depletion of SIRT1 leads to cell death in GSCs but not in GSC-derived differentiated cells. (A) The morphology of Cont (dimethyl sulfoxide)- or EX527 (5 μM)-treated GSC 528NS cells (scale bars, 200 m). Size of the sphere was measured using ImageJ. The levels of SIRT1, active caspase-3, and cleaved poly(ADP-ribose) polymerase 1 (PARP-1) were determined by immunoblotting. (B) The levels of SIRT1 and active caspase-3 were determined by immunoblotting. (C) 528NS cells were transfected with siRNA negative control (N.C) or TP53 (encoding p53) and then treated with EX527 starting the next day for 2 days. The levels of p53, acetyl-p53 (K382), PARP-1, and active caspase-3 were determined by immunoblotting. (D) Morphological image of GSC 528NS and their differentiated cells (upper panel). GSC 528NS and their differentiated cells were immunostained with an NSC marker (hNestin) and astrocyte marker (glial fibrillary acidic protein [GFAP]). 4′,6′-diamidino-2-phenylindole was used for nuclear counterstain (scale bars, 200 m). (E) The mRNA levels of CD133, nestin, Sox2, GFAP, and SIRT1 in GSC 528NS and their differentiated cells were determined by real-time PCR. (F) GSC 528NS and their differentiated cells were treated with EX527 in a dose-dependent manner and harvested after 2 days. The levels of SIRT1, PARP-1, and Sox2 were determined by immunoblotting. Beta-actin was loading control.
Discussion
Due to the high tumorigenic potential and chemo/radioresistance of stemlike cancer cells—cancer cells with stemness (referred to as cancer stem cells or tumor initiating cells)—extensive efforts have been made to understand the molecular characteristics of cancer cells with stemness or the molecular mechanisms that confer cancer stemness. On the other hand, high throughput screening of small molecules in chemical libraries targeting the specific molecular entities of cancer cells with stemness to achieve selective cell death would be a promising strategy in the process of cancer drug development.
In the present studies, we took advantage of F3.Ras.CNSCs that were derived from human NSCs, maintaining neural stemness and forming the heterogeneous primitive neuroectodermal tumor–like cancer in vivo, for characterizing the molecular targets of cancer cells with neural stemness. We reasoned that the attenuated p53 response in F3.NSCs upon oncogenic challenge with H-Ras, which allowed cellular transformation,11 may result from the high expression of a negative regulator of p53, such as SIRT1.11 As predicted, SIRT1 expression in F3.NSCs was critical for oncogenic susceptibility (Fig. 2) and was also important for maintaining neural cancer stemness of F3.Ras.CNSCs (Fig. 3). Considering that the glioma cell lines were segregated from cells with neural stemness such as F3.NSCs, F3.Ras.CNSCs, and GSCs by plotting the level of SIRT1 and Sox2 (Fig. 4A), apoptosis in F3.Ras.CNSCs but not in U87 cells on the loss of SIRT1 would imply a possible significant role of SIRT1 in cancer cells with neural stemness. The notion that SIRT1 may be required for acquiring neural cancer stemness was also verified by apoptosis in GSCs by either SIRT1 depletion or enzymatic inhibition (Fig. 6A and B). More importantly, the susceptibility to the enzymatic inhibitor of SIRT1 was significantly diminished when GSCs were differentiated into a glial type of cells (Fig. 6F). These results suggest that SIRT1 expression, which appeared to be high in cancer cells with neural stemness, would be a putative therapeutic target to induce selective apoptosis of cancer cells with neural stemness. The role of SIRT1 in the survival of cancer cells with stemness has also been demonstrated in CD133-positive GSCs33 and chronic myeloid leukemia stem cells.34 Recent studies in a glioma cell line model (U251) revealed that the deacetylase activity of SIRT1 might also contribute to maintaining proliferation and survival in malignant glioma by regulating acetylation of primitive neuroectodermal tumor,35 although the biological significance remains unclear. Alteration of p53 acetylation status other than phosphorylation is important for its role as both a transactivator36 and transcription-independent cell death inducer.37 Thereby, inhibition of p53 deacetylation would be a strategy to regain p53-dependent cell death in certain types of cancer cells. For example, the pharmacological inhibition of SIRT1 in chronic myelogenous leukemia stem cells induces reactivation of p53 to cause both growth inhibition and defects in tumor engraftment through p53 acetylation.34 Similarly, inhibition of SIRT2 promotes cell death in lung cancer cells by inducing p53 acetylation and consequent p53-dependent apoptotic gene response.38 We also demonstrated that the selective cell death of F3.Ras.CNSCs by SIRT1 depletion resulted from increased acetylation of p53, and p53 codepletion partly rescued cell death in F3.Ras.CNSCs (Fig. 5D and E). Moreover, considering that p53 frequently functions as a barrier for “acquiring stemness”39 or maintaining stemness,40 cancer dedifferentiation, generating cancer cells with neural stemness (additional defects in p53 such as premature deacetylation of p53 by the induction of SIRT1 in this case), may be a prerequisite event for acquiring or maintaining cancer stemness in GSCs. Thus, the negative regulation of p53 by direct41 or indirect mechanisms18 would be favorable for the growth of cancer cells with stemness. Thereby, regaining the p53 response in cancer cells with stemness by abrogating a set of negative regulators toward p53 would be a promising therapeutic strategy to cancer cells with stemness. In conclusion, SIRT1 is critical for not only cancerous transformation of F3.NSCs but also survival of F3.Ras.CNSCs and in abrogating the p53 response in GSCs, so that it would be a promising target for selectively targeting cancer cells with stemness. In addition, a comparison of the common molecular characteristics of F3.Ras.CNSCs with GSCs could provide an important set of information to understand cancer cells with stemness and to serve as a feasible model system for high throughput screening targeting cancer cells with stemness.
Supplementary Material
Funding
This work was supported by a grant for Bio & Medical Technology Development Program (no. 2010-0020232), by a National Research Foundation of Korea (NRF) grant (nos. 2013-023601 and 2011-0030043) and by Korea Foundation for Cancer Research (KFCR) grant (no. 2012-004) funded by the Korean government.
Conflict of interest statement. None declared.
Supplementary Material
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Collado M, Serrano M. The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer. 2006;6(6):472–476. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- 3.Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27(20):2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- 4.Kaye FJ. RB and cyclin dependent kinase pathways: defining a distinction between RB and p16 loss in lung cancer. Oncogene. 2002;21(45):6908–6914. doi: 10.1038/sj.onc.1205834. [DOI] [PubMed] [Google Scholar]
- 5.Bringold F, Serrano M. Tumor suppressors and oncogenes in cellular senescence. Exp Gerontol. 2000;35(3):317–329. doi: 10.1016/s0531-5565(00)00083-8. [DOI] [PubMed] [Google Scholar]
- 6.Vonlanthen S, Heighway J, Altermatt HJ, et al. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br J Cancer. 2001;84(10):1372–1376. doi: 10.1054/bjoc.2001.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe J, Cha H, Lee MO, et al. Regulation of the Wip1 phosphatase and its effects on the stress response. Front Biosci. 2012;17:1480–1498. doi: 10.2741/3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker N, Ridgway RA, van Es JH, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 9.Kim HS, Woolard K, Lai C, et al. Gliomagenesis arising from Pten- and Ink4a/Arf-deficient neural progenitor cells is mediated by the p53-Fbxw7/Cdc4 pathway, which controls c-Myc. Cancer Res. 2012;72(22):6065–6075. doi: 10.1158/0008-5472.CAN-12-2594. [DOI] [PubMed] [Google Scholar]
- 10.Rich JN, Guo C, McLendon RE, et al. A genetically tractable model of human glioma formation. Cancer Res. 2001;61(9):3556–3560. [PubMed] [Google Scholar]
- 11.Lee JS, Lee HJ, Moon BH, et al. Generation of cancerous neural stem cells forming glial tumor by oncogenic stimulation. Stem Cell Rev. 2012;8(2):532–545. doi: 10.1007/s12015-011-9280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng C. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5(2):147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prozorovski T, Schulze-Topphoff U, Glumm R, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10(4):385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 14.Hisahara S, Chiba S, Matsumoto H, et al. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc Natl Acad Sci U S A. 2008;105(40):15599–15604. doi: 10.1073/pnas.0800612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67(9):4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 16.Flax JD, Aurora S, Yang C, et al. Engraftable human neural stem cells respond to developmental cues, replace neurons, and express foreign genes. Nat Biotechnol. 1998;16(11):1033–1039. doi: 10.1038/3473. [DOI] [PubMed] [Google Scholar]
- 17.Joshi K, Banasavadi-Siddegowda Y, Mo X, et al. MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem Cells. 2013;31(6):1051–1063. doi: 10.1002/stem.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu C, Banasavadi-Siddegowda YK, Joshi K, et al. Tumor-specific activation of the C-JUN/MELK pathway regulates glioma stem cell growth in a p53-dependent manner. Stem Cells. 2013;31(5):870–881. doi: 10.1002/stem.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao P, Joshi K, Li J, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110(21):8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurita R, Suda N, Sudo K, et al. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One. 2013;8(3):e59890. doi: 10.1371/journal.pone.0059890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Y, Zhao W, Chen Y, et al. Acetylation is indispensable for p53 activation. Cell. 2008;133(4):612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi H, Woods N, Piluso L, et al. p53 acetylation is crucial for its transcription-independent proapoptotic functions. J Biol Chem. 2009;284(17):11171–11183. doi: 10.1074/jbc.M809268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis P, Fagan BM, Magness ST, et al. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26(2–4):148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 24.Luo J, Nikolaev AY, Imai S, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 25.Kim EJ, Kho JH, Kang MR, et al. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28(2):277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 26.Kamel C, Abrol M, Jardine K, et al. SirT1 fails to affect p53-mediated biological functions. Aging Cell. 2006;5(1):81–88. doi: 10.1111/j.1474-9726.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- 27.Peck B, Chen CY, Ho KK, et al. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. 2010;9(4):844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- 28.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao P, Joshi K, Li J, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110(21):8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwahara N, Hisahara S, Hayashi T, et al. Transcriptional activation of NAD+-dependent protein deacetylase SIRT1 by nuclear receptor TLX. Biochem Biophys Res Commun. 2009;386(4):671–675. doi: 10.1016/j.bbrc.2009.06.103. [DOI] [PubMed] [Google Scholar]
- 31.Guo W, Qian L, Zhang J, et al. Sirt1 overexpression in neurons promotes neurite outgrowth and cell survival through inhibition of the mTOR signaling. J Neurosci Res. 2011;89(11):1723–1736. doi: 10.1002/jnr.22725. [DOI] [PubMed] [Google Scholar]
- 32.Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 33.Chang C, Hsu C, Yung M, et al. Enhanced radiosensitivity and radiation-induced apoptosis in glioma CD133-positive cells by knockdown of SirT1 expression. Biochem Biophys Res Commun. 2009;380(2):236–242. doi: 10.1016/j.bbrc.2009.01.040. [DOI] [PubMed] [Google Scholar]
- 34.Li L, Wang L, Li L, et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21(2):266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu Y, Zhang J, Wu S, et al. SIRT1 promotes proliferation and inhibits apoptosis of human malignant glioma cell lines. Neurosci Lett. 2012;525(2):168–172. doi: 10.1016/j.neulet.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 36.Mellert HS, Stanek TJ, Sykes SM, et al. Deacetylation of the DNA-binding domain regulates p53-mediated apoptosis. J Biol Chem. 2011;286(6):4264–4270. doi: 10.1074/jbc.M110.184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sykes SM, Mellert HS, Holbert MA, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24(6):841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann G, Breitenbucher F, Schuler M, et al. A novel sirtuin 2 (SIRT2) inhibitor with p53-dependent pro-apoptotic activity in non-small cell lung cancer. J Biol Chem. 2014;289(8):5208–5216. doi: 10.1074/jbc.M113.487736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong H, Takahashi K, Ichisaka T, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460(7259):1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stecca B, Ruiz i Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J. 2009;28(6):663–676. doi: 10.1038/emboj.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandolfi S, Montagnani V, Penachioni JY, et al. WIP1 phosphatase modulates the Hedgehog signaling by enhancing GLI1 function. Oncogene. 2013;32(40):4737–4747. doi: 10.1038/onc.2012.502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.