Abstract
Despite 6 decades of research, only 3 drugs have been approved for astrocytomas, the most common malignant primary brain tumors. However, clinical drug development is accelerating with the transition from empirical, cytotoxic therapy to precision, targeted medicine. Preclinical animal model studies are critical for prioritizing drug candidates for clinical development and, ultimately, for their regulatory approval. For decades, only murine models with established tumor cell lines were available for such studies. However, these poorly represent the genomic and biological properties of human astrocytomas, and their preclinical use fails to accurately predict efficacy in clinical trials. Newer models developed over the last 2 decades, including patient-derived xenografts, genetically engineered mice, and genetically engineered cells purified from human brains, more faithfully phenocopy the genomics and biology of human astrocytomas. Harnessing the unique benefits of these models will be required to identify drug targets, define combination therapies that circumvent inherent and acquired resistance mechanisms, and develop molecular biomarkers predictive of drug response and resistance. With increasing recognition of the molecular heterogeneity of astrocytomas, employing multiple, contemporary models in preclinical drug studies promises to increase the efficiency of drug development for specific, molecularly defined subsets of tumors.
Keywords: astrocytoma, drug development, genomics, glioblastoma, mouse models
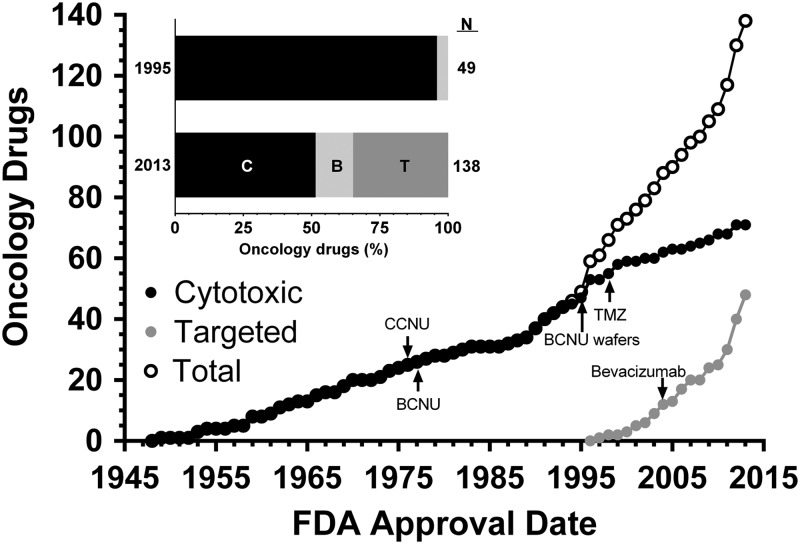
Oncology drug development is an inherently long and expensive process. The average time required from the initial filing of an investigational new drug application to marketing approval by the US Food and Drug Administration (FDA) is ∼9 years.1 Most oncology drugs fail late in the clinical trial process from lack of efficacy. Only 5% ultimately receive FDA approval after a typical cost of $400 million.2 In addition to length and cost, this inefficiency significantly limits the number of drugs with proven clinical benefit. Since nitrogen mustard (mechlorethamine) became the first cytotoxic anticancer agent in 1949, only 138 oncology drugs have received FDA approval (an average of 1.4 per year). The approval rate was even lower (∼1 drug per year) during the era of empirical therapy with cytotoxic drugs (1949–1996). However, the rate of new drug approvals has increased dramatically (∼4 per year) since the dawn of the precision-medicine era of oncology3 marked by the 1996 approval of rituximab, the first targeted anticancer agent. Targeted agents now constitute 36% of all FDA-approved oncology drugs. Twenty-three were approved in the last 4 years alone (Fig. 1). Dozens more are currently in clinical trials, and hundreds are in preclinical development at pharmaceutical companies and academic centers worldwide.
Fig. 1.
FDA approved oncology drugs. The cumulative number of cytotoxic (C), biological (B), and targeted (T) drugs approved by the FDA is shown. Dates correspond to the first indication approved. Approvals for subsequent indications are not shown. Drugs used for astrocytomas are indicated. Data were compiled from http://www.drugs.com (accessed September 12, 2014) and http://www.medilexicon.com/drugs-list/cancer.php (accessed September 12, 2014).
Only 3 drugs have been approved specifically for the treatment of astrocytomas, the most common malignant primary brain tumors.4 These include 2 cytotoxic agents, carmustine wafers and temozolomide (TMZ), and one targeted agent, bevacizumab. Carmustine wafers were approved for recurrent and newly diagnosed glioblastoma (GBM), a WHO grade IV astrocytoma, in 1997 and 2003. TMZ was approved for recurrent anaplastic astrocytomas (WHO grade III) and newly diagnosed GBM in 1999 and 2005. The targeted agent bevacizumab, a humanized monoclonal antibody to vascular endothelial growth factor, was approved for recurrent GBM in 2009. Thus, the therapeutic armamentarium for astrocytomas remains severely limited despite the accelerated pace of oncology drug development over the past 2 decades.
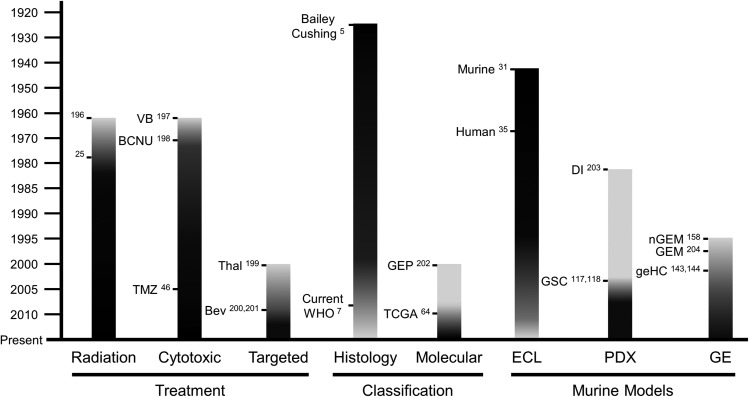
Histopathological classification has served as the foundation for diagnosis and management of astrocytomas for nearly a century.5 Periodic refinement of the initial 1926 classification culminated in the current scheme, published in 2007 by the World Health Organization (WHO).6,7 This system utilizes cytological evidence of astrocytic differentiation and the presence of morphological features, including mitotic activity, angiogenesis, and necrosis, to stratify patients into prognostically distinct diagnostic entities with increasingly poor survival. Low grade (WHO grade II) astrocytomas have a 10-year median overall survival. High-grade astrocytomas, including anaplastic astrocytomas and GBMs, feature elevated mitotic activity and angiogenesis and/or necrosis. These tumors have dismal prognoses of ∼3 years and 15 months, respectively.7 Despite their classification into distinct diagnostic entities, comprehensive genomics analyses have shown that astrocytomas of all grades are molecularly diverse.8,9 Recognition of this fact has fueled efforts to develop a molecular classification scheme to further accelerate the shift from empirical, cytotoxic therapies to precision medicine with targeted agents in molecularly defined tumor subsets (Fig. 2).3,8
Fig. 2.
Evolution of astrocytoma treatment, classification, and murine models. Shading in black depicts increased emphasis over time. Major developments in each category are noted. Abbreviations: BCNU, bis-chloroethylnitrosourea; Bev, bevacizumab; DI, direct injection; ECL, established cell line; GE, genetically engineered; geHC, genetically engineered human cells; GEM, genetically engineered mice; GEP, gene expression profiling; GSC, glioma stem cell; nGEM, non-germline genetically engineered mice; PDX, patient-derived xenografts; TCGA, The Cancer Genome Atlas; Thal, thalidomide; TMZ, temozolomide; VB, vinblastine; WHO, World Health Organization.
Although astrocytomas contribute significantly to cancer-related death and disability, they are relatively rare. GBMs account for 86% of all astrocytomas but only affect 3.2 in 100 000 individuals in the United States. Its incidence is age dependent and ranges from 0.14 in children to a peak of 14.9 in 100 000 75–84 year-old adults.4 Moreover, <20% of adult astrocytoma patients enroll in clinical trials.10 The low prevalence and limited clinical trial participation represent significant challenges for astrocytoma drug development.
As the diagnostic and treatment landscape evolves from empirical to precision medicine, now is an opportune time to reassess the way drugs are developed for adult patients with astrocytomas (Fig. 2). The reality that astrocytomas are rare relative to other cancer types and that trial participation is limited makes this issue even more acute. A number of innovative approaches to clinical trial design, including adaptive biomarker-based trials, are currently under investigation and will not be the focus of this review.11,12 Rather, we focus on the role of murine models in preclinical drug development for adult astrocytoma patients and will argue that fundamental changes in their use are required to expand the therapeutic armamentarium and improve outcomes for these devastating malignancies.
Given the number of promising targeted agents that deserve clinical testing, preclinical astrocytoma modeling will be critical for validating drug targets and prioritizing candidates for clinical studies. These models will also be critical for the discovery and development of novel predictive biomarkers that can be used to stratify patients into biologically meaningful disease subtypes and identify likely responders. Finally, these models will be critical for defining the molecular mechanisms of drug sensitivity and resistance so that rational combination therapies and molecular diagnostic tests to guide their use can be developed.
The last 20 years have witnessed major improvements in the preclinical modeling of astrocytomas. A number of recent reviews have described these in detail.13–24 Here we examine the role of conventional mouse models in the development of cytotoxic drugs commonly used to treat adult astrocytoma patients. We then examine their use in the development of select targeted agents that have recently failed in late stage clinical trials. We will then describe contemporary mouse models and how they may be best utilized to improve clinical drug development in the future.
Development of DNA Alkylating Agents for Glioblastoma
Three cytotoxic, DNA alkylating agents, carmustine (bis-chloroethylnitrosourea [BCNU]), lomustine (1-[2-chloroethyl]-3-cyclohexyl-1-nitrosourea [CCNU]), and temozolomide (TMZ) have been the cornerstones of astrocytoma chemotherapy for the past 5 decades. The nitrosoureas BCNU and CCNU entered clinical practice in the 1960s and were studied in a number of clinical trials through the 1990s. TMZ was developed in the late 1980s and entered clinical practice in the mid 1990s. How were animal model studies utilized to inform their clinical development, and what lessons can be learned?
Randomized phase III clinical trials of nitrosoureas in adult astrocytoma patients were published in the late 1970s.25,26 BCNU and CCNU were initially chosen for clinical development based on their excellent ability to cross the blood-brain barrier (BBB) and their efficacy in multiple preclinical models. However, individual trial results from 1976 to 2001 were inconclusive, and a meta-analysis of > 3000 patients from 12 trials was required to definitively demonstrate their efficacy (ie, an ∼2-month increase in median survival and a 5% increase in 2-year survival).27 Although this study established chemotherapy as a valuable adjuvant to surgical resection and radiation therapy, the clinical benefits of nitrosoureas were marginal, and systemic toxicity was not infrequent.
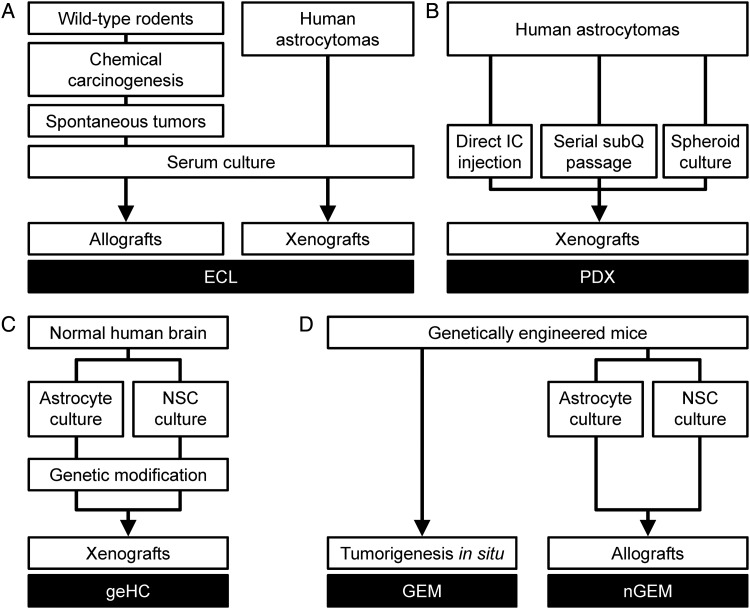
The successful culture of spontaneous tumors from chemically mutagenized rodents as established cell lines (ECLs) transformed the cancer research landscape in the 1940s (Fig. 3A). Development of the P388 and L1210 models of leukemia in particular were critical in development of dozens of cytotoxic drugs by the Developmental Therapeutics Program at the National Cancer Institute from the 1950s through the 1980s.28,29 Their rapid, reproducible growth, high penetrance, and short latency when injected into syngeneic hosts made these models particularly attractive for preclinical drug studies. It is therefore not surprising, in retrospect, that the nitrosoureas were first tested and found to have efficacy in intracranial leukemia models.30 While chemical mutagenesis had been shown to produce gliomas in rodents during the 1940s,31 ECL and allograft models of murine gliomas, including GL26, GL261, 9L, and C6, became widespread during the late 1960s and early 1970s.13,19,31–34 ECL cultures from human astrocytomas and xenotransplantation into immunodeficient mice were developed contemporaneously.19,35–39 Thus, preclinical testing of nitrosoureas in glioma models occurred much later during their clinical development, with the first results being published in 1973.33 A meta-analysis of preclinical studies employing either murine allograft or human xenograft ECL models showed highly variable efficacy that was significantly influenced by experimental design. Overall effect sizes were small (0.19-fold and 0.43-fold increases in median survival for BCNU and CCNU, respectively), and no statistically significant beneficial effect was found.40
Fig. 3.
Conventional and contemporary murine astrocytoma models. Conventional murine and human astrocytoma models utilized established cell lines (ECL) (A). Murine ECL models were generated by serum culture of cells harvested from spontaneous rodent astrocytomas induced by chemical carcinogens and transplantation into immunocompetent rodents. Human ECL were similarly generated from human astrocytomas and xenografted into immunodeficient mice. Contemporary human models (B) consist of patient-derived xenografts (PDX) whereby tumor cells harvested from human astrocytomas are directly injected or culture as nonadherent spheroids in defined, serum-free medium prior to engraftment into immunodeficient mice. Genetically engineered models include genetically engineered human cells (geHC), whereby astrocytes or neural stem cells (NSC) are harvested from normal human brains that have been genetically modified with oncogenic mutations and xenografted into immunodeficient mice (C); GEM, whereby induction of oncogenic mutations produces tumorigenesis in situ (D); and non-germline genetically engineered mouse (nGEM) models, whereby astrocytes or NSCs are harvested from GEM, cultured in vitro, and allografted into immunocompetent or immunodeficient mice (D).
TMZ, a mono-alkylating agent developed in the 1980s, was clinically investigated due to its broad antitumor activity and favorable toxicity profile in preclinical models, particularly L1210 leukemias (Fig. 4).41 It was found to have excellent oral bioavailability in a phase I study. Fortunately, this trial included several high-grade astrocytoma patients who experienced partial sustained responses.42 Subsequent clinical experience in recurrent and newly diagnosed high-grade astrocytoma patients was similarly favorable.43 Phase II trials in recurrent high-grade astrocytomas and in combination with radiation in newly diagnosed GBM patients showed 58% radiographic response rates and 16-month median survival, respectively.44,45 The definitive phase III trial published in 2005 established adjuvant TMZ in combination with fractionated radiation as the standard of care for newly diagnosed GBM based on a 21% increase in median survival and a 5-fold increase in 5-year survival compared with radiation alone, which produced only 12.1-month median and 1.9% 5-year survivals.46,47
Fig. 4.
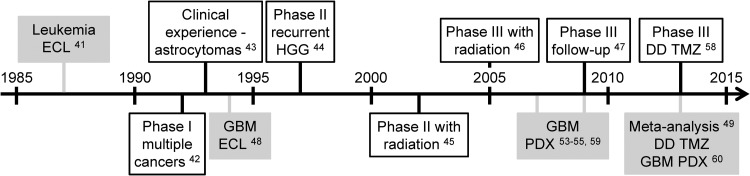
Developmental timeline for temozolomide (TMZ). Clinical studies outlined in black were conducted with newly diagnosed GBM patients, unless otherwise noted. Preclinical studies are highlighted in gray. Abbreviations: DD, dose-dense; ECL, established cell line models; HGG, high-grade gliomas; PDX, patient-derived xenograft models.
The first preclinical study of TMZ efficacy in GBM ECL models was published in 1994, 7 years after its first description and 2 years after entering clinical trials.48 A 2013 meta-analysis of TMZ in murine allograft and human xenograft ECL models of GBM showed that it was consistently efficacious in both, producing 50% decreases in tumor volume and ∼2-fold increases in median survival on average.49 This contrasts with the meta-analysis of nitrosoureas in similar preclinical models that showed far smaller and inconsistent effect sizes.40
The biological effects of TMZ are largely mediated by its ability to methylate the O6 position of guanine.50 Repair of this lesion is carried out by a single enzyme, encoded by the methylguanine-DNA methyltransferase (MGMT) gene. MGMT expression is regulated through promoter methylation, and ∼50% of GBMs have methylated MGMT.51 Based upon this knowledge, a retrospective companion study to the definitive phase III trial that established TMZ as the standard of care for GBM showed that MGMT promoter methylation was a favorable prognostic, and likely predictive, marker for TMZ benefit in GBM patients.52 Subsequent studies using more contemporary human GBM models, whereby patient-derived tumors are directly xenografted into immunodeficient mice without prior serum-based culture, showed that MGMT promoter methylation was an important predictor of TMZ efficacy.53,54 TMZ, given concurrently with radiation, produced a survival benefit only in a subset of GBM patient-derived xenograft (PDX) models with methylated MGMT.55 Moreover, TMZ showed a wider response range in PDX models (0.2–5.9-fold increases in median survival) than in human ECL models (0.3–2.5-fold) of GBMs, suggesting that these newer models may more accurately reflect its clinical efficacy, particularly in molecularly defined subsets of tumors.49,54,55
Based on the role of MGMT in TMZ resistance and the fact that protracted, dose-dense TMZ depleted MGMT activity in peripheral blood mononuclear cells, the hypothesis that dose-dense TMZ would enhance its therapeutic benefits, particularly in GBM with unmethylated MGMT, was explored in a randomized, phase III clinical trial.56–58 Although this recently published trial prospectively confirmed the prognostic significance of MGMT promoter methylation, dose-dense TMZ failed to improve survival in either MGMT unmethylated or methylated GBM. These results are consistent with a preclinical study in GBM PDX models showing that TMZ induced MGMT expression, even in MGMT-unmethylated GBM.59 A similar lack of efficacy and correlation with MGMT methylation status was found in a preclinical study of dose-dense TMZ with 7 GBM PDX models published during trial accrual.60
Development of Targeted Agents for Glioblastoma
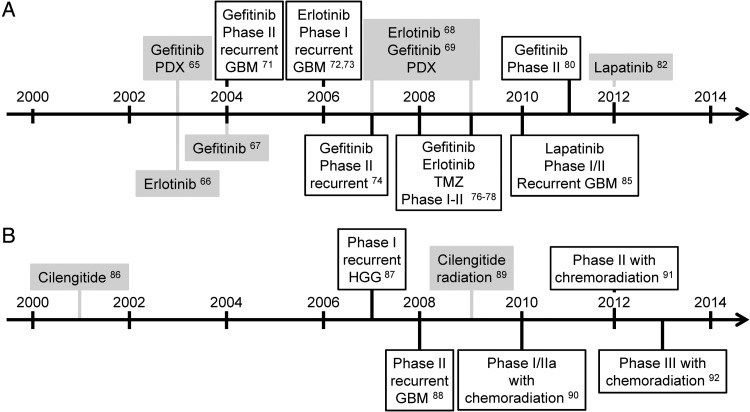
The promise of small molecule inhibitors that target the dysregulated signaling pathways driving gliomagenesis has fueled neuro-oncology drug development since the late 1990s. The epidermal growth factor receptor (EGFR) had long been known to be a mutated target in GBM, where amplifications and activating truncation mutations are among the most common genetic abnormalities.61–64 Based upon this knowledge, their antitumor activity in preclinical models,65–69 and their efficacy in EGFR-mutant non–small cell lung cancers (NSCLCs),70 the first-generation EGFR tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib entered clinical trials in the 2000s (Fig. 5A). However, no significant activity was found in a number of phase II GBM studies, and no reliable biomarkers to predict their efficacy could be identified in retrospective molecular analyses.71–81 Reasons for their failure remain poorly understood, but signaling pathway redundancy and molecular heterogeneity were likely contributers.82 Subsequent pharmacokinetic (PK) studies in patients with NSCLC brain metastases showed limited CSF penetration of both gefitinib and erlotinib.83 Moreover, expression of ABC transporters, including P-glycoprotein and breast cancer resistance protein, were shown to significantly limit brain penetration of erlotinib.84
Fig. 5.
Developmental timelines for EGFR tyrosine kinase inhibitors (A) and cilengitide (B). Clinical studies outlined in black were conducted with newly diagnosed GBM patients, unless otherwise noted. Preclinical studies are highlighted in gray and were performed with established GBM xenograft models, unless otherwise noted.
Newer EGFR TKIs showed broader activity spectra and targeted multiple EGFR family receptors. Whereas first-generation EGFR TKIs bound only the active conformation of the EGFR TK domain, second-generation TKIs, such as lapatinib, bound the inactive conformation. Studies using GBM ECL and neurosphere culture models showed that GBM-specific EGFR extracellular domain mutations were poorly inhibited by first-generation TKI, but effectively inhibited by lapatinib, in contrast to TK domain mutations in NSCLC.82 However, lapatinib showed minimal activity in recurrent GBM clinical trials.85 Subsequent retrospective PK studies using NABTC 04-01 trial material showed intratumoral lapatinib concentrations to be well below its predicted therapeutic threshold.82
In contrast to EGFR TKI, data from preclinical glioma models significantly influenced the design of clinical trials with cilengitide, an alpha v-integrin antagonist and putative antiangiogenic agent (Fig. 5B). Cilengitide showed efficacy in subcutaneous GBM ECL xenografts in a 2001 study.86 A subsequent phase I trial in recurrent GBM patients reported promising biological activity and demonstrated a correlation between PK parameters and radiographic response.87 Moderate antitumor activity in the recurrent setting was confirmed in a single-agent phase II trial, supporting its continued investigation in combination regimens.88 Prior to those trials, preclinical studies in GBM ECL xenograft models demonstrated the radiosensitization effects of cilengitide and revealed an unanticipated dependence on schedule.89 These preclinical data informed the design of subsequent phase I–III trials of cilengitide in combination with chemoradiation in newly diagnosed GBM.90–92 Retrospective molecular analysis of phase II specimens suggested that MGMT methylation was associated with cilengitide benefit in this clinical setting;90 however, cilengitide failed to prolong survival in newly diagnosed GBM patients with methylated MGMT in a randomized phase III trial.92
Lessons From the Development of Cytotoxic and Targeted Agents for Glioblastoma
What lessons can be gleaned from the development of alkylating and targeted agents for GBM that might improve future drug development efforts? Clinical trials of nitrosoureas and TMZ were initiated based on preclinical data from murine leukemia models. Data from glioma models came later. With the benefit of hindsight and decades of research conclusively demonstrating that neoplasms from different tissues are molecularly and biologically distinct, it is now clear that the decision to initiate clinical studies should be based on preclinical data in the tumor type of interest. Preclinical data from models that do not accurately reflect the tumor histology or its native organ-based microenvironmental interactions are likely to produce misleading results.28,93
Comparison of alkylating agent efficacy in preclinical glioma models and clinical studies demonstrates striking similarities. Nitrosoureas produced small, but significant benefits in some murine models and astrocytoma patients. In contrast, TMZ was consistently effective in both ECL model studies and clinical trials, with relatively large effect sizes. These data suggest that large effect sizes in multiple preclinical models may be required to accurately predict efficacy in clinical trials, particularly those that enroll molecularly heterogeneous, unselected patient populations. We therefore recommend that the bar for future preclinical drug studies be set well beyond small, but statistically significant prolongations of survival in single model systems, particularly ECL models. Rather, consistent demonstration of large effect sizes in newer model systems that more accurately recapitulate the genomic and biological properties of human astrocytomas may have increased ability to predict clinical efficacy in unselected patient populations.
How do newer model systems, such as PDXs, fare in predicting clinical success relative to conventional ECL models? Preclinical studies with nitrosoureas and cilengitide were only conducted in ECL models (Table 1). Many of the same ECL models were also used in the initial preclinical development of TMZ (Fig. 4), as well as the EGFR TKIs gefitinib and erlotinib (Fig. 5). However, more recent preclinical TMZ and EGFR TKI studies have utilized PDX models to characterize genetic mechanisms of response and resistance and discover predictive biomarkers.53–55,59,60,68,69,94–96 Thus, the fact that TMZ succeeded clinically, while EGFR TKIs and cilengitide failed, cannot be attributed simply to use of newer model systems in preclinical development. Comprehensive comparison of drug efficacy in newer PDX versus conventional ECL models is limited to TMZ. Such data suggest that PDX models may more accurately reflect the heterogeneity of response seen in GBM patients. Therefore, systematic drug efficacy screening in multiple genomically characterized PDX models might be useful for prospectively identifying sets of tumors that are likely to respond.18,97 In order to evaluate the predictive accuracy of these models and maximize their utility for biomarker discovery and development, preclinical studies in PDX models should be performed earlier in the drug development process, ideally prior to initiating clinical trials or concurrently as “co-clinical trials.”98
Table 1.
GBM models used in preclinical development of alkylating and targeted agents
| Chemotherapeutic Agent |
Conventional ECL Models |
Contemporary Models |
|||
|---|---|---|---|---|---|
| Murine Allografts | Human Xenografts | PDX | nGEM | ||
| Alkylating agents | Carmustine (BCNU) | 9L40 GL26, GL261 VMDk 497-P(1) |
D54MG40 U251MG |
||
| Lomustine (CCNU) | G XII40 G XIII GL26, GL261 VMDk 497-P(1) |
U251MG40 | |||
| Temozolomide (TMZ) | 9L49 C6 F98 T98 |
A17249 D54MG, Hs683 SNB-75, SF295 U251MG, U373MG U87MG |
GBM649,53–55,59,60,94–96 GBM8, GBM10, GBM12 GBM14, GBM22, GBM 26 GBM34, GBM36, GBM39 GBM43, GBM44 |
TRP127,185,186 | |
| Targeted agents | Gefitinib | U87MG67 | ODA-4-GEN69 GBM-1-HAM, GBM-17-ROM GBM-14-RAV, TG-17-GIR GBM-9-THI |
||
| Erlotinib | U87MG66 | GBM6, GBM868 GBM12, GBM14, GBM15 GBM22, GBM 28 GBM34, GBM36, GBM39 GBM44 |
|||
| Lapatinib | GS676, GS60082 | ||||
| Celingitide | U87MG86 U251MG89 |
||||
Rather than relying on data from preclinical models, predictive biomarker discovery for TMZ and EGFR TKIs was largely conducted by retrospective molecular analyses of clinical trial specimens. Of the above examples, only the phase III cilengitide trial utilized prospective molecular stratification of MGMT-methylated GBM to increase molecular homogeneity and enrich for likely responders. While this approach is critical for clinical biomarker validation and required for its eventual incorporation as an inclusion criterion in prospective trials, retrospective analyses are inefficient because they require completion of the trial. Given the limited incidence and trial participation of astrocytoma patients, we would argue that a more efficient approach for the discovery of predictive biomarkers would be an increased reliance on preclinical drug studies in genomically and biologically faithful murine models. Such markers could then be validated in retrospective molecular studies of clinical trial specimens. Indeed, this approach has been applied to investigate the ability of a prognostic gene expression signature for predicting bevacizumab efficacy in mesenchymal GBM.99–102 Increased use of this approach for future drug development is likely to reduce time and costs and improve the efficiency of clinical trials.
Nitrosoureas and TMZ were attractive clinical candidates due to their lipophilic chemical structures and favorable brain PK profiles. Indeed, their ability to penetrate the BBB and reach diffusely infiltrative tumor cells is likely one reason for their clinical efficacy.103 In contrast, the failure of first- and second-generation TKIs to fulfill the promise of EGFR-targeted therapy was likely due in part to their poor brain PKs and the activity of BBB drug efflux pumps. Unfortunately, CNS neoplasms are a frequent exclusion criterion in phase I studies, and brain PKs are not routinely analyzed during early development of many targeted agents. The unfortunate failure of EGFR TKIs clearly demonstrates the importance of such data. Preclinical PK studies in both glioma models and mice with genetically engineered defects in BBB efflux pumps have the potential to predict clinical failure on the basis of poor PKs if used prior to, or concurrent with, the initiation of advanced clinical trials.
In addition to poor brain PKs, receptor tyrosine kinase (RTK) signaling pathway redundancy and intertumoral molecular heterogeneity likely contributed to the clinical failures of first-generation EGFR TKIs. Although they share EGFR mutations in common, the divergent efficacy of these drugs in NSCLC and GBM demonstrates that mutational status of a biologically attractive target gene alone is insufficient to predict efficacy of inhibitors that specifically target its activity. Rather, preclinical and clinical data with these drugs in NSCLC and GBM demonstrate that mutation location, within specific functional domains, and its impact on protein structure and catalytic activity is equally important for determining efficacy. Moreover, these data suggest that current precision medicine initiatives that utilize next-generation sequencing to identify “actionable” somatic mutations in oncogenic kinases may require more nuance to fulfill their potential of targeted therapy. Indeed, success of such efforts rests on 3 critical assumptions: (i) that the identified mutation activates downstream signaling and promotes tumorigenesis in the specific tumor type in which it is found; (ii) that the mutated kinase is sensitive to drug inhibition in the appropriate anatomical context (eg, lung vs brain); and (iii) that inhibition results in clinical benefit in the specific tumor type of interest. Because the biological function and druggability of mutational targets are likely tissue specific, it stands to reason that experimental evidence, such as that provided by contemporary preclinical model studies, is necessary to prove the actionability of drug-targetable gene mutations in the specific clinical context to be investigated.
Astrocytoma Genomic Heterogeneity and its Impact on Drug Development
Over the last 15 years, advances in genomics and DNA sequencing technologies have revolutionized cancer research. Studies have conclusively demonstrated that significant intertumoral heterogeneity exists on multiple molecular levels, both within and among the 3 diagnostic categories of astrocytomas.9,104,105 The transcriptome profiles of lower-grade astrocytomas (WHO grades II and III) and GBM are distinct,102,103 and each consists of 3 or 4 transcriptomal subtypes.61,64,106,107 Particular patterns of somatic mutations, chromosomal alterations, and DNA methylation are evident not only within each grade but also within grade-specific subtypes.61,64,108 For example, the IDH1 and IDH2 genes are mutated in ∼60%–80% of lower-grade astrocytomas and ∼50%–80% of the secondary GBMs into which they inevitably progress but are only found in ∼3%–7% of primary GBMs that arise de novo without a clinically detectable, lower-grade antecedent.109 Lower-grade astrocytomas that lack IDH mutations have transcriptome and copy number profiles similar to GBM. A molecular classification system that supplements histological classifiers with layers of molecular information promises to provide a diagnostic framework that not only reflects the intertumoral heterogeneity present in these neoplasms but also facilitates more accurate prognostic stratification and prediction of therapeutic response to targeted therapies.8,9
Many of the putative oncogenic driver mutations in astrocytomas occur in genes that comprise 3 core signaling pathways: (i) the G1/S cell cycle checkpoint controlled by the Rb family of pocket proteins, (ii) RTKs and their downstream RAS-mitogen activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K) effector pathways; and (iii) the TP53 pathway; and (iii) the TP53 pathway.63 Dozens of drugs are currently in development to inhibit kinases in these pathways, and many are actively being investigated in astrocytoma clinical trials. However, given limited trial participation, low disease prevalence, and the number of promising targeted agents that deserve clinical testing, a more rational approach to preclinical drug development for astrocytomas is required.
In addition to intertumoral heterogeneity, genomic studies have shown significant molecular heterogeneity within individual astrocytomas as well.110–113 Sequencing data from multiple samples of individual tumors have demonstrated coexistence of spatially distinct clones with divergent mutational and transcriptomal profiles. Phylogenetic reconstruction showed patient-specific patterns of evolution within each tumor.111,112 Moreover, treatment of lower-grade astrocytomas with DNA-damaging agents such as TMZ may modify the evolutionary path to high-grade disease by inducing alternative mutational spectra.112 Similarly, comprehensive fluorescence in situ hybridization studies have shown that multiple RTK genes, including EGFR, MET, and PDGFRA, can be simultaneously amplified not only within spatially distinct subpopulations of tumor cells but within individual tumor cells as well.114 This mosaic gene amplification can lead to coactivation of multiple redundant RTK signaling pathways, limiting the effectiveness of inhibitors targeting individual kinases and suggesting the need to develop rational combination therapies.115
In addition to molecular heterogeneity, murine modeling studies of human GBM have suggested that individual cells within the tumor may be functionally heterogeneous.116 The cancer stem hypothesis posits that a small subpopulation of tumor cells, termed cancer stem cells (CSCs), are uniquely capable of tumor maintenance and hierarchical differentiation into multiple tumor cell lineages.117–119 CSCs have been proposed as a cause of therapeutic resistance and tumor recurrence.120 Their implications in astrocytoma biology and drug development have been previously reviewed in detail.121–125
GBM subtypes may have distinct treatment responses.64 Although transcriptome profiling was recently evaluated in the trial of bevacizumab in combination with standard-of-care therapy for newly diagnosed GBM,99,102 it remains unclear how this measure of intertumoral genomic heterogeneity should be incorporated in future clinical trials. It is likely that novel drug efficacy will be restricted to specific molecular subtypes with unique mutational, epigenetic, or CSC profiles. However, the ideal molecular diagnostic approach to prospectively identify likely responders has yet to be developed. How should molecular profiles be incorporated into clinical trial design to account for the heterogeneity present in astrocytomas? In the absence of definitive data on their prognostic and predictive significance, a prudent approach would be to retrospectively characterize genomic heterogeneity in clinical trial specimens on as many molecular levels as is economically feasible. An even more cost-effective approach may be to conduct preclinical drug studies, either before or parallel with clinical trials, using biologically diverse panels of contemporary murine models that have also been comprehensively profiled.
Conventional and Contemporary Murine Models of Astrocytomas
Like diagnosis and therapy, the last 2 decades have witnessed major improvements in preclinical modeling of astrocytomas (Figs. 2 and 3). A number of recent reviews have described these improvements in detail.13–24 Here we compare contemporary with conventional ECL models and discuss how they may be utilized to improve preclinical astrocytoma drug development.
Established Cell Line Models
Established cell lines (ECLs) were cultured from rodent astrocytomas induced by chemical mutagenesis and transformed the preclinical drug development landscape in the late 1960s (Fig. 3A).13,19,31–38 These technically straightforward, highly penetrant models were widely disseminated and developed tumors with rapid, uniform growth kinetics and short latency when transplanted subcutaneously or orthotopically into the brains of murine hosts (Table 2). Many recapitulated the histopathological features of human high-grade astrocytomas, including their diffuse invasion of normal brain. Orthotopic murine allograft models were particularly attractive for the development of drugs targeting tumor-stroma interactions or immunomodulatory therapies because tumorigenesis could be induced in the native brain microenvironment in immunocompetent, syngeneic mice.13,19
Table 2.
Comparison of conventional and contemporary murine astrocytoma models
| Characteristic |
Conventional ECL Models |
Contemporary Models |
|||||
|---|---|---|---|---|---|---|---|
| Mouse Allografts |
Human Xenografts |
Human Xenografts |
Engineered Mice |
||||
| PDX | geHC | GEM | nGEM | ||||
| Host | Intact immune system | Ya | Y | Y | |||
| Faithful microenvironment | Y | Y | Y | ||||
| Intact DNA repair | Y | Yb | Yb | Yb | Y | Y | |
| Host and tumor genomes differ | Y | Y | Y | ||||
| Tumor | In vitro culture possible | Y | Y | Y | Y | Y | |
| Subcutaneous growth | Y | Y | Y | Y | Y | ||
| Orthotopic growth | Y | Y | Y | Y | Y | Y | |
| Histologically faithful | Yc | Y | Y | Y | Y | ||
| Rapid growth kinetics | Y | Y | Y | Y | d | d | |
| High penetrance | Y | Y | Y | Y | d | d | |
| Short latency | Y | Y | Y | Y | d | d | |
| Defined oncogenic mutations | e | e | e | Y | Y | Y | |
| Straightforward genotype-phenotype comparisons | Y | Y | Y | ||||
| Complex genome landscapes | Yf | Y | Y | Yf | |||
| Defined cellular origin | Y | Yg | Y | ||||
| Low grade astrocytomas develop | h | Y | Y | ||||
| Stochastic malignant progression | Y | ||||||
aSome murine ECL are immunogenic and xenografting requires immunodeficient hosts.19
bImmunodeficient severe combined immunodeficiency, but not nude mice have genetic DNA repair defects.187
cSome murine ECL fail to invade normal brain.19
dGrowth kinetics, penetrance, and latency in GEM and nGEM models vary greatly depending on oncogenic mutations and targeted cell type.
eMutational profiles can be defined by genomic analyses, but genomic complexity renders direct genotype-phenotype correlations difficult.
fComplex gene rearrangements occur less frequently in murine compared to human tumors.
gConventional knockout GEM models do not have a defined cellular origin.
Xenograft models using ECLs cultured from human astrocytomas, which were developed from the late 1960s through the 1980s, shared many of the attractive features of murine allograft models. Because ECLs originated from human tumors, extrapolation of experimental results were uncomplicated by molecular and physiological differences between mice and humans. However, many failed to recapitulate the brain invasive histopathology of human astrocytomas.20,126,127 The requirement for immunodeficient hosts also rendered examination of microenvironmental and immune influences on drug response impossible.
Comprehensive genomic analyses of both murine and human ECL models identified a number of additional limitations. Phenotypic and genotypic drift due to clonal selection upon serial culture of adherent cells in serum-containing media rendered ECLs markedly different from their original tumor.19 ECLs cultured under these nonphysiological conditions adapted to the presence of abundant nutrients by increasing metabolic and proliferation pathways and to their artificial microenvironment by decreasing cell adhesion. Thus, ECLs frequently developed uncharacteristic and complex chromosomal abnormalities, and their molecular profiles differed significantly from acutely isolated GBM samples.128–131 Although genomic analyses have defined the mutational landscapes of many astrocytoma ECLs, their abundance of mutations and complex chromosomal alterations render genotype-phenotype comparisons difficult.
Subcutaneous ECL models remain a popular method of assessing both in vivo tumorigenesis and drug efficacy due to the technical ease of monitoring growth kinetics in this anatomic compartment. Nevertheless, these models fail to account for native microenvironmental influences on tumor pathogenesis and drug response and do not accurately model PK effects of the BBB. Targeted agents, such as palbociclib, that show efficacy in subcutaneous xenografts have failed when tested in orthotopic models due to drug efflux pumps at the BBB.132 The molecular profiles of ECLs xenografted subcutaneously differ markedly from corresponding orthotopic xenografts.133 Tumor location can also significantly impact molecular and biological responses to cytotoxic therapies such as radiation.134 Because subcutaneous ECL models can overestimate the therapeutic potential of novel agents, their use should be restricted to validation of therapeutic targets in biological proof-of-principle experiments, and their role in prioritizing drugs for clinical investigation should be minimized.
Patient-derived Xenograft Models
Many of the shortcomings of ECL models can be directly attributed to their serial culture as adherent cells in serum-containing medium. Some of these have subsequently been overcome through development of PDX (Fig. 3B), whereby fresh tumor fragments are directly injected into the brain or serially passaged subcutaneously in immunodeficient mice. Alternatively, PDX can be cultured as nonadherent spheroids in growth factor-defined, serum-free medium prior to orthotopic transplantation.15,19,135 Development of these techniques has been critical for defining the functional heterogeneity present in human astrocytoma cells and exploring the biological and therapeutic implications of the CSC hypothesis in these tumors.121–125
PDXs share many of the advantages of ECL models for preclinical drug development including high penetrance, short latency, and rapid, uniform growth kinetics in vivo. However, unlike ECL, PDXs maintain the genomic features of the tumors from which they were derived and faithfully recapitulate the molecular profiles and histopathological features of GBM, including diffuse brain invasion.19,136–141 Although their cellular origin is undefined and their genomic complexity renders elucidating the phenotypic consequences of individual oncogenic mutations difficult when studied in multimodel panels, PDXs more broadly recapitulate the intertumoral genomic heterogeneity evident in GBM.137 As such, systematic drug screening in these models has the potential to more accurately reflect clinical activity of novel drugs and more readily identify predictive molecular characteristics. The multi-institutional Ivy Genomics-based Medicine Project is currently utilizing this approach to investigate both novel and conventional cytotoxic agents and develop predictive biomarkers in a genomically diverse panel of PDX models.18,97
Genetically Engineered Human Cell Models
Models using genetically engineered normal human brain cells (geHCs) have been recently developed to overcome some of the limitations of human astrocytoma ECL and PDX models (Fig. 3C). These models were generated by purifying specific cell types, such as astrocytes or neural stem cells, from normal human brains and using standard molecular biology techniques to engineer their expression of specific oncogenic mutations.142–145 By virtue of their design, these genetically defined models permit direct determination of the phenotypic consequences of astrocytoma-associated mutations in specific neural cell types. Their serial culture in vitro is generally unaccompanied by additional genomic abnormalities.144 While penetrance, growth kinetics, and latency vary based on mutations and cellular origin, many geHC models give rise to diffusely infiltrative astrocytomas when orthotopically injected into the brains of immunodeficient mice.
Genetically Engineered Mouse Models
Genetically engineered mouse (GEM) models revolutionized basic cancer research in the 1990s. Over the past 2 decades, dozens of astrocytoma GEM models have been developed to dissect the genetics of de novo tumorigenesis in the native brain microenvironment (Fig. 3D).19,20 Because knockout and transgenic GEMs harbored engineered mutations in all cell types, embryonic lethality precluded study of genes critical for development.20 GEMs that utilized conditional alleles were subsequently developed to overcome this limitation and to spatially restrict oncogenic mutations to defined cell types within the brain. Conditional, inducible, and somatic gene transfer GEM models, including the RCAS-tva system, were designed to facilitate temporal as well as spatial control of mutations. The value of these models in basic astrocytoma research has been reviewed extensively elsewhere.16–24 Here we focus on their use in astrocytoma drug development.
A number of factors inherent in the design of astrocytoma GEM models has limited their use in preclinical drug development, particularly for studies evaluating drug efficacy by conventional clinical endpoints (eg, radiographic response and overall survival).146–149 Whereas single oncogenic alleles are sufficient to induce tumorigenesis in medulloblastoma models, multiple mutations are typically required to induce astrocytoma tumorigenesis in GEMs.20,150,151 Conditional GEM models require complex and often inefficient breeding schemes to generate sufficiently large cohorts for preclinical drug studies. RCAS/tva GEMs are much more amenable because multiple predefined, oncogenic alleles can be simultaneously introduced into specific neural cell types using a single transgenic mouse line engineered to express tva receptors.20,152 However, because RCAS retroviral vectors are required, these models are limited to the transformation of endogenously proliferative cell types.152 High-grade astrocytoma tumorigenesis typically occurs with variable penetrance after relatively long periods of latency in these model systems. Moreover, GBMs develop in a temporally heterogeneous, stochastic manner. Therefore, the presence and location of tumors in individual mice must be confirmed by radiographic imaging prior to treatment initiation and intermittently thereafter to monitor drug response in vivo.20,152–154 Taken together, these features make preclinical drug studies in astrocytoma GEMs long, cumbersome, and expensive.
Non-germline Genetically Engineered Mouse Models
Non-germline GEM (nGEM) models overcome many of the limitations of GEMs and may be more amenable to preclinical drug studies. Like geHC, nGEM models utilize cultures of specific cell types harvested from GEM brains, including astrocytes and neural stem cells (Fig. 3D).127,155–159 They harbor defined genetic mutations, and their serial culture is generally unaccompanied by additional genomic abnormalities (unpublished observations). While penetrance and latency vary with mutations and cellular origin, nGEM astrocytomas developed with uniform growth kinetics in vivo when injected into syngeneic, immunocompetent hosts, precluding the need for radiographic screening.127 Like ECLs, PDXs, and geHCs, these cells can be readily modified genetically to express luminescent proteins that facilitate monitoring of disease burden and drug response with bioluminescence imaging.53,160 Both geHC and nGEM models enable direct determination of genotype-phenotype relationships. Unlike ECLs and PDXs, these models can be used to define the oncogenic roles of single mutations and the cooperative roles of multiple mutations during tumorigenesis. They are thus uniquely suited for the systematic validation of putative oncogenic driver mutations identified in large-scale genome characterization projects.161 Because geHC and nGEM grow in vitro and in vivo and feature defined genomic landscapes, these models can also be used to unambiguously define the genetics of drug response and resistance.
nGEMs may be useful for dissecting the role of individual mutations and their cellular origin in generating genomic diversity of human astrocytomas. As such, subtype-specific nGEM models of GBM may be developed for drug development. We have recently published a nGEM model derived from G1/S defective astrocytes with activated MAPK and PI3K signaling that molecularly mimics proneural human GBM.64,127 However, in contrast to ECL, PDX, and geHC models, nGEM models utilize syngeneic, immunocompetent hosts and may be useful in development of drugs targeting tumor-stroma interactions or immunomodulatory therapies.
Modeling Low-grade Astrocytomas
Despite advances in modeling techniques, murine models that mimic the natural history of low-grade astrocytomas (WHO grade II) in humans have been difficult to develop. These tumors generally fail to become established when cultured in vitro or grow when transplanted into immunodeficient mice. In fact, in vivo tumorigenesis has long been known to correlate with histological grade and poor prognosis.162 Thus, human ECL and PDX models of low-grade astrocytomas are virtually nonexistent. Their absence has significantly impeded study of the genetics of malignant progression and the development of effective drugs for these tumors. However, several GEM models have recently been described that develop as clinically silent but histopathologically detectable low-grade astrocytomas.154,163 These tumors progressively expand over time, spontaneously acquire additional mutations, and undergo malignant progression to lethal, high-grade disease.164 Despite their initiation by a limited number of oncogenic mutations, malignant progression in these GEMs results in GBMs with transcriptomes that recapitulate the full spectrum of human subtypes.153,154 Moreover, we have genomically characterized multiple, spatially distinct GBMs that developed in different brain regions of individual mice and found that their genomic landscapes differed, suggesting that divergent genetic evolution occurs in these models.164 These GEM models therefore may be uniquely suited for defining the genetics of malignant progression, the prognostic impact of TMZ-induced hypermutation in low grade astrocytomas,112,165 and the development of novel treatments to prevent or delay their progression.
IDH1 and IDH2 mutations are subtype-defining genetic features of lower-grade astrocytomas.107,109 However, preclinical models for the development of IDH-mutant astrocytoma therapies are scarce. Most studies published to date have used stably transfected ECLs or geHCs to investigate the biological effects of IDH mutations in vitro.166–174 Adherent serum cultures of IDH-mutant astrocytomas have been shown to lose the mutant allele upon serial passage, and mutant-containing clones have failed to become ECLs.175 In contrast, 4 IDH1R132H-mutant anaplastic gliomas (WHO grade III) have been successfully cultured as neurospheres in vitro.176–179 However, only 2 of these, both from anaplastic oligodendroglial neoplasms, formed serially transplantable gliomas in the immunodeficient mouse brain. Thus, only 2 potential PDX models are currently available for preclinical IDH-mutant glioma drug development.178,179
The initial attempt to develop a GEM model of IDH-mutant gliomas was also disappointing.180 Conditional activation of a heterozygous, floxed IDH1R132H mutant allele using Nestin-cre or Gfap-cre drivers failed to elicit tumorigenesis in the developing mouse brain, despite production of the oncometabolite D-2-hydroxyglutarate (D2HG). Rather, D2HG blocked collagen maturation and altered vascular basement membranes, leading to brain hemorrhage and embryonic lethality. These results suggest that IDH1 mutations alone are not sufficient to induce tumorigenesis, at least in the developing mouse brain. However, the effects of IDH1R132H have not been examined in the adult brain. It therefore remains possible that temporal control of IDH1R132H induction in the adult mouse brain using drug-inducible Cre drivers may be more successful in modeling IDH-mutant gliomas.
Alternatively, successful culture and xenografting of IDH-mutant human gliomas as well as GEM modeling may require the presence of cooperative oncogenic mutations. geHCs and nGEMs represent attractive model systems for exploring this hypothesis. In this regard, the IDH1R132H mutation has been shown to impair histone demethylation in immortalized normal human astrocytes.173 IDH1R132H also remodeled the DNA methylome of these cells and was sufficient to induce the glioma CpG island methylator (G-CIMP) phenotype. Moreover, IDH1R132H blocked astrocytic differentiation in neurosphere cultures of neural stem cells harvested from neonatal Ink4a/Arf null GEMs.174 When engineered to express mutations in the RAS-MAPK and PI3K pathways, immortalized human and murine astrocytes have been shown to induce tumorigenesis upon transplantation into mouse brains.127,143,144,181 Thus, IDH-mutant geHC or nGEM brain cells with additional engineered mutations may represent promising preclinical systems for development of IDH targeted therapies.
Contemporary Murine Models in Preclinical Astrocytoma Drug Development
Murine models can address a number of issues important in the development of clinical drugs for astrocytomas. These include validating molecular targets, defining the role of cellular origin in drug response, prioritizing drugs for clinical development, and developing predictive markers to identify potential responders (Table 3). The ideal model(s) to address these issues differs based on the inherent strengths and weaknesses of their design.
Table 3.
The role of contemporary murine models in preclinical astrocytoma drug development
| Clinical Issue | Role of Preclinical Models | Ideal Model(s) | References |
|---|---|---|---|
| Target validation | Define role in tumorigenesis | GEM, nGEM, geHC | 127,143,144,154,188 |
| Cellular origin | Define role in tumorigenesis | GEM, nGEM, geHC | 127,143,145,153,154,189,190 |
| Drug prioritization | Characterize CNS penetration | PDX, GEM | 132,183 |
| Define effective dose and schedule | PDX, nGEM, geHC | 53,60,89 | |
| Define resistance mechanisms | PDX, nGEM, geHC | 82,191,192 | |
| Test combination therapies | PDX, nGEM, geHC | 95,156,193–195 | |
| Patient selection | Developing predictive biomarkers | PDX, nGEM | 53–55,59,60,68,69,94–96 |
GEMs have established roles in the validation of molecular targets, particularly in defining the role of putative oncogenic drivers in the initiation and progression of tumorigenesis. Because of their genetic tractability, nGEM and geHC models are poised to supplement GEMs in future target validation efforts. Indeed, we utilized nGEM models to establish that cooperation between MAPK and PI3K signaling is required for GBM pathogenesis in vivo, suggesting that simultaneous inhibition of both pathways may be required for effective therapeutic design.127 Moreover, geHC models with human astrocytes proved critical in defining the effects of IDH mutations on the epigenetic control of tumor cell differentiation.173
In addition to target validation, GEM, nGEM, and geHC models may be useful for defining the impact of cellular origin on astrocytoma tumorigenesis and drug sensitivity.20 Like astrocytomas, multiple genomic subtypes of medulloblastoma with distinct mutations exist.182 GEM models have shown that different oncogenic mutations in specific cells of origin in the developing mouse cerebellum lead to distinct genomic subtypes of medulloblastoma that mimic their human counterparts. GEM models of sonic hedgehog-associated medulloblastoma in particular are currently being utilized for the preclinical evaluation of subtype-specific targeted therapies.150
PDX and GEM models with defective ABC transporters have established roles in characterizing CNS PKs.132,183 The advantages of PDX, nGEM, and geHC over ECL models promise to replace their use in defining dose and schedule dependencies of combination therapies for GBM.89 Due to the redundancies of RTK signaling, mosaic amplification of multiple RTKs in GBM and significant inter- and intratumoral molecular heterogeneity, monotherapy with single targeted agents will likely prove ineffective for GBM.105 Increasing use of contemporary PDX, nGEM, and geHC models in preclinical development of these agents is likely to aid in defining and further characterizing these mechanisms of drug resistance. Because these cells can be cultured in vitro, synthetic lethality screens or kinome profiling promise to aid definition of rational combination therapies to combat drug resistance.184 Moreover, more systematic use of multiple genomically diverse models in preclinical drug efficacy screens promises to aid development of predictive genomic biomarkers.18,97
Conclusion
Despite decades of research, the therapeutic armamentarium of approved drugs for astrocytomas remains limited. The field of neuro-oncology has yet to benefit from the accelerated pace of oncology drug development due to issues of prevalence, trial participation, and biological complexity of the disease. However, comprehensive genomic characterization and changes in clinical trial design promise to improve disease classification and increase the number of targeted agents that can be clinically evaluated. Improvements in preclinical murine models and their systematic integration by the neuro-oncology community during early drug development promises to further accelerate therapeutic advances for these devastating malignancies.
Funding
CRM is a Damon Runyon-Genentech Clinical Investigator supported by a Clinical Investigator Award from the Damon Runyon Cancer Research Foundation (CI-45-09). This work was supported by grants from the UNC University Cancer Research Fund and the Department of Defense (W81XWH-09-2-0042). RSM is a Robert H. Wagner Scholar in the Molecular and Cellular Pathology Graduate Program and the Howard Hughes Medical Institute-supported Graduate Training Program in Translational Medicine.
Acknowledgments
Conflict of interest statement. The authors declare no conflicts of interest.
References
- 1.DiMasi JA, Grabowski HG. Economics of new oncology drug development. J Clin Oncol. 2007;25(2):209–216. doi: 10.1200/JCO.2006.09.0803. [DOI] [PubMed] [Google Scholar]
- 2.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5(9):741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 3.Mendelsohn J. Personalizing oncology: perspectives and prospects. J Clin Oncol. 2013;31(15):1904–1911. doi: 10.1200/JCO.2012.45.3605. [DOI] [PubMed] [Google Scholar]
- 4.Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl 2):ii1–i56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey P, Cushing H. A Classification of the Tumors of the Glioma Group on a Histogenetic Basis With a Correlated Study of Prognosis. Philadelphia: J.B. Lippincott; 1926. [Google Scholar]
- 6.Miller CR, Perry A. Glioblastoma. Arch Pathol Lab Med. 2007;131(3):397–406. doi: 10.5858/2007-131-397-G. [DOI] [PubMed] [Google Scholar]
- 7.Louis DN, Ohgaki H, Wiestler OD, et al. WHO Classification of Tumours of the Central Nervous System. 4th ed. Lyon: IARC; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riemenschneider MJ, Louis DN, Weller M, et al. Refined brain tumor diagnostics and stratified therapies: the requirement for a multidisciplinary approach. Acta Neuropathol. 2013;126(1):21–37. doi: 10.1007/s00401-013-1127-4. [DOI] [PubMed] [Google Scholar]
- 9.Vitucci M, Hayes DN, Miller CR. Gene expression profiling of gliomas: merging genomic and histopathological classification for personalised therapy. Br J Cancer. 2011;104(4):545–553. doi: 10.1038/sj.bjc.6606031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang SM, Barker FG, 2nd, Schmidt MH, et al. Clinical trial participation among patients enrolled in the Glioma Outcomes Project. Cancer. 2002;94(10):2681–2687. doi: 10.1002/cncr.10536. [DOI] [PubMed] [Google Scholar]
- 11.Trippa L, Lee EQ, Wen PY, et al. Bayesian adaptive randomized trial design for patients with recurrent glioblastoma. J Clin Oncol. 2012;30(26):3258–3263. doi: 10.1200/JCO.2011.39.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander BM, Wen PY, Trippa L, et al. Biomarker-based adaptive trials for patients with glioblastoma--lessons from I-SPY 2. Neuro Oncol. 2013;15(8):972–978. doi: 10.1093/neuonc/not088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barth RF, Kaur B. Rat brain tumor models in experimental neuro-oncology: the C6, 9L, T9, RG2, F98, BT4C, RT-2 and CNS-1 gliomas. J Neurooncol. 2009;94(3):299–312. doi: 10.1007/s11060-009-9875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Candolfi M, Curtin JF, Nichols WS, et al. Intracranial glioblastoma models in preclinical neuro-oncology: neuropathological characterization and tumor progression. J Neurooncol. 2007;85(2):133–148. doi: 10.1007/s11060-007-9400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garber K. From human to mouse and back: ‘tumorgraft’ models surge in popularity. J Natl Cancer Inst. 2009;101(1):6–8. doi: 10.1093/jnci/djn481. [DOI] [PubMed] [Google Scholar]
- 16.de Vries NA, Beijnen JH, van Tellingen O. High-grade glioma mouse models and their applicability for preclinical testing. Cancer Treat Rev. 2009;35(8):714–723. doi: 10.1016/j.ctrv.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Fomchenko EI, Holland EC. Mouse models of brain tumors and their applications in preclinical trials. Clin Cancer Res. 2006;12(18):5288–5297. doi: 10.1158/1078-0432.CCR-06-0438. [DOI] [PubMed] [Google Scholar]
- 18.Gutmann DH, Stiles CD, Lowe SW, et al. Report from the fifth National Cancer Institute Mouse Models of Human Cancers Consortium Nervous System Tumors Workshop. Neuro Oncol. 2011;13(7):692–699. doi: 10.1093/neuonc/nor080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huszthy PC, Daphu I, Niclou SP, et al. In vivo models of primary brain tumors: pitfalls and perspectives. Neuro Oncol. 2012;14(8):979–993. doi: 10.1093/neuonc/nos135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmid RS, Vitucci M, Miller CR. Genetically engineered mouse models of diffuse gliomas. Brain Res Bull. 2012;88(1):72–79. doi: 10.1016/j.brainresbull.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149(1):36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hambardzumyan D, Parada LF, Holland EC, et al. Genetic modeling of gliomas in mice: new tools to tackle old problems. Glia. 2011;59(8):1155–1168. doi: 10.1002/glia.21142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huse JT, Holland EC. Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathol. 2009;19(1):132–143. doi: 10.1111/j.1750-3639.2008.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westphal M, Lamszus K. The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nat Rev Neurosci. 2011;12(9):495–508. doi: 10.1038/nrn3060. [DOI] [PubMed] [Google Scholar]
- 25.Walker MD, Alexander E, Jr., Hunt WE, et al. Evaluation of BCNU and/or radiotherapy in the treatment of anaplastic gliomas. A cooperative clinical trial. J Neurosurg. 1978;49(3):333–343. doi: 10.3171/jns.1978.49.3.0333. [DOI] [PubMed] [Google Scholar]
- 26.Weir B, Band P, Urtasun R, et al. Radiotherapy and CCNU in the treatment of high-grade supratentorial astrocytomas. J Neurosurg. 1976;45(2):129–134. doi: 10.3171/jns.1976.45.2.0129. [DOI] [PubMed] [Google Scholar]
- 27.Stewart LA. Chemotherapy in adult high-grade glioma: a systematic review and meta-analysis of individual patient data from 12 randomised trials. Lancet. 2002;359(9311):1011–1018. doi: 10.1016/s0140-6736(02)08091-1. [DOI] [PubMed] [Google Scholar]
- 28.Sausville EA, Burger AM. Contributions of human tumor xenografts to anticancer drug development. Cancer Res. 2006;66(7):3351–3354. doi: 10.1158/0008-5472.CAN-05-3627. discussion 3354. [DOI] [PubMed] [Google Scholar]
- 29.Dykes DJ, Waud WR. Murine L1210 and P388 leukemias. In: Teicher BA, editor. Tumor Models in Cancer Research. Totowa, NJ: Humana Press; 2011. pp. 23–41. [Google Scholar]
- 30.Venditti JM, Kline I, Goldin A. Evaluation of antileukemic agents employing advanced leukemia L1210 in mice. 8. Cancer Res. 1964;24:827–879. [PubMed] [Google Scholar]
- 31.Zimmerman HM, Arnold H. Experimental brain tumors I. Tumors produced with methylcholanthrene. Cancer Res. 1941;1(12):919–938. [Google Scholar]
- 32.Ausman JI, Shapiro WR, Rall DP. Studies on the chemotherapy of experimental brain tumors: development of an experimental model. Cancer Res. 1970;30(9):2394–2400. [PubMed] [Google Scholar]
- 33.Barker M, Hoshino T, Gurcay O, et al. Development of an animal brain tumor model and its response to therapy with 1,3-bis(2-chloroethyl)-1-nitrosourea. Cancer Res. 1973;33(5):976–986. [PubMed] [Google Scholar]
- 34.Benda P, Lightbody J, Sato G, et al. Differentiated rat glial cell strain in tissue culture. Science. 1968;161(3839):370–371. doi: 10.1126/science.161.3839.370. [DOI] [PubMed] [Google Scholar]
- 35.Ponten J, Macintyre EH. Long term culture of normal and neoplastic human glia. Acta Pathol Microbiol Scand. 1968;74(4):465–486. doi: 10.1111/j.1699-0463.1968.tb03502.x. [DOI] [PubMed] [Google Scholar]
- 36.Giard DJ, Aaronson SA, Todaro GJ, et al. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51(5):1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 37.Owens RB, Smith HS, Nelson-Rees WA, et al. Epithelial cell cultures from normal and cancerous human tissues. J Natl Cancer Inst. 1976;56(4):843–849. doi: 10.1093/jnci/56.4.843. [DOI] [PubMed] [Google Scholar]
- 38.Bigner SH, Bullard DE, Pegram CN, et al. Relationship of in vitro morphologic and growth characteristics of established human glioma-derived cell lines to their tumorigenicity in athymic nude mice. J Neuropathol Exp Neurol. 1981;40(4):390–409. doi: 10.1097/00005072-198107000-00004. [DOI] [PubMed] [Google Scholar]
- 39.Rygaard J, Povlsen CO. Heterotransplantation of a human malignant tumour to “Nude” mice. Acta Pathol Microbiol Scand. 1969;77(4):758–760. doi: 10.1111/j.1699-0463.1969.tb04520.x. [DOI] [PubMed] [Google Scholar]
- 40.Amarasingh S, Macleod MR, Whittle IR. What is the translational efficacy of chemotherapeutic drug research in neuro-oncology? A systematic review and meta-analysis of the efficacy of BCNU and CCNU in animal models of glioma. J Neurooncol. 2009;91(2):117–125. doi: 10.1007/s11060-008-9697-z. [DOI] [PubMed] [Google Scholar]
- 41.Stevens MF, Hickman JA, Langdon SP, et al. Antitumor activity and pharmacokinetics in mice of 8-carbamoyl-3-methyl-imidazo[5,1-d]-1,2,3,5-tetrazin-4(3H)-one (CCRG 81045; M & B 39831), a novel drug with potential as an alternative to dacarbazine. Cancer Res. 1987;47(22):5846–5852. [PubMed] [Google Scholar]
- 42.Newlands ES, Blackledge GR, Slack JA, et al. Phase I trial of temozolomide (CCRG 81045: M&B 39831: NSC 362856) Br J Cancer. 1992;65(2):287–291. doi: 10.1038/bjc.1992.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Reilly SM, Newlands ES, Glaser MG, et al. Temozolomide: a new oral cytotoxic chemotherapeutic agent with promising activity against primary brain tumours. Eur J Cancer. 1993;29A(7):940–942. doi: 10.1016/s0959-8049(05)80198-4. [DOI] [PubMed] [Google Scholar]
- 44.Bower M, Newlands ES, Bleehen NM, et al. Multicentre CRC phase II trial of temozolomide in recurrent or progressive high-grade glioma. Cancer Chemother Pharmacol. 1997;40(6):484–488. doi: 10.1007/s002800050691. [DOI] [PubMed] [Google Scholar]
- 45.Stupp R, Dietrich PY, Ostermann Kraljevic S, et al. Promising survival for patients with newly diagnosed glioblastoma multiforme treated with concomitant radiation plus temozolomide followed by adjuvant temozolomide. J Clin Oncol. 2002;20(5):1375–1382. doi: 10.1200/JCO.2002.20.5.1375. [DOI] [PubMed] [Google Scholar]
- 46.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 47.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 48.Plowman J, Waud WR, Koutsoukos AD, et al. Preclinical antitumor activity of temozolomide in mice: efficacy against human brain tumor xenografts and synergism with 1,3-bis(2-chloroethyl)-1-nitrosourea. Cancer Res. 1994;54(14):3793–3799. [PubMed] [Google Scholar]
- 49.Hirst TC, Vesterinen HM, Sena ES, et al. Systematic review and meta-analysis of temozolomide in animal models of glioma: was clinical efficacy predicted? Br J Cancer. 2013;108(1):64–71. doi: 10.1038/bjc.2012.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Margison GP, Santibanez Koref MF, Povey AC. Mechanisms of carcinogenicity/chemotherapy by O6-methylguanine. Mutagenesis. 2002;17(6):483–487. doi: 10.1093/mutage/17.6.483. [DOI] [PubMed] [Google Scholar]
- 51.Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J Clin Oncol. 2002;20(9):2388–2399. doi: 10.1200/JCO.2002.06.110. [DOI] [PubMed] [Google Scholar]
- 52.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 53.Dinca EB, Sarkaria JN, Schroeder MA, et al. Bioluminescence monitoring of intracranial glioblastoma xenograft: response to primary and salvage temozolomide therapy. J Neurosurg. 2007;107(3):610–616. doi: 10.3171/JNS-07/09/0610. [DOI] [PubMed] [Google Scholar]
- 54.Kitange GJ, Carlson BL, Mladek AC, et al. Evaluation of MGMT promoter methylation status and correlation with temozolomide response in orthotopic glioblastoma xenograft model. J Neurooncol. 2009;92(1):23–31. doi: 10.1007/s11060-008-9737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carlson BL, Grogan PT, Mladek AC, et al. Radiosensitizing effects of temozolomide observed in vivo only in a subset of O6-methylguanine-DNA methyltransferase methylated glioblastoma multiforme xenografts. Int J Radiat Oncol Biol Phys. 2009;75(1):212–219. doi: 10.1016/j.ijrobp.2009.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wick W, Platten M, Weller M. New (alternative) temozolomide regimens for the treatment of glioma. Neuro Oncol. 2009;11(1):69–79. doi: 10.1215/15228517-2008-078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tolcher AW, Gerson SL, Denis L, et al. Marked inactivation of O6-alkylguanine-DNA alkyltransferase activity with protracted temozolomide schedules. Br J Cancer. 2003;88(7):1004–1011. doi: 10.1038/sj.bjc.6600827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilbert MR, Wang M, Aldape KD, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. J Clin Oncol. 2013;31(32):4085–4091. doi: 10.1200/JCO.2013.49.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kitange GJ, Carlson BL, Schroeder MA, et al. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro Oncol. 2009;11(3):281–291. doi: 10.1215/15228517-2008-090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cen L, Carlson BL, Pokorny JL, et al. Efficacy of protracted temozolomide dosing is limited in MGMT unmethylated GBM xenograft models. Neuro Oncol. 2013;15(6):735–746. doi: 10.1093/neuonc/not010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2(87):re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 63.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guillamo JS, Leuraud P, De Bouard S, et al. Anti-proliferative and anti-invasive EGFR amplification dependent and anti-angiogenic EGFR independent activity of ZD1839 (‘Iressa’) tyrosine kinase inhibitor on human glioblastomas [abstract] Proc Am Assoc Cancer Res. 2003;44:1009. [Google Scholar]
- 66.Vogelbaum MA, Goldlust S, Kanner A. The EGFR tyrosine kinase inhibitor Tarceva (OSI-774) shows activity against both wild-type and mutant EGFR function [abstract] Neuro Oncol. 2003;5(4):309. [Google Scholar]
- 67.Learn CA, Hartzell TL, Wikstrand CJ, et al. Resistance to tyrosine kinase inhibition by mutant epidermal growth factor receptor variant III contributes to the neoplastic phenotype of glioblastoma multiforme. Clin Cancer Res. 2004;10(9):3216–3224. doi: 10.1158/1078-0432.ccr-03-0521. [DOI] [PubMed] [Google Scholar]
- 68.Sarkaria JN, Yang L, Grogan PT, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6(3):1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 69.Guillamo JS, de Bouard S, Valable S, et al. Molecular mechanisms underlying effects of epidermal growth factor receptor inhibition on invasion, proliferation, and angiogenesis in experimental glioma. Clin Cancer Res. 2009;15(11):3697–3704. doi: 10.1158/1078-0432.CCR-08-2042. [DOI] [PubMed] [Google Scholar]
- 70.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10(11):760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rich JN, Reardon DA, Peery T, et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol. 2004;22(1):133–142. doi: 10.1200/JCO.2004.08.110. [DOI] [PubMed] [Google Scholar]
- 72.Krishnan S, Brown PD, Ballman KV, et al. Phase I trial of erlotinib with radiation therapy in patients with glioblastoma multiforme: results of North Central Cancer Treatment Group protocol N0177. Int J Radiat Oncol Biol Phys. 2006;65(4):1192–1199. doi: 10.1016/j.ijrobp.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 73.Prados MD, Lamborn KR, Chang S, et al. Phase 1 study of erlotinib HCl alone and combined with temozolomide in patients with stable or recurrent malignant glioma. Neuro Oncol. 2006;8(1):67–78. doi: 10.1215/S1522851705000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Franceschi E, Cavallo G, Lonardi S, et al. Gefitinib in patients with progressive high-grade gliomas: a multicentre phase II study by Gruppo Italiano Cooperativo di Neuro-Oncologia (GICNO) Br J Cancer. 2007;96(7):1047–1051. doi: 10.1038/sj.bjc.6603669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brandes AA, Franceschi E, Tosoni A, et al. Epidermal growth factor receptor inhibitors in neuro-oncology: hopes and disappointments. Clin Cancer Res. 2008;14(4):957–960. doi: 10.1158/1078-0432.CCR-07-1810. [DOI] [PubMed] [Google Scholar]
- 76.Brown PD, Krishnan S, Sarkaria JN, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J Clin Oncol. 2008;26(34):5603–5609. doi: 10.1200/JCO.2008.18.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prados MD, Yung WK, Wen PY, et al. Phase-1 trial of gefitinib and temozolomide in patients with malignant glioma: a North American brain tumor consortium study. Cancer Chemother Pharmacol. 2008;61(6):1059–1067. doi: 10.1007/s00280-007-0556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prados MD, Chang SM, Butowski N, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27(4):579–584. doi: 10.1200/JCO.2008.18.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peereboom DM, Shepard DR, Ahluwalia MS, et al. Phase II trial of erlotinib with temozolomide and radiation in patients with newly diagnosed glioblastoma multiforme. J Neurooncol. 2010;98(1):93–99. doi: 10.1007/s11060-009-0067-2. [DOI] [PubMed] [Google Scholar]
- 80.Uhm JH, Ballman KV, Wu W, et al. Phase II evaluation of gefitinib in patients with newly diagnosed Grade 4 astrocytoma: Mayo/North Central Cancer Treatment Group Study N0074. Int J Radiat Oncol Biol Phys. 2011;80(2):347–353. doi: 10.1016/j.ijrobp.2010.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353(19):2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 82.Vivanco I, Robins HI, Rohle D, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2(5):458–471. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Togashi Y, Masago K, Masuda S, et al. Cerebrospinal fluid concentration of gefitinib and erlotinib in patients with non-small cell lung cancer. Cancer Chemother Pharmacol. 2012;70(3):399–405. doi: 10.1007/s00280-012-1929-4. [DOI] [PubMed] [Google Scholar]
- 84.de Vries NA, Buckle T, Zhao J, et al. Restricted brain penetration of the tyrosine kinase inhibitor erlotinib due to the drug transporters P-gp and BCRP. Invest New Drugs. 2012;30(2):443–449. doi: 10.1007/s10637-010-9569-1. [DOI] [PubMed] [Google Scholar]
- 85.Thiessen B, Stewart C, Tsao M, et al. A phase I/II trial of GW572016 (lapatinib) in recurrent glioblastoma multiforme: clinical outcomes, pharmacokinetics and molecular correlation. Cancer Chemother Pharmacol. 2010;65(2):353–361. doi: 10.1007/s00280-009-1041-6. [DOI] [PubMed] [Google Scholar]
- 86.MacDonald TJ, Taga T, Shimada H, et al. Preferential susceptibility of brain tumors to the antiangiogenic effects of an alpha(v) integrin antagonist. Neurosurgery. 2001;48(1):151–157. doi: 10.1097/00006123-200101000-00026. [DOI] [PubMed] [Google Scholar]
- 87.Nabors LB, Mikkelsen T, Rosenfeld SS, et al. Phase I and correlative biology study of cilengitide in patients with recurrent malignant glioma. J Clin Oncol. 2007;25(13):1651–1657. doi: 10.1200/JCO.2006.06.6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reardon DA, Fink KL, Mikkelsen T, et al. Randomized phase II study of cilengitide, an integrin-targeting arginine-glycine-aspartic acid peptide, in recurrent glioblastoma multiforme. J Clin Oncol. 2008;26(34):5610–5617. doi: 10.1200/JCO.2008.16.7510. [DOI] [PubMed] [Google Scholar]
- 89.Mikkelsen T, Brodie C, Finniss S, et al. Radiation sensitization of glioblastoma by cilengitide has unanticipated schedule-dependency. Int J Cancer. 2009;124(11):2719–2727. doi: 10.1002/ijc.24240. [DOI] [PubMed] [Google Scholar]
- 90.Stupp R, Hegi ME, Neyns B, et al. Phase I/IIa study of cilengitide and temozolomide with concomitant radiotherapy followed by cilengitide and temozolomide maintenance therapy in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28(16):2712–2718. doi: 10.1200/JCO.2009.26.6650. [DOI] [PubMed] [Google Scholar]
- 91.Nabors LB, Mikkelsen T, Hegi ME, et al. A safety run-in and randomized phase 2 study of cilengitide combined with chemoradiation for newly diagnosed glioblastoma (NABTT 0306) Cancer. 2012;118(22):5601–5607. doi: 10.1002/cncr.27585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stupp R, Hegi ME, Gorlia T, et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma and methylated O6-methylguanine-DNA methyltransferase (MGMT) gene promoter: Key results of the multicenter, randomized, open-label, controlled, phase III CENTRIC study [abstract] Proc Annu Meet Am Soc Clin Oncol. 2013;31:LBA2009. [Google Scholar]
- 93.Becher OJ, Holland EC. Genetically engineered models have advantages over xenografts for preclinical studies. Cancer Res. 2006;66(7):3355–3358. doi: 10.1158/0008-5472.CAN-05-3827. discussion 3358–3359. [DOI] [PubMed] [Google Scholar]
- 94.Agnihotri S, Gajadhar AS, Ternamian C, et al. Alkylpurine-DNA-N-glycosylase confers resistance to temozolomide in xenograft models of glioblastoma multiforme and is associated with poor survival in patients. J Clin Invest. 2012;122(1):253–266. doi: 10.1172/JCI59334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kitange GJ, Mladek AC, Carlson BL, et al. Inhibition of histone deacetylation potentiates the evolution of acquired temozolomide resistance linked to MGMT upregulation in glioblastoma xenografts. Clin Cancer Res. 2012;18(15):4070–4079. doi: 10.1158/1078-0432.CCR-12-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oliva CR, Nozell SE, Diers A, et al. Acquisition of temozolomide chemoresistance in gliomas leads to remodeling of mitochondrial electron transport chain. J Biol Chem. 2010;285(51):39759–39767. doi: 10.1074/jbc.M110.147504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nasser S, Kiefer J, Armstrong B, et al. Aligning xenograft models to glioblastoma patient tumors to assess chemovulnerability of patients [abstract] Proc Am Assoc Cancer Res. 2012;52:4917. [Google Scholar]
- 98.Herter-Sprie GS, Kung AL, Wong KK. New cast for a new era: preclinical cancer drug development revisited. J Clin Invest. 2013;123(9):3639–3645. doi: 10.1172/JCI68340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Colman H, Zhang L, Sulman EP, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12(1):49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bhat KP, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Agarwal S, Sane R, Oberoi R, et al. Delivery of molecularly targeted therapy to malignant glioma, a disease of the whole brain. Expert Rev Mol Med. 2011;13:e17. doi: 10.1017/S1462399411001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huse JT, Holland E, Deangelis LM. Glioblastoma: molecular analysis and clinical implications. Annu Rev Med. 2013;64:59–70. doi: 10.1146/annurev-med-100711-143028. [DOI] [PubMed] [Google Scholar]
- 105.Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 106.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 107.Gorovets D, Kannan K, Shen R, et al. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin Cancer Res. 2012;18(9):2490–2501. doi: 10.1158/1078-0432.CCR-11-2977. [DOI] [PubMed] [Google Scholar]
- 108.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dunn GP, Rinne ML, Wykosky J, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Burrell RA, McGranahan N, Bartek J, et al. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 111.Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Snuderl M, Fazlollahi L, Le LP, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 115.Stommel J, Kimmelman A, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318(5848):287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 116.Zhou BB, Zhang H, Damelin M, et al. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8(10):806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 117.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 118.Yuan X, Curtin J, Xiong Y, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23(58):9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 119.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 120.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 121.Singh SK, Clarke ID, Hide T, et al. Cancer stem cells in nervous system tumors. Oncogene. 2004;23(43):7267–7273. doi: 10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- 122.Nduom EK, Hadjipanayis CG, Van Meir EG. Glioblastoma cancer stem-like cells: implications for pathogenesis and treatment. Cancer J. 2012;18(1):100–106. doi: 10.1097/PPO.0b013e3182452e0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huang Z, Cheng L, Guryanova OA, et al. Cancer stem cells in glioblastoma--molecular signaling and therapeutic targeting. Protein Cell. 2010;1(7):638–655. doi: 10.1007/s13238-010-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang J, Ma Y, Cooper MK. Cancer stem cells in glioma: challenges and opportunities. Transl Cancer Res. 2013;2(5):429–441. doi: 10.3978/j.issn.2218-676X.2013.08.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yan K, Yang K, Rich JN. The evolving landscape of glioblastoma stem cells. Curr Opin Neurol. 2013;26(6):701–707. doi: 10.1097/WCO.0000000000000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Miller CR, Williams CR, Buchsbaum DJ, et al. Intratumoral 5-fluorouracil produced by cytosine deaminase/5-fluorocytosine gene therapy is effective for experimental human glioblastomas. Cancer Res. 2002;62(3):773–780. [PubMed] [Google Scholar]
- 127.Vitucci M, Karpinich NO, Bash RE, et al. Cooperativity between MAPK and PI3 K signaling activation is required for glioblastoma pathogenesis. Neuro Oncol. 2013;15(10):1317–1329. doi: 10.1093/neuonc/not084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Clark MJ, Homer N, O'Connor BD, et al. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6(1):e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stein WD, Litman T, Fojo T, et al. A Serial Analysis of Gene Expression (SAGE) database analysis of chemosensitivity: comparing solid tumors with cell lines and comparing solid tumors from different tissue origins. Cancer Res. 2004;64(8):2805–2816. doi: 10.1158/0008-5472.can-03-3383. [DOI] [PubMed] [Google Scholar]
- 130.Szakacs G, Gottesman MM. Comparing solid tumors with cell lines: implications for identifying drug resistance genes in cancer. Mol Interv. 2004;4(6):323–325. doi: 10.1124/mi.4.6.5. [DOI] [PubMed] [Google Scholar]
- 131.Li A, Walling J, Kotliarov Y, et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res. 2008;6(1):21–30. doi: 10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- 132.Parrish KE, Pokorny JL, Mittapalli RK, et al. BBB efflux pump activity limits brain penetration of palbociclib (PD0332991) in glioblastoma [abstract] Mol Cancer Ther. 2013;12(S11):C81. [Google Scholar]
- 133.Camphausen K, Purow B, Sproull M, et al. Influence of in vivo growth on human glioma cell line gene expression: convergent profiles under orthotopic conditions. Proc Natl Acad Sci USA. 2005;102(23):8287–8292. doi: 10.1073/pnas.0502887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Camphausen K, Purow B, Sproull M, et al. Orthotopic growth of human glioma cells quantitatively and qualitatively influences radiation-induced changes in gene expression. Cancer Res. 2005;65(22):10389–10393. doi: 10.1158/0008-5472.CAN-05-1904. [DOI] [PubMed] [Google Scholar]
- 135.Tentler JJ, Tan AC, Weekes CD, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9(6):338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Giannini C, Sarkaria JN, Saito A, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol. 2005;7(2):164–176. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Joo KM, Kim J, Jin J, et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 2013;3(1):260–273. doi: 10.1016/j.celrep.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 138.Yost SE, Pastorino S, Rozenzhak S, et al. High-resolution mutational profiling suggests the genetic validity of glioblastoma patient-derived pre-clinical models. PLoS One. 2013;8(2):e56185. doi: 10.1371/journal.pone.0056185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hodgson JG, Yeh RF, Ray A, et al. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol. 2009;11(5):477–487. doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 141.Lottaz C, Beier D, Meyer K, et al. Transcriptional profiles of CD133+ and CD133- glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res. 2010;70(5):2030–2040. doi: 10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- 142.Rich JN, Guo C, McLendon RE, et al. A genetically tractable model of human glioma formation. Cancer Res. 2001;61(9):3556–3560. [PubMed] [Google Scholar]
- 143.Sonoda Y, Ozawa T, Aldape KD, et al. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001;61(18):6674–6678. [PubMed] [Google Scholar]
- 144.Sonoda Y, Ozawa T, Hirose Y, et al. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 2001;61(13):4956–4960. [PubMed] [Google Scholar]
- 145.Mao XG, Hutt-Cabezas M, Orr BA, et al. LIN28A facilitates the transformation of human neural stem cells and promotes glioblastoma tumorigenesis through a pro-invasive genetic program. Oncotarget. 2013;4(7):1050–1064. doi: 10.18632/oncotarget.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhu H, Woolfenden S, Bronson RT, et al. The novel Hsp90 inhibitor NXD30001 induces tumor regression in a genetically engineered mouse model of glioblastoma multiforme. Mol Cancer Ther. 2010;9(9):2618–2626. doi: 10.1158/1535-7163.MCT-10-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pitter KL, Galban CJ, Galban S, et al. Perifosine and CCI 779 co-operate to induce cell death and decrease proliferation in PTEN-intact and PTEN-deficient PDGF-driven murine glioblastoma. PLoS One. 2011;6(1):e14545. doi: 10.1371/journal.pone.0014545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jun HJ, Acquaviva J, Chi D, et al. Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene. 2012;31(25):3039–3050. doi: 10.1038/onc.2011.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.de Vries NA, Bruggeman SW, Hulsman D, et al. Rapid and robust transgenic high-grade glioma mouse models for therapy intervention studies. Clin Cancer Res. 2010;16(13):3431–3441. doi: 10.1158/1078-0432.CCR-09-3414. [DOI] [PubMed] [Google Scholar]
- 150.Lau J, Schmidt C, Markant SL, et al. Matching mice to malignancy: molecular subgroups and models of medulloblastoma. Childs Nerv Syst. 2012;28(4):521–532. doi: 10.1007/s00381-012-1704-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Remke M, Ramaswamy V, Taylor MD. Medulloblastoma molecular dissection: the way toward targeted therapy. Curr Opin Oncol. 2013;25(6):674–681. doi: 10.1097/CCO.0000000000000008. [DOI] [PubMed] [Google Scholar]
- 152.von Werder A, Seidler B, Schmid RM, et al. Production of avian retroviruses and tissue-specific somatic retroviral gene transfer in vivo using the RCAS/TVA system. Nat Protoc. 2012;7(6):1167–1183. doi: 10.1038/nprot.2012.060. [DOI] [PubMed] [Google Scholar]
- 153.Chow LM, Endersby R, Zhu X, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19(3):305–316. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Song Y, Zhang Q, Kutlu B, et al. Evolutionary etiology of high-grade astrocytomas. Proc Natl Acad Sci USA. 2013;110(44):17933–17938. doi: 10.1073/pnas.1317026110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kim HS, Woolard K, Lai C, et al. Gliomagenesis arising from Pten- and Ink4a/Arf-deficient neural progenitor cells is mediated by the p53-Fbxw7/Cdc4 pathway, which controls c-Myc. Cancer Res. 2012;72(22):6065–6075. doi: 10.1158/0008-5472.CAN-12-2594. [DOI] [PubMed] [Google Scholar]
- 156.McEllin B, Camacho CV, Mukherjee B, et al. PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res. 2010;70(13):5457–5464. doi: 10.1158/0008-5472.CAN-09-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Radke J, Bortolussi G, Pagenstecher A. Akt and c-Myc induce stem-cell markers in mature primary p53(-)/(-) astrocytes and render these cells gliomagenic in the brain of immunocompetent mice. PLoS One. 2013;8(2):e56691. doi: 10.1371/journal.pone.0056691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yahanda AM, Bruner JM, Donehower LA, et al. Astrocytes derived from p53-deficient mice provide a multistep in vitro model for development of malignant gliomas. Mol Cell Biol. 1995;15(8):4249–4259. doi: 10.1128/mcb.15.8.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Blouw B, Song H, Tihan T, et al. The hypoxic response of tumors is dependent on their microenvironment. Cancer Cell. 2003;4(2):133–146. doi: 10.1016/s1535-6108(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 160.McNeill RS, Van Swearingen AED, Bash RE, et al. Efficacy of mono and dual PI3 K and MAPK inhibition in glioblastoma and triple-negative breast cancer brain metastasis models [abstract] J Neuropathol Exp Neurol. 2014;73(6):587. [Google Scholar]
- 161.Frattini V, Trifonov V, Chan JM, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45(10):1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shapiro WR, Basler GA, Chernik NL, et al. Human brain tumor transplantation into nude mice. J Natl Cancer Inst. 1979;62(3):447–453. doi: 10.1093/jnci/62.3.447. [DOI] [PubMed] [Google Scholar]
- 163.Shannon P, Sabha N, Lau N, et al. Pathological and molecular progression of astrocytomas in a GFAP:12 V-Ha-Ras mouse astrocytoma model. Am J Pathol. 2005;167(3):859–867. doi: 10.1016/S0002-9440(10)62057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Schmid RS, Irvin DM, Vitucci M, et al. The role of regional astrocyte identity in astrocytoma genomic heterogeneity [abstract] Neuro Oncol. 2013;15(S3):iii213. [Google Scholar]
- 165.Huse JT, Wallace M, Aldape KD, et al. Where are we now? And where are we going? A report from the Accelerate Brain Cancer Cure (ABC2) Low-grade Glioma Research Workshop. Neuro Oncol. 2014;16(2):173–178. doi: 10.1093/neuonc/not229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Should "models" be inserted after "gene transfer GEM"? [Google Scholar]
- 167.Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324(5924):261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Duncan CG, Barwick BG, Jin G, et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res. 2012;22(12):2339–2355. doi: 10.1101/gr.132738.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jin G, Pirozzi CJ, Chen LH, et al. Mutant IDH1 is required for IDH1 mutated tumor cell growth. Oncotarget. 2012;3(8):774–782. doi: 10.18632/oncotarget.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li S, Chou AP, Chen W, et al. Overexpression of isocitrate dehydrogenase mutant proteins renders glioma cells more sensitive to radiation. Neuro Oncol. 2013;15(1):57–68. doi: 10.1093/neuonc/nos261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Piaskowski S, Bienkowski M, Stoczynska-Fidelus E, et al. Glioma cells showing IDH1 mutation cannot be propagated in standard cell culture conditions. Br J Cancer. 2011;104(6):968–970. doi: 10.1038/bjc.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jin G, Reitman ZJ, Duncan CG, et al. Disruption of wild-type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res. 2013;73(2):496–501. doi: 10.1158/0008-5472.CAN-12-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kelly JJ, Blough MD, Stechishin OD, et al. Oligodendroglioma cell lines containing t(1;19)(q10;p10) Neuro Oncol. 2010;12(7):745–755. doi: 10.1093/neuonc/noq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Klink B, Miletic H, Stieber D, et al. A novel, diffusely infiltrative xenograft model of human anaplastic oligodendroglioma with mutations in FUBP1, CIC, and IDH1. PLoS One. 2013;8(3):e59773. doi: 10.1371/journal.pone.0059773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Luchman HA, Stechishin OD, Dang NH, et al. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro Oncol. 2012;14(2):184–191. doi: 10.1093/neuonc/nor207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sasaki M, Knobbe CB, Itsumi M, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26(18):2038–2049. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.McNeill RS, Schmid RS, Bash RE, et al. Modeling astrocytoma pathogenesis in vitro and in vivo using cortical astrocytes or neural stem cells from conditional, genetically engineered mice. J Vis Exp. 2014;(90):e51763. doi: 10.3791/51763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Northcott PA, Korshunov A, Pfister SM, et al. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol. 2012;8(6):340–351. doi: 10.1038/nrneurol.2012.78. [DOI] [PubMed] [Google Scholar]
- 183.Salphati L, Heffron TP, Alicke B, et al. Targeting the PI3 K pathway in the brain--efficacy of a PI3 K inhibitor optimized to cross the blood-brain barrier. Clin Cancer Res. 2012;18(22):6239–6248. doi: 10.1158/1078-0432.CCR-12-0720. [DOI] [PubMed] [Google Scholar]
- 184.Duncan JS, Whittle MC, Nakamura K, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149(2):307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bash R, Karpinich NO, Vitucci M, et al. Concurrent temozolomide-external-beam radiation therapy is effective for experimental glioblastomas in an orthotopic, genetically engineered syngeneic mouse allograft model system [abstract] Neuro Oncol. 2009;11(5):638. [Google Scholar]
- 186.Miller CR, Bash RE, Vitucci M, et al. A genetically-defined, orthotopic allograft model system of glioblastoma: Pathological features and experimental therapeutics [abstract] J Neuropathol Exp Neurol. 2010;69(5):522. [Google Scholar]
- 187.Fulop GM, Phillips RA. The scid mutation in mice causes a general defect in DNA repair. Nature. 1990;347(6292):479–482. doi: 10.1038/347479a0. [DOI] [PubMed] [Google Scholar]
- 188.Holmen SL, Williams BO. Essential role for Ras signaling in glioblastoma maintenance. Cancer Res. 2005;65(18):8250–8255. doi: 10.1158/0008-5472.CAN-05-1173. [DOI] [PubMed] [Google Scholar]
- 189.Holland EC, Celestino J, Dai C, et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25(1):55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 190.Liu C, Sage JC, Miller MR, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146(2):209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kim YW, Liu TJ, Koul D, et al. Identification of novel synergistic targets for rational drug combinations with PI3 kinase inhibitors using siRNA synthetic lethality screening against GBM. Neuro Oncol. 2011;13(4):367–375. doi: 10.1093/neuonc/nor012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Piao Y, Liang J, Holmes L, et al. Acquired resistance to anti-VEGF therapy in glioblastoma is associated with a mesenchymal transition. Clin Cancer Res. 2013;19(16):4392–4403. doi: 10.1158/1078-0432.CCR-12-1557. [DOI] [PubMed] [Google Scholar]
- 193.Gruber Filbin M, Dabral SK, Pazyra-Murphy MF, et al. Coordinate activation of Shh and PI3 K signaling in PTEN-deficient glioblastoma: new therapeutic opportunities. Nat Med. 2013;19(11):1518–1523. doi: 10.1038/nm.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Dinca EB, Lu KV, Sarkaria JN, et al. p53 Small-molecule inhibitor enhances temozolomide cytotoxic activity against intracranial glioblastoma xenografts. Cancer Res. 2008;68(24):10034–10039. doi: 10.1158/0008-5472.CAN-08-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Clarke MJ, Mulligan EA, Grogan PT, et al. Effective sensitization of temozolomide by ABT-888 is lost with development of temozolomide resistance in glioblastoma xenograft lines. Mol Cancer Ther. 2009;8(2):407–414. doi: 10.1158/1535-7163.MCT-08-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Taveras JM, Thompson HG, Jr, Pool JL. Should we treat glioblastoma multiforme? A study of survival in 425 cases. Am J Roentgenol Radium Ther Nucl Med. 1962;87:473–479. [PubMed] [Google Scholar]
- 197.Mealey J., Jr Treatment of malignant cerebral astrocytomas by intra-arterial infusion of vinblastine. Cancer Chemother Rep. 1962;20:121–126. [PubMed] [Google Scholar]
- 198.Walker MD, Hurwitz BS. BCNU (1,3-bis(2-chloroethyl)-1-nitrosourea; NSC-409962) in the treatment of malignant brain tumor--a preliminary report. Cancer Chemother Rep. 1970;54(4):263–271. [PubMed] [Google Scholar]
- 199.Fine HA, Figg WD, Jaeckle K, et al. Phase II trial of the antiangiogenic agent thalidomide in patients with recurrent high-grade gliomas. J Clin Oncol. 2000;18(4):708–715. doi: 10.1200/JCO.2000.18.4.708. [DOI] [PubMed] [Google Scholar]
- 200.Friedman HS, Prados MD, Wen PY, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27(28):4733–4740. doi: 10.1200/JCO.2008.19.8721. [DOI] [PubMed] [Google Scholar]
- 201.Kreisl TN, Kim L, Moore K, et al. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J Clin Oncol. 2009;27(5):740–745. doi: 10.1200/JCO.2008.16.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Caskey LS, Fuller GN, Bruner JM, et al. Toward a molecular classification of the gliomas: histopathology, molecular genetics, and gene expression profiling. Histol Histopathol. 2000;15(3):971–981. doi: 10.14670/HH-15.971. [DOI] [PubMed] [Google Scholar]
- 203.Horten BC, Basler GA, Shapiro WR. Xenograft of human malignant glial tumors into brains of nude mice. A histopatholgical study. J Neuropathol Exp Neurol. 1981;40(5):493–511. doi: 10.1097/00005072-198109000-00002. [DOI] [PubMed] [Google Scholar]
- 204.Ding H, Roncari L, Shannon P, et al. Astrocyte-specific expression of activated p21-ras results in malignant astrocytoma formation in a transgenic mouse model of human gliomas. Cancer Res. 2001;61(9):3826–3836. [PubMed] [Google Scholar]