Promoter region methylation of O6-methylguanine DNA methyltransferase (MGMT) is a critical biomarker in neuro-oncology. Introduction of temozolomide (TMZ) and concomitant radiation as a standard of care (Stupp protocol) for glioblastoma multiforme (GBM) has improved survival for some patients. Hegi et al. suggested that methylation status might be predictive of benefit from TMZ,1 and multiple studies ever since have confirmed that MGMT methylation status is predictive of TMZ response.2 However, debate persists regarding the best method for its evaluation. Consequently, MGMT status in the clinic has been underutilized, and TMZ remains a staple in caring for GBM patients regardless of their MGMT methylation status.
The MGMT promoter consists of 97 cytosine-guanine (CpG) dinucleotide sites, which can be grouped into 3 regions: R1, R2 and R3, where R1 is the most distal to the transcription start site and R3 is adjacent to the transcription start site. In our previous study,3 we comprehensively characterized methylation across the entire MGMT promoter and showed that all 3 regions correlate to mRNA expression, protein expression, and progression-free survival. The most widely used test that measures MGMT methylation is the qMSP test.4 The qMSP test measures methylation in region R3, which is the most strongly correlated to patient outcome. However, the success of the test depends upon the co-methylation of at least 4 out of the 7 CpG sites in R3. Another test, the MS-MLPA, can measure methylation of 3 individual CpG sites, one in each of the aforementioned regions.5 Hence, both tests are complementary in nature.
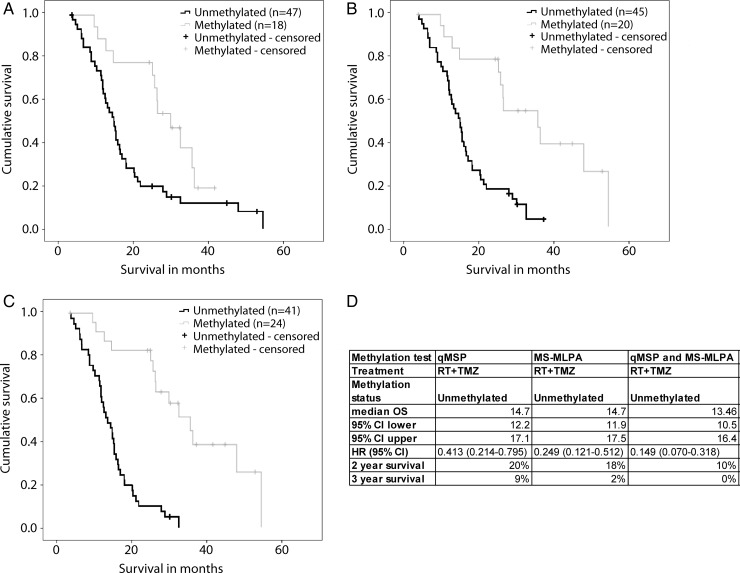
It has been suggested that GBM patients with and without MGMT methylation benefit from TMZ.1 The tail in the unmethylated group (>3-year survival, Fig. 1A and B) seemingly indicated that even patients with unmethylated MGMT benefit from TMZ treatment. This survival tail in our cohort is consistent with other large-scale studies.2 However, when we performed both qMSP and MS-MLPA tests together on 65 GBM samples, we observed increased detection sensitivity, and the tail in the unmethylated group disappeared (Fig. 1C and D). All 5 patients comprising the tail, when using just one of the tests, were indeed methylated by the other test. These data suggest that accurate MGMT characterization is grounds for questioning the alleged benefit of TMZ for all GBM patients and strengthen the case for needing new treatment regimens because the current standard of care does not provide benefit for MGMT-unmethylated patients.
Fig. 1.
Kaplan–Meier survival curves for MGMT methylated versus unmethylated patients. (A) MGMT methylation tested using qMSP (log-rank P = .006). (B) MGMT methylation tested using MS-MLPA (log-rank P < .001). (C) MGMT methylation determined using both MS-MLPA and qMSP (log-rank P < .001). A sample was considered methylated if either of the test results was methylated (D). Survival statistics for MGMT unmethylated GBM patients using different testing techniques.
References
- 1.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert MR, Wang M, Aldape KD, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. J Clin Oncol. 2013;31(32):4085–4091. doi: 10.1200/JCO.2013.49.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah N, Lin B, Sibenaller Z, et al. Comprehensive analysis of MGMT promoter methylation: correlation with MGMT expression and clinical response in GBM. PloS One. 2011;6(1):e16146. doi: 10.1371/journal.pone.0016146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vlassenbroeck I, Califice S, Diserens AC, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn. 2008;10(4):332–337. doi: 10.2353/jmoldx.2008.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeuken JW, Cornelissen SJ, Vriezen M, et al. MS-MLPA: an attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest. 2007;87(10):1055–1065. doi: 10.1038/labinvest.3700664. [DOI] [PubMed] [Google Scholar]