Abstract
Background
Atypical teratoid rhabdoid tumor (ATRT) is an aggressive pediatric brain tumor with limited therapeutic options. The hypothesis for this study was that the Wnt pathway triggered by the Wnt5B ligand plays an important role in ATRT biology. To address this hypothesis, the role of WNT5B and other Wnt pathway genes was analyzed in ATRT tissues and ATRT primary cell lines.
Methods
Transcriptome-sequencing analyses were performed using nanoString platforms, immunohistochemistry, Western blotting, quantitative reverse transcriptase PCR, immunoprecipitation, short interference RNA studies, cell viability studies, and drug dose response (DDR) assays.
Results
Our transcriptome-sequencing results of Wnt pathway genes from ATRT tissues and cell lines indicated that the WNT5B gene is significantly upregulated in ATRT samples compared with nontumor brain samples. These results also indicated a differential expression of both canonical and noncanonical Wnt genes. Imunoprecipitation studies indicated that Wnt5B binds to Frizzled1 and Ryk receptors. Inhibition of WNT5B by short interference RNA decreased the expression of FRIZZLED1 and RYK. Cell viability studies a indicated significant decrease in cell viability by inhibiting Frizzled1 receptor. DDR assays showed promising results with some inhibitors.
Conclusions
These promising therapeutic options will be studied further before starting a translational clinical trial. The success of these options will improve care for these patients.
Keywords: ATRT, Frizzled 1, Ryk, Wnt pathway, Wnt5B
Atypical teratoid rhabdoid tumor (ATRT) is an aggressive tumor comprising <5% of CNS tumors in children aged <18 years and up to 20% of CNS tumors in children aged <3 years.1,2 A characteristic feature of ATRTs is an aberration of chromosome 22,3 which results in a loss of the gene SMARCB1.4,5,6 Whole-genome sequencing of one ATRT patient sample performed in our laboratory as part of an earlier study demonstrated a complete loss of the locus encompassing SMARCB1 gene.7
In this study, transcriptome-sequencing results indicate that WNT5B gene is significantly upregulated in ATRT patient tissues compared with nontumor brain tissue samples. This was a novel finding for ATRT tumors, and Wnt5B signaling was analyzed in ATRT. Wnt proteins are a family of 19 secreted glycoproteins that act as ligands and have crucial roles in the regulation of diverse cellular processes.8,9 Perturbation of Wnt ligands can result in abnormal Wnt signaling and disease etiology. A role for Wnt proteins in cancer was first described in mouse mammary cancer and human and mouse colon cancer.10,11 It is now clear that Wnt ligands modulate both β-catenin dependent (often referred to as “canonical”) Wnt signaling and β-catenin-independent (often referred to as “noncanonical”) Wnt signaling pathways.8 The precise mechanisms by which a Wnt ligand stimulates cellular responses are not fully elucidated but probably involve a specific Wnt ligand bound to a specific receptor. The known receptors of the Wnt pathway include 10 Frizzled receptors, Ryk, Ror1, Ror2, and Lrp receptors.9 These receptors have been studied as therapeutic targets for other cancers.12 Wnt5B is often implicated as an activator of the noncanonical pathway.13 Although Wnt signaling has been implicated in other cancers (eg, colon cancer) in which β-catenin (CTNNB1) gene is overexpressed.14; among the group of brain cancers, Wnt signaling has been implicated in medulloblastomas and aggressive meningiomas. Four distinct molecular subgroups of medulloblastoma have been identified including, wingless (WNT), sonic hedgehog (SHH), Group 3, and Group 4. Profiling these subgroups may confer prognostic impact along with targeted therapy.15,16 Studies have indicated that CTNNB1 is upregulated and localized to the nucleus in the menigiomas.17 Studies have also shown that the SMARCA4 subunit of SWI/SNF complex interacts with CTNNB1.18,19 Loss of SNF5 from the SWI/SNF complex has been shown to activate β-catenin/TCF targets.20
In this study, the Wnt pathway triggered by the Wnt5B ligand was hypothesized as playing an important role in ATRT biology. Differential expression of Wnt pathway genes by transcriptome analyses was first investigated in 20 ATRT tissue samples and 3 primary ATRT cell lines. Since downstream events of Wnt pathway are determined by ligand-receptor binding, Wnt5B receptor-binding studies were performed. Inhibition of WNT5B by short interference RNA (siRNA) was performed to study expression of other Wnt genes that would be simultaneously inhibited. Cell viability assays were performed to study the effects of inhibiting the Wnt5B-bound receptor. Finally, drug-dose response (DDR) assays were performed with known Wnt inhibitors to study ATRT cell viability. The results from these studies are discussed, and we have demonstrated that the Wnt5B pathway may play an important role in ATRT biology. These important findings may lead to the discovery of alternative therapeutic options that can be further tested in children with ATRT.
Materials and Methods
Cell Lines
CHLA-04-ATRT was obtained from a 20-month old male, CHLA-05-ATRT from a 2-year old male, and CHLA-06-ATRT from a 4 month-old female. Informed consent was signed by the guardians of the children. Tissues were prepared as described21 and were initially cultured as neurospheres in modified neurobasal medium consisting of 1:1 Dulbecco's modified Eagle's medium:F12, HEPES (15 mM), sodium pyruvate (110 mg/L), sodium bicarbonate (1.2 g/L), 1xB27 (Invitrogen), epidermal growth factor (20 ng/mL; Invitrogen), and bovine fibroblast growth factor (20 ng/mL; Cell Sciences). Gentamycin 25 µg/ml was used during the first 2 weeks of growth. Passaging was at a ratio of 1:2–3 with 25% conditioned medium.22 Loss of SMARCB1 was confirmed by immunohistochemistry, Western blotting, quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), and G-band karyotyping.
Patient Samples
Six brain regions from 2 age-matched patients' nontumor brain tissues (in good pathological condition) were used as controls. (Three regions from each patient included frontal cortex, temporal cortex, and white matter.) The nontumor brain tissues were from NICHD Brain and Tissue Bank for Developmental disorders at the University of Maryland, Baltimore. Twenty ATRT patient tissues were from Children's Hospital Westmead (Australia), Phoenix Children's Hospital, and St. Joseph's Hospital (USA). Informed patient consent was obtained; IRB guidelines and Materials Transfer Agreement protocols were followed prior to transfer of ATRT tissues.
RNA Sequencing Analysis by nanoString
Total RNA (250 ng) from each sample was analyzed using the nanoString nCounter Analysis System with a custom miRGE CodeSet. All procedures related to mRNA quantification were performed according to manufacturer's recommendations. Data were normalized using positive control normalization. Raw counts were normalized using the geometric mean of 6 control probes present in each assay. These values were further corrected using the geometric mean of the top upregulated 80 genes, then log2-transformed and used as input for prediction analysis. The negative control average was subtracted from all normalized data points before log2 transformation. P values were generated using an ANOVA of log2 normalized values.
Quantitative Reverse Transcriptase PCR
RNA was isolated (Qiagen), and cDNA was synthesized using Superscript II® cDNA kit (Life Technologies). For quantitative PCR, SYBr green I master mix (Roche) was used. One set of forward and reverse primers for the genes studied were used (detailed primer information in Supplementary Methods). Quantitative PCR was run on a real-time PCR system (Roche Light Cycler 480 system). Each sample was run in triplicate, and Roche software was used to analyze data. The expression of each gene was compared with nontumor brain expression and normalized against GAPDH. Expression was calculated as fold changes using the standard ΔCp value calculation. The Student t test was used to calculate significance.
Short Interference RNA Treatment by Reverse Transfection
A dose response analysis over time was performed for WNT5B siRNA inhibition (Qiagen). The siRNA was transiently transfected using RNAi max (Life Technologies) by reverse transfection. For siRNA qRT-PCR studies, a total of 1 × 106 cells were treated with 100 ng WNT5B siRNA. After 110 hours, cells were subjected to qRT-PCR analysis using the appropriate primers. These siRNA studies were repeated using WNT5B siRNA from Santa Cruz Biotechnology.
For cell viability studies using both FRIZZLED1 siRNA (Santa Cruz Biotechnology) and Frizzled1 (Fz1) Mab (R&D Systems, glutamine72 – histidine 248 epitope), 5000 cells per well in a 96-well plate were first treated with 100 ng of FRIZZLED1 siRNA followed by Fz1Mab (5 µg/100 µL after dose-response study). In order to spike cells with siRNA, 100 ng of siRNA were added with RNAi max reagent at 50 hours. The plate was read at 110 hours using CellTiter-Glo ATP assay reagents (Promega). Controls included scrambled siRNA (Qiagen), AllStars death control siRNA (Qiagen), and IgG2A (R&D Systems). Cell viability studies were repeated using FRIZZLED1 siRNA from Origene Inc.
Immunoprecipitation
The Catch and Release Reversible Immunoprecipitation System v2.0 (Millipore) was used. For every affinity column, lysate from a T75-flask was used. After the appropriate treatment, cell lysate was made in 500 μL of cold RIPA buffer (Cell Signaling Technology) with 0.1 M PMSF (10 µL/mL of lysate). Lysate was added to the affinity column along with 2 μg of appropriate antibody and 10 μL of the affinity ligand and gently rocked for 1 hour at room temperature. Elutes were isolated and analyzed by Western blotting using appropriate primary and secondary antibodies.
Western Blotting
Samples were subject to SDS-PAGE using 4%–12% Bis-Tris protein gels (Life Technologies) at 30 µg/mL in each lane after a bicinchoninic acid assay. Standard blotting procedure was followed with a 1:1000 dilution of the appropriate primary antibody and a 1:2000 dilution of the secondary antibody. Supersignal West Dura Extended Duration Substrate (ThermoScientific) was used for detection.
Immunohistochemistry
Sample blocks were sectioned at 4 mm thickness, and slides were dewaxed, rehydrated, and antigen-retrieved on the Bond Max autostainer (Leica Microsystems). All slides were subjected to heat-induced epitope retrieval using a citrate-based retrieval solution. All slides were incubated for 30 minutes with INI-1 (MRQ-27) and Wnt5B antibody. Sections were visualized using the Bond Polymer Refine Detection kit (Leica). Antibody detection levels were compared with the detection levels of control tissues and given a numerical score of either 2 or 3. This scoring was performed at a core laboratory facility.
Drug Studies
Cells were seeded 1000 cells/well in opaque 96-well plates and given 4 hours to condition with 6 replicates of each treatment. A serial dilution series covering 8 concentrations centered on the expected IC50 concentration (from scientific literature) were spiked onto cells along with vehicle control and positive kill control (MG132, Selleck Chemicals). After 72 hours, viability was measured using the CellTiter-Glo ATP assay (Promega,). IC50 values were calculated using GraphPad Prism 6.1.
Results
Transcriptome Analyses
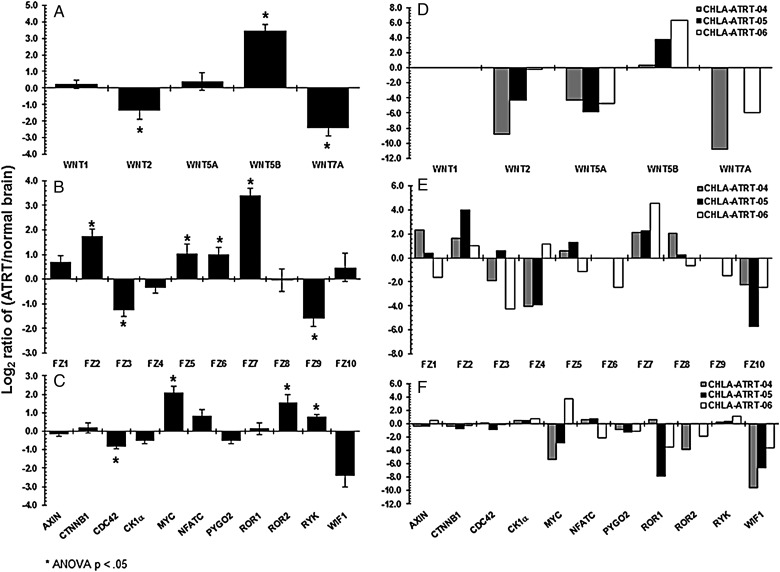
A probe-specific transcriptome analysis was performed using the nanoString platform to study differential expression of Wnt pathway genes in 20 ATRT tissues and 3 ATRT primary cell lines compared with nontumor brain control samples (see Methods). An earlier study indicated that the transcriptome varies in the human brain by brain location and age.23 Figure 1A shows a 4-fold upregulation of WNT5B in ATRT tissues. Supplementary Fig. S1 shows the upregulation of WNT5B in 19 of 20 tumor tissues. Figure 1D shows a 4–6-fold upregulation of WNT5B in 2 cell lines. In Fig. 1B and E, FRIZZLED receptors 1, 2, 5, and 7 are upregulated in tumors and cell lines. Figure 1C and F show differential expression of other Wnt pathway genes in tumors and cell lines (see Supplementary Figs S2A, S2B, S3A, and S3B for other Wnt genes in tumors and cell lines, respectively). All ATRT tissue samples were pathologically validated at diagnosis as being negative for Smarcb1 protein by immunohistochemistry. SMARCB1 mRNA expression in these ATRT samples was further confirmed as part of this study when 18 of the 20 ATRT samples (Supplementary Fig. S4A), and 3 ATRT cell lines (Supplementary Fig. S4B) indicated a downregulation of SMARCB1.
Fig. 1.
Transcriptome sequencing analyses using the nanoString platform on 20 ATRT tissues and 3 ATRT cell lines with Wnt pathway genes as probes. The differential expression of Wnt genes shown in this figure was calculated as logarithmic ratios of fold changes in ATRT samples compared with nontumor brain samples (Y-axis). For the 20 tumors, average values for each gene were calculated. (A) Expression of Wnt ligands in tumor tissues. (B) Expression of frizzled receptors in tumor tissues. (C) Expression of other Wnt genes in tumor tissues. (D) Expression of Wnt ligands in cell lines. (E) Expression of frizzled receptors in cell lines. (F) Expression of some of the other Wnt genes in cell lines. An asterisk above each bar represents a significant fold change with the P value <.05.
A preliminary transcriptome analysis was performed using the Illumina platform (Illumina Inc.) using one ATRT patient sample along with nontumor brain samples. The top 100 upregulated and 100 downregulated genes were identified (see Supplementary methods); among those, upregulated Wnt genes included WNT5B, CK1γ, MYC, NFATC, and PYGOPUS, while MAPRE2 was downregulated (Supplementary Fig. S5). SMARCB1 was among the 100 downregulated genes.
Validation of Transcriptome Analyses
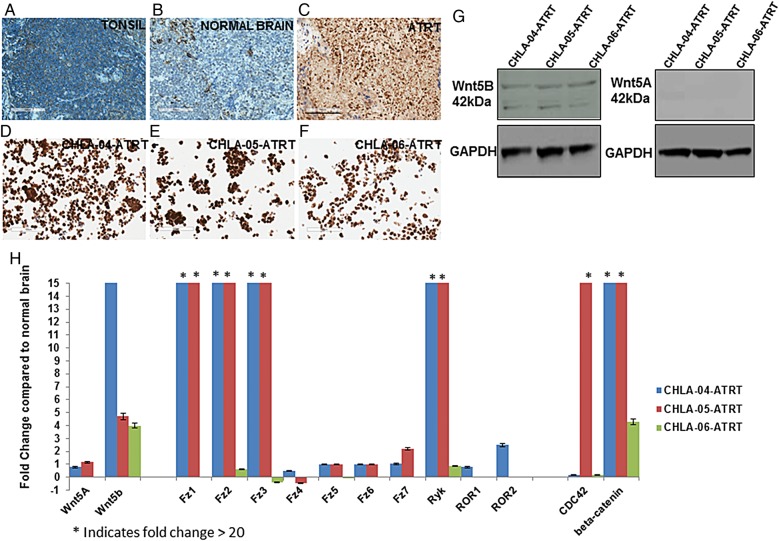
The transcriptome analyses from above were further validated by immunohistochemistry using Wnt5B antibody in 10 of the 20 ATRT tissues used for transcriptome-sequencing analyses and 3 primary ATRT cell lines. Nontumor brain and pharyngeal tonsil tissue were used as controls. Figure 2A is a representative image of tonsil tissue, while Fig. 2B is a representative image of normal brain tissue with an absence of Wnt5B protein (blue). A representative image of ATRT tissue (Fig. 2C) indicates the presence of Wnt5B protein (brown). Figure 2D–F illustrates corresponding representative images of cell lines CHLA-04-ATRT, CHLA-05-ATRT, and CHLA-06-ATRT, indicating the presence of Wnt5B protein. In ATRT tissues, Wnt5B antibody detection levels were compared with the Wnt5B detection levels of control tissues and scored based on the staining intensity levels (see Supplementary Fig. S6A for the score numbers). These tissues were also treated with a Smarcb1 antibody and scored in the same manner as Wnt5B scoring (Supplementary Fig. S6B). Western blotting was performed to study Wnt5B and Wnt5A protein expression in the cell lines (Fig. 2G). Wnt5B expression can be seen in the cell lines at 42 kD, but there is an absence of Wnt5A protein expression.
Fig. 2.
(A–F) Immunohistochemical analysis with a Wnt5B antibody. The presence of Wnt5B is indicated by brown, while the absence of Wnt5B is indicated by blue. The samples used were (A) tonsil control, (B) nontumor brain control, (C) ATRT tissue, (D) CHLA-04-ATRT cells, (E) CHLA-05-ATRT cells, and (F) CHLA-06-ATRT cells. Images are at a 20× magnification. (G) Western blotting to detect Wnt5B and Wnt5A proteins as 42 kDa protein bands. GAPDH was the loading control. (H) Real-time qRT-PCR analyses using cell lines to investigate differential expression of Wnt pathway genes (shown on the X-axis). Differential expression of these genes was calculated as a fold change compared with nontumor brain samples (Y-axis). An asterisk above a bar indicates fold change >20. Scale bar in C represents 100 μm.
In order to validate the differential expression of some of the Wnt pathway genes, qRT-PCR was performed using total RNA from the cell lines compared with nontumor brain samples (Fig. 2H). The results in Fig. 2H indicate that WNT5B was significantly upregulated between 4-fold to greater than 20-fold. Some other differentially expressed Wnt pathway genes, indicating similar expression as seen from nanoString analyses, were FRIZZLED1, 2, 3, and RYK, which are upregulated 20-fold or higher than nontumor brain expression. However, there are differences in expression levels of some mRNAs between these 2 analyses (eg, CDC42, CTNNB1, FRIZZLED3) as seen from the qRT-PCR results that may be due to differences in the probe sequences used to detect these mRNAs.
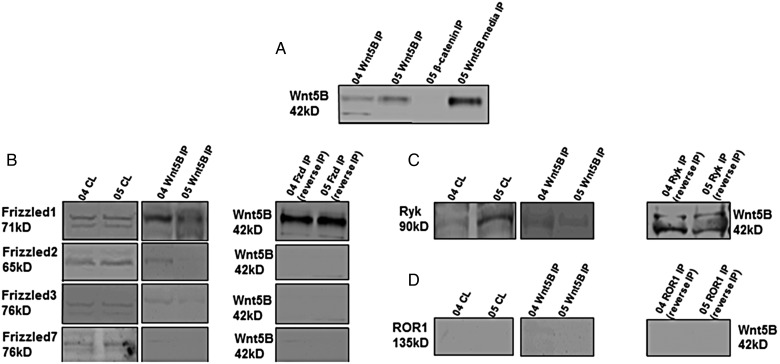
Wnt5B-Receptor Interactions
One therapeutic target that could be investigated was the receptor to which Wnt5B binds. Iimmunoprecipitation (IP) analyses were performed to determine the receptor(s) that bind Wnt5B. As control IP experiments (Fig. 3A), lysates from cell lines CHLA-04-ATRT and CHLA-05-ATRT were first passed through an IP column bound with either Wnt5B antibody or β-catenin antibody. The column eluates were tested for the presence of Wnt5B protein. The column eluates from Wnt5B IP of each cell line indicated the presence of Wnt5B protein, while eluates from β-catenin IP showed an absence of Wnt5B protein because Wnt5B ligand does not bind to intracellular β-catenin. In the “05 Wnt5B media IP” lane in Fig. 3A, cellular media (collected from CHLA-05-ATRT cells 24 h after cell passage) was passed through the IP column instead of passing the lysate through the IP column. Because Wnt5B is a secreted protein ligand, this lane indicates the presence of Wnt5B protein. These results also serve as control IP tests. In Fig. 3B, IP and reverse IP analyses were performed to test the binding of Wnt5B with Frizzled 1, 2, 3, and 7 receptors. The presence of basal protein levels, using the corresponding antibody indicated by the side of each Western blot, can be seen (“CL” lanes). The Wnt5B IP lanes in this figure indicate the presence of bound frizzled proteins in the Wnt5B IP eluate samples using the corresponding antibodies. The Fzd IP lanes indicate the presence of bound Wnt5B protein in the eluate from a Frizzled IP column performed as a “reverse IP” to validate our IP analyses. The frizzled antibody used in each Fzd IP column is shown by the corresponding frizzled label on the left side of each blot. The frizzled receptor antibodies used for IP analyses were chosen based on the frizzled receptor expression levels from our nanoString sequencing analyses and qRT-PCR analyses. As seen in Fig. 3B, Frizzled1 receptor binds Wnt5B stronger in cell lines than the other Frizzled receptors tested in the IP lanes.
Fig. 3.
Immunoprecipitation (IP), reverse IP, followed by Western blotting analyses were performed on the ATRT cell lines, CHLA-04-ATRT (04), and CHLA-05-ATRT (05). The appropriate proteins that were detected for these analyses are indicated beside the blots along with their molecular weights. For each cell line, a cell lysate (CL) sample and immunoprecipitated (IP) sample were used as indicated above each lane in the blots. (A) As controls, Wnt5B IP samples from each cell line were analyzed, along with a β-catenin IP and a Wnt5B IP with media. (B) IPs with 2 cell lines using Wnt5B antibody. The CL and IP samples were blotted using antibodies against Frizzled 1, 2, 3, and 7. A reverse IP with cell lines using Frizzled 1, 2, 3, and 7 antibodies each bound to an IP column. These IP samples were blotted to detect Wnt5B protein as indicated. (C) IPs with 2 cell lines using Wnt5B antibody. The CL and IP samples were blotted with a Ryk antibody. (D) IPs with 2 cell lines using Wnt5B antibody. The CL and IP samples were blotted with a Ror1 antibody.
Since Wnt5B was considered a noncanonical Wnt13 and Ryk receptor was upregulated in this study, the noncanonical Ryk (Fig. 3C) and ROR1 (Fig. 3D) receptors24 were also subject to the same IP analyses described above. Figure 3C indicates that Wnt5B does not bind to Ryk receptor in the IP lanes. The reverse IP lanes, however, indicate that Wnt5B may bind weakly with Ryk. Figure 3D indicates that Wnt5B does not bind to the ROR1 receptor.
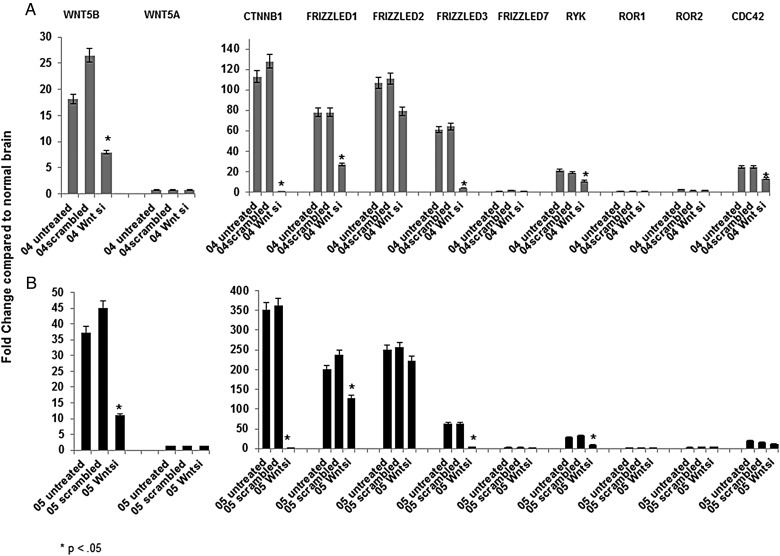
WNT5B Short Interference RNA Studies
The effects of inhibiting WNT5B transcription by a WNT5B siRNA, on expression of some of the Wnt pathway genes, were studied in CHLA-04-ATRT (Fig. 4A) and CHLA-05-ATRT (Fig. 4B). After the treatments, RNA from each cell line was subjected to real-time qRT-PCR analyses using the appropriate primers. Cells untreated with WNT5B siRNA and cells treated with scrambled siRNA were used as negative and positive controls. It can be seen from these results that WNT5B expression was inhibited significantly (P <.05) in both cell lines after siRNA treatment, while the expression of WNT5A was unaffected. Inhibition of WNT5B reduced the expression of CTNNB1, FRIZZLED1, FRIZZLED3, and RYK genes significantly in both cell lines tested, while the mRNA expression of CDC42 gene was significantly reduced in CHLA-04-ATRT cells only. These siRNA studies were repeated using WNT5B siRNA from a different manufacturer with similar results as before. Results of the repeat siRNA study are shown in Supplementary Fig. S7.
Fig. 4.
Wnt5B inhibition studies using siRNA (Wntsi) followed by qRT-PCR on ATRT cell lines, (A) CHLA-04-ATRT (04) and (B) CHLA-05-ATRT (05). The mRNA detected by qRT-PCR is indicated above the graphs. Controls used for these experiments included untreated cells and scrambled siRNA treated cells. Gene expression was calculated as a fold change compared with nontumor brain samples indicated on the Y-axis. An asterisk above each bar indicates significance of P < .05.
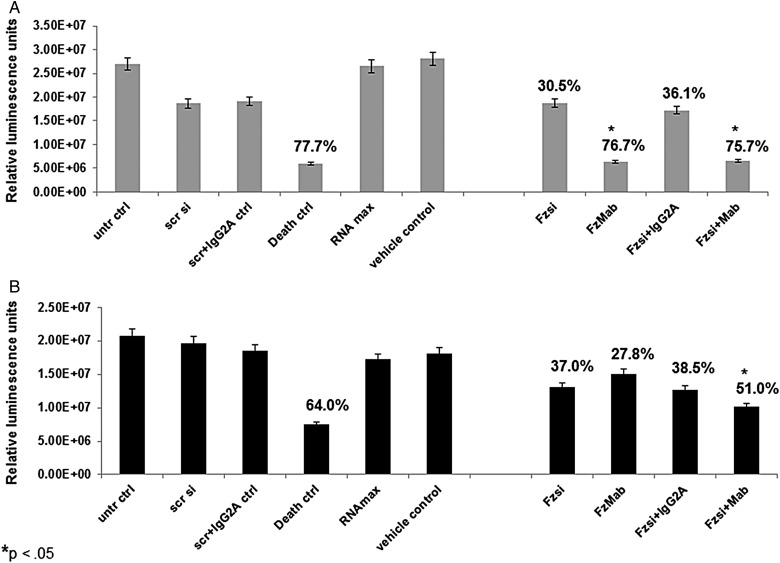
Cell Viability Studies to Test Frizzled1 Receptor as a Potential Therapeutic Target
Based on our results, cell viability studies were performed with Frizzled1 receptor as a target. The luminescence readout from the CellTitre-Glo assay was used to study cell viability. FRIZZLED1 siRNA and a commercial Frizzled1 monoclonal antibody (FzMab) were used for these studies. Reverse transcription was used to treat cells with FRIZZLED1 siRNA. CHLA-04-ATRT (Fig. 5A) and CHLA-05-ATRT (Fig. 5B) cells were treated with separate treatments as triplicates. Appropriate controls were also tested, as indicated in the figures. Any cell death of treated cells was compared with untreated control cells and calculated as an average cell death percentage indicated above each corresponding bar in the graphs. From the results in Fig. 5A. it can be seen that the death siRNA treatment showed a 77.7% cell death in CHLA-04-ATRT cells while in CHLA-05-ATRT cells there was 64% cell death, indicative of the transfection efficiency. In Fig. 5A, FRIZZLED1 siRNA treatment indicated a 30.5% cell death, FzMab treatment indicated a significant 76.7% cell death, and treatment with a combination of FRIZZLED1 siRNA and FzMab indicated a significant 75.7% cell death. A 36.1% cell death was seen in the FRIZZLED1 siRNA added with IgG2A control, indicating that there was no effect of IgG2A on cell death. In CHLA-05-ATRT cells (Fig. 5B), while FRIZZLED1 siRNA showed a 37% cell death and FzMab showed a 27.8% cell death, a significant 51% cell death was seen when cells were treated with a combination of FRIZZLED1 siRNA and FzMab. A 38.5% cell death was seen in the FRIZZLED1 siRNA added with IgG2A control. These siRNA studies were repeated using FRIZZLED1 siRNA from a different manufacturer, and the results concurred with the first study. Results are shown in Supplementary Fig. S8.
Fig. 5.
Frizzled1 receptor was inhibited, and cell viability studies were performed on (A) CHLA-04-ATRT and (B) CHLA-05-ATRT cell lines. The treatments included: (1) a FRIZZLED1 siRNA (Fzsi) treatment, (2) a monoclonal antibody treatment against Frizzled1 receptor (FzMab), and (3) a combination of Fzsi with FzMab treatment. The controls used were untreated control (untr ctrl), scrambled siRNA control (scr si), scrsi with antibody isotype control (scr + IgG2A ctrl), death siRNA control (Death ctrl), siRNA reagent control (RNAmax), and the antibody vehicle control. The percent decrease in cell viability is indicated above the bars. An asterisk above each bar represents a significant change compared with untreated cells and the death siRNA control with the P value <.05.
Testing Known Wnt Inhibitors as Potential Therapeutic Options
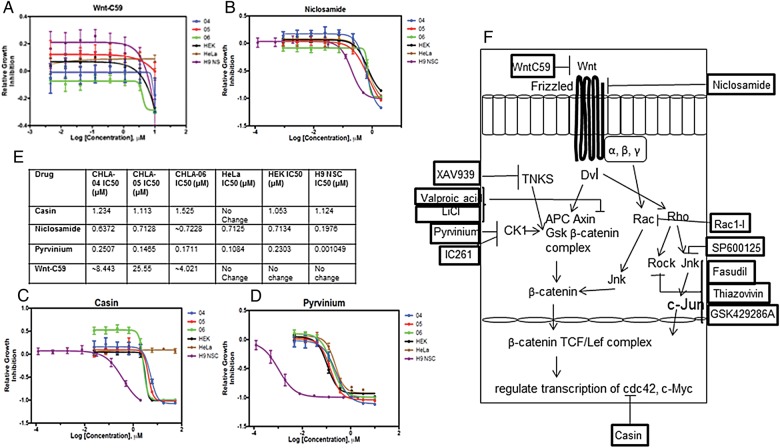
Thirteen chemical compounds, all known Wnt inhibitors from the scientific literature, were tested on ATRT cell lines. Non-ATRT cell lines such as human neural stem cells (H9-derived, Life Technologies), HeLa cells, and HEK cells were also included in these tests. CellTitre-Glo® cell viability assays were performed to test the cell death effects of these compounds, and the DDR curves were calculated. Figure 6A–D shows the DDR curves for compounds such as WntC59, niclosamide, casin, and pyrvinium, with the IC50 values of these compounds shown in Fig. 6E. Figure 6F shows a schematic indicating all the compounds used in this study and their known targets in the Wnt pathway. Supplementary Table S1 shows the results from other known Wnt inhibitors used in this study. Interestingly, WntC59 showed significant decrease in cell viability specifically in ATRT cells but not NSC, HEK, and HeLa cells. An independent preliminary chemical compound screen was conducted using the CHLA-05-ATRT cell line only, where >2500 compounds from 2 commercial libraries (Actable library and the Prestwick library) were applied to cells. Subsequently, pyrvinium pamoate and IC261 were identified as hits. This independent screen validates some of the Wnt inhibitor results.
Fig. 6.
DDR studies with known Wnt inhibitors using cell lines CHLA-04-ATRT, CHLA-05-ATRT, CHLA-06-ATRT, NSC, HEK, and HeLa. The representative DDR curves shown are (A) WntC59, (B) niclosamide, (C) casin, and (D) pyrvinium. The IC50 values for these inhibitors are shown in (E). (F) shows a schematic indicating the Wnt inhibitors used in this study and their known targets.
Discussion
Earlier studies have shown the importance of Wnt genes in the biology of primitive neuroectodermal tumors and medulloblastomas.25,26 In the Wnt pathway, ligand-receptor interactions play a crucial role in determining the expression of downstream genes of this pathway. Our studies indicate that in ATRT biology, Wnt5B binds to Frizzled1 receptor and may play a crucial role in determining the differential expression and functioning of the downstream pathway genes. Wnt5B bound to Frizzled1 receptor, and its subsequent functional role has not been elucidated yet. In our study, CHLA-04-ATRT cells indicated a higher percentage of cell death when Frizzled1 receptor was blocked by FzMab and when both FRIZZLED1 siRNA and FzMab were added. CHLA-05-ATRT, however, indicated a higher percentage of cell death when Frizzled1 receptor was blocked both at the cell membrane level and the mRNA level. This difference in cell death numbers between the ATRT cell lines can be attributed to the higher expression of Frizzled1 receptor in CHLA-04-ATRT compared with the expression in CHLA-05-ATRT, as seen from nanoString analyses. The effects of Fz1Mab, either alone or combined with FRIZZLED1 siRNA, on ATRT cells will be studied further as a potential therapeutic option by investigating the downstream effects.
Wnt pathway is typically segregated as canonical Wnt pathway, PCP pathway, and the Wnt calcium pathway.9,27,28 In some cancers, a distinction can be made about the specific Wnt pathway that is crucial to the biology of the cancer. Examples are colorectal cancer29 and colon cancer,30 in which the canonical Wnt/β-catenin pathway has been shown to play an important role. In ATRT biology, however, our results from tumor tissues indicated that there is no clear distinction about the specific Wnt pathway that plays an important role. CTNNB1 is upregulated, while AXIN, APC, and GSK3β upstream of CTNNB131,32 are downregulated. DISHEVELLED (DVL) and CASEIN KINASE 1α (CK1α), which function as both canonical and noncanonical Wnt genes33,34 are both downregulated. DIKKOPF, a negative regulator of the Wnt pathway,35,36 and canonical MYC gene,37 are upregulated. The noncanonical receptors RYK, ROR1, and ROR238–40 are upregulated, while PCP pathway genes JNK, c-JUN, DAAM, RAC, and ROCK29 are downregulated. The NFAT gene of the Wnt calcium pathway41 is upregulated, while CaMKII is downregulated. Thus Wnt pathway genes need to be investigated further to elucidate their functions in ATRT biology. Wnt inhibitory factor (WIFI) is known to bind Wnt proteins and inhibit their functions and thus acts as a tumor suppressor in cancers.42 Studies have shown that in 75% of glioblastoma type of brain tumors and in astrocytomas, WIF1 is downregulated and these tumor cells are susceptible to Wnt inhibitors.43 WIF1 is downregulated in ATRT samples; thus, these cells may be susceptible to Wnt inhibitors.
Our study has indicated that some Wnt inhibitors (eg, WntC59, casin, pyrvinium pamoate, and niclosamide) may be promising therapeutic candidates that need to be further elucidated. WntC59 prevents palmitylation of Wnt proteins by Porcupine (Porcn, a membrane-bound O-acyltransferase), thereby blocking Wnt secretion and activity,44 while casin is a potent inhibitor of Cdc42.45 Pyrvinium pamoate, an FDA-approved drug, has been shown to be a potent inhibitor of Wnt signaling by activating protein kinase CK1α.46 This drug has also been shown to target β-catenin through the Akt/PKB/GSK3β pathway.47 Niclosamide has been shown to inhibit Wnt3A-Frizzled1 signaling48 that functions as part of the Wnt pathway.
In summary, our results demonstrate for the first time that components in the Wnt signaling pathway play a central role in ATRT biology and may thus constitute attractive therapeutic targets. Future studies will need to further determine the role of Wnt signaling in ATRT as well as provide in vitro and in vivo preclinical data on the therapeutic potential of Wnt pathway inhibitors in ATRT.
Supplementary Material
Funding
Phoenix Children's Hospital (PCH) Foundation, Barrow Neurological Institute at PCH, the Jaydie Lynn King family, and Students supporting brain tumor research, Grayson's Gift Foundation, “The Kids Cancer Project”, in Sydney Australia were funding sources.
Supplementary Material
Acknowledgments
Conflict of interest statement. None declared.
References
- 1.Buscariollo DL, Park HS, Roberts KB, et al. Survival outcomes in atypical teratoid rhabdoid tumor for patients undergoing radiotherapy in a Surveillance, Epidemiology, and End Results analysis. Cancer. 2012;118(17):4212–4219. doi: 10.1002/cncr.27373. [DOI] [PubMed] [Google Scholar]
- 2.Lafay-Cousin L, Hawkins C, Carret AS, et al. Central nervous system atypical teratoid rhabdoid tumours: the Canadian Paediatric Brain Tumour Consortium experience. Eur J Cancer. 2012;48(3):353–359. doi: 10.1016/j.ejca.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Coccé MC, Lubieniecki F, Kordes U, et al. A complex karyotype in an atypical teratoid/rhabdoid tumor: case report and review of the literature. J Neurooncol. 2011;104(1):375–380. doi: 10.1007/s11060-010-0478-0. [DOI] [PubMed] [Google Scholar]
- 4.Biegel JA, Burk CD, Parmiter AH, et al. Molecular analysis of a partial deletion of 22q in a central nervous system rhabdoid tumor. Genes Chromosomes Cancer. 1992;5(2):104–108. doi: 10.1002/gcc.2870050203. [DOI] [PubMed] [Google Scholar]
- 5.Lee RS, Stewart C, Carter SL, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122(8):2983–2988. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee MC, Park SK, Lim JS, et al. Atypical teratoid/rhabdoid tumor of the central nervous system: clinico-pathological study. Neuropathology. 2002;22(4):252–260. doi: 10.1046/j.1440-1789.2002.00458.x. [DOI] [PubMed] [Google Scholar]
- 7.Chakravadhanula M, Tembe W, Legendre C, et al. Detection of an atypical teratoid rhabdoid brain tumor gene deletion in circulating blood using next-generation sequencing. J Child Neurol. 2014;29(9):NP81–85. doi: 10.1177/0883073813503904. [DOI] [PubMed] [Google Scholar]
- 8.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 9.Rao TP, Kühl M. An updated overview on Wnt signaling pathways: a prelude for more. Circ Res. 2010;106(12):1798–1806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- 10.Nusse R, Van Ooyen A, Cox D, et al. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome15. Nature. 1984;307(5947):131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- 11.Korinek V, Barker N, Morin PJ, et al. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275(5307):1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 12.Gurney A, Axelrod F, Bond CJ, et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci USA. 2012;109(29):11717–11722. doi: 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S, Baye LM, Westfall TA, et al. Wnt5b-Ryk pathway provides directional signals to regulate gastrulation movement. J Cell Biol. 2010;190(2):263–278. doi: 10.1083/jcb.200912128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krausova M, Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal. 2014;26(3):570–579. doi: 10.1016/j.cellsig.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 15.Ellison DW, Dalton J, Kocak M, et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121(3):381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Remke M, Ramaswamy V, Taylor MD. Medulloblastoma molecular dissection: the way toward targeted therapy. Curr Opin Oncol. 2013;25(6):674–681. doi: 10.1097/CCO.0000000000000008. [DOI] [PubMed] [Google Scholar]
- 17.Pfister SM, Korshunov A, Kool M, et al. Molecular diagnostics of CNS embryonal tumors. Acta Neuropathol. 2010;120(5):553–566. doi: 10.1007/s00401-010-0751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin CT, Curtis CD, Davis RB, et al. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proc Natl Acad Sci USA. 2011;108(6):2282–2287. doi: 10.1073/pnas.1013751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtis CD, Griffin CT. The chromatin-remodeling enzymes BRG1 and CHD4 antagonistically regulate vascular Wnt signaling. Mol Cell Biol. 2012;32(7):1312–1320. doi: 10.1128/MCB.06222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mora-Blanco EL, Mishina Y, Tillman EJ, et al. Activation of β- catenin/TCF targets following loss of the tumor suppressor SNF5. Oncogene. 2014;33(7):933–938. doi: 10.1038/onc.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Erdreich-Epstein A, Gonzalez-Gomez I, et al. Novel cell lines established from pediatric brain tumors. J Neurooncol. 2012;107(2):269–280. doi: 10.1007/s11060-011-0756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erdreich-Epstein A, Robison N, Ren X, et al. PID1 (NYGGF4), a new growth-inhibitory gene in embryonal brain tumors and gliomas. Clin Cancer Res. 2014;20(4):827–836. doi: 10.1158/1078-0432.CCR-13-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang HJ, Kawasawa YI, Cheng F, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morioka K, Tanikawa C, Ochi K, et al. Orphan receptor tyrosine kinase ROR2 as a potential therapeutic target for osteosarcoma. Cancer Sci. 2009;100(7):1227–1233. doi: 10.1111/j.1349-7006.2009.01165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Utsuki S, Oka H, Sato Y, et al. E, N-cadherins and beta-catenin expression in medulloblastoma and atypical teratoid/rhabdoid tumor. Neurol Med Chir. 2004;44(8):402–406. doi: 10.2176/nmc.44.402. [DOI] [PubMed] [Google Scholar]
- 26.Rogers HA, Ward JH, Miller S, et al. The role of the WNT/β-catenin pathway in central nervous system primitive neuroectodermal tumours (CNS PNETs) Br J Cancer. 2013;108(10):2130–2141. doi: 10.1038/bjc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willert K, Nusse R. Wnt proteins. Cold Spring Harb Perspect Biol. 2012;4(9):a007864. doi: 10.1101/cshperspect.a007864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmerman ZF, Kulikauskas RM, Bomsztyk K, et al. Activation of Wnt/β-catenin signaling increases apoptosis in melanoma cells treated with trail. PLoS One. 2013;8(7):e69593. doi: 10.1371/journal.pone.0069593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larriba MJ, González-Sancho JM, Barbáchano A, et al. Vitamin D is a multilevel repressor of Wnt/b-catenin signaling in cancer cells. Cancers (Basel) 2013;5(4):1242–1260. doi: 10.3390/cancers5041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boman BM, Fields JZ. An APC: WNT counter-current-like mechanism regulates cell division along the human colonic crypt axis: A mechanism that explains how APC mutations induce proliferative abnormalities that drive colon cancer development. Front Oncol. 2013;3:e244. doi: 10.3389/fonc.2013.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee E, Salic A, Krüger R, et al. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 2003;1(1):E10. doi: 10.1371/journal.pbio.0000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitz Y, Rateitschak K, Wolkenhauer O. Analysing the impact of nucleo-cytoplasmic shuttling of β-catenin and its antagonists APC, Axin and GSK3 on Wnt/β-catenin signaling. Cell Signal. 2013;25(11):2210–2221. doi: 10.1016/j.cellsig.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Sinnberg T, Menzel M, Kaesler S, et al. Suppression of casein kinase 1alpha in melanoma cells induces a switch in beta-catenin signaling to promote metastasis. Cancer Res. 2010;70(17):6999–7009. doi: 10.1158/0008-5472.CAN-10-0645. [DOI] [PubMed] [Google Scholar]
- 34.González-Sancho JM, Greer YE, Abrahams CL, et al. Functional consequences of Wnt-induced dishevelled 2 phosphorylation in canonical and noncanonical Wnt signaling. J Biol Chem. 2013;288(13):9428–9437. doi: 10.1074/jbc.M112.448480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caricasole A, Copani A, Caraci F, et al. Induction of Dickkopf-1, a negative modulator of the Wnt pathway, is associated with neuronal degeneration in Alzheimer's brain. J Neurosci. 2004;24(26):6021–6027. doi: 10.1523/JNEUROSCI.1381-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niida A, Hiroko T, Kasai M, et al. DKK1, a negative regulator of Wnt signaling, is a target of the beta-catenin/TCF pathway. Oncogene. 2004;23(52):8520–8526. doi: 10.1038/sj.onc.1207892. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Li Y, Wu Y, et al. Wnt/β-catenin signaling pathway upregulates c-Myc expression to promote cell proliferation of P19 teratocarcinoma cells. Anat Rec (Hoboken) 2012;295(12):2104–2113. doi: 10.1002/ar.22592. [DOI] [PubMed] [Google Scholar]
- 38.Forrester WC. The Ror receptor tyrosine kinase family. Cell Mol Life Sci. 2002;59(1):83–96. doi: 10.1007/s00018-002-8407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoda A, Oishi I, Minami Y. Expression and function of the Ror-family receptor tyrosine kinases during development: lessons from genetic analyses of nematodes, mice, and humans. J Recept Signal Transduct Res. 2003;23(1):1–15. doi: 10.1081/rrs-120018757. [DOI] [PubMed] [Google Scholar]
- 40.Li W, Yamamoto V, Ortega B, et al. Mammalian Ryk is a Wnt coreceptor required for stimulation of neurite outgrowth. Cell. 2004;119(1):97–108. doi: 10.1016/j.cell.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 41.Huang T, Xie Z, Wang J, et al. Nuclear factor of activated T cells (NFAT) proteins repress canonical Wnt signaling via its interaction with Dishevelled (Dvl) protein and participate in regulating neural progenitor cell proliferation and differentiation. J Biol Chem. 2011;286(43):37399–37405. doi: 10.1074/jbc.M111.251165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malinauskas T, Aricescu AR, Lu W, et al. Modular mechanism of Wnt signaling inhibition by Wnt inhibitory factor 1. Nat Struct Mol Biol. 2011;18(8):886–893. doi: 10.1038/nsmb.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambiv WL, Vassallo I, Delorenzi M, et al. The Wnt inhibitory factor 1 (WIF1) is targeted in glioblastoma and has a tumor suppressing function potentially by induction of senescence. Neuro Oncol. 2011;13(7):736–747. doi: 10.1093/neuonc/nor036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Proffitt KD, Madan B, Ke Z, et al. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res. 2013;73(2):502–507. doi: 10.1158/0008-5472.CAN-12-2258. [DOI] [PubMed] [Google Scholar]
- 45.Peterson JR, Lebensohn AM, Pelish HE, et al. Biochemical suppression of small-molecule inhibitors: a strategy to identify inhibitor targets and signaling pathway components. Chem Biol. 2006;13(4):443–452. doi: 10.1016/j.chembiol.2006.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saraswati S, Alfaro MP, Thorne CA, et al. Pyrvinium, a potent small molecule Wnt inhibitor, promotes wound repair and post-MI cardiac remodeling. PLoS ONE. 2010;5(11):e15521. doi: 10.1371/journal.pone.0015521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venerando A, Girard C, Ruzzene M, et al. Pyrvinium pamoate does not activate protein kinase CK1, but promotes Akt/PKB down-regulation and GSK3 activation. Biochem J. 2013;452(1):131–137. doi: 10.1042/BJ20121140. [DOI] [PubMed] [Google Scholar]
- 48.Chen M, Wang J, Lu J, et al. The anti-helminthic niclosamide inhibits Wnt/Frizzled1 signaling. Biochemistry. 2009;48(43):10267–10274. doi: 10.1021/bi9009677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.