Abstract
Background
Glioblastomas remain ominous tumors that almost invariably escape treatment. Connexins are a family of transmembrane, gap junction-forming proteins, some members of which were reported to act as tumor suppressors and to modulate cellular metabolism in response to cytotoxic stress.
Methods
We analyzed the copy number and expression of the connexin (Cx)30 gene gap junction beta-6 (GJB6), as well as of its protein immunoreactivity in several public and proprietary repositories of glioblastomas, and their influence on patient survival. We evaluated the effect of the expression of this gap junction protein on the growth, DNA repair and energy metabolism, and treatment resistance of these tumors.
Results
The GJB6 gene was deleted in 25.8% of 751 analyzed tumors and mutated in 15.8% of 158 tumors. Cx30 immunoreactivity was absent in 28.9% of 145 tumors. Restoration of Cx30 expression in human glioblastoma cells reduced their growth in vitro and as xenografts in the striatum of immunodeficient mice. Cx30 immunoreactivity was, however, found to adversely affect survival in 2 independent retrospective cohorts of glioblastoma patients. Cx30 was found in clonogenic assays to protect glioblastoma cells against radiation-induced mortality and to decrease radiation-induced DNA damage. This radioprotection correlated with a heat shock protein 90–dependent mitochondrial translocation of Cx30 following radiation and an improved ATP production following this genotoxic stress.
Conclusion
These results underline the complex relationship between potential tumor suppressors and treatment resistance in glioblastomas and single out GJB6/Cx30 as a potential biomarker and target for therapeutic intervention in these tumors.
Keywords: connexin 30, glioblastoma, mitochondria, proliferation, radiation resistance
Glioblastoma multiforme (GBM) is the most prevalent and dismal central nervous system tumor.1 It thrives due to rapid proliferation, widespread brain invasion, and resistance to conventional treatments. While the biological events and key signaling pathway disruptions that drive its growth are being progressively deciphered,2,3 the tumor's mechanisms of resistance to conventional DNA-damaging agents and ionizing radiation—the mainstays of current therapies4—remain largely unknown.5
Connexins, the major constituents of gap junction channels, constitute a family of 21 transmembrane proteins6 that contribute to intercellular communication, help regulate cell proliferation and survival, and are frequently underexpressed in the early stages of oncogenesis.7 Astrocytes can express connexin (Cx)43, Cx30, and Cx26.8–10 Astrocytic gliomas, by contrast, frequently underexpress Cx43,11 which can act as a tumor suppressor in these tumors.12,13 Studies also suggest that astrocytomas may lack the expression of Cx30,14 and forced expression of this protein could reduce the tumorigenicity of rodent glioma cells.14
The tumor suppressive effects of connexins do not exclusively stem from the restoration of functional gap junction structures.14–16 Cx43, for instance, interacts with zona occludens (ZO)–1, ZO-2, Src, tubulin, and beta-catenin to modulate the growth control pathways of ZO-1–associated nucleic acid-binding protein, mitogen-activated protein kinase/extracellular signal-regulated kinase, and Wnt beta-catenin17–21 and to regulate the expression of cell cycle regulators like p27Kip, cyclins D1 and D3, and Skp2.15,22,23 Cx43 can also shuttle to the mitochondria and help protect cells against ischemia-reperfusion stresses.24 In addition, the carboxy terminus of Cx43 can shuttle to the nucleus, where it is believed to directly regulate gene expression and help inhibit cell growth.25,26 In contrast to Cx43, however, little is known of the mechanisms of Cx30-dependent cell growth inhibition, even though this protein can also associate with ZO-2 and alpha- and beta-catenin and can shuttle to the nucleus upon forced expression.20,27
In this study, we analyzed the tumor suppressive effects of Cx30 in malignant astrocytic gliomas. We demonstrated its growth inhibitory potential in these tumors and dissected the involved signaling cascades. In addition, however, Cx30 protected human gliomas against radiation-induced DNA damage via a heat shock protein (HSP)90–dependent translocation into the mitochondria that favors ATP production, DNA repair, and cell survival. As a consequence, Cx30 expression adversely affected patient survival in 2 independent cohorts of radiation-treated glioblastoma patients.
These results demonstrate the central regulatory role of Cx30 in the biology of human gliomas. Acting as both a tumor growth suppressor and a radioprotective agent, this protein is a potential biomarker and an attractive target for specific therapeutic interventions against these tumors.
Materials and Methods
Glioma Cells, Tissue Samples, and Patient Cohorts
Human U87 malignant glioma cells were obtained from the American Type Culture Collection, and their comparative genomic hybridization (CGH) genetic profiles were confirmed using Affymetrix single nucleotide polymorphism (SNP) 6.0 arrays and TP53 sequencing to ascertain their conformity. GM1 primary glioblastoma cells were derived from a fresh sample of human glioblastoma, as published previously,82 and characterized using glial fibrillary acidic protein immunohistochemistry, TP53 sequencing, and Affymetrix SNP 6.0 array CGH analysis. Cells were grown in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% fetal bovine serum (Gibco), and 1% 5 mg/mL penicillin–streptomycin solution (Gibco). Cells were grown at 37°C in 5% CO2. Biopsies and clinical data from patients treated at the University Hospital of Liège and at the University Medical Center of Utrecht were included in the study after institutional review board and ethic committee approval. Cohort 1 consisted of patients accrued between 1999 and 2001 at the University of Liège when the standard of care consisted of radiation therapy alone following surgery or biopsy, while cohort 2 included patients treated with surgery or biopsy followed by radiation therapy in combination with temozolomide4 between 2005 and 2008 at the University Medical Center of Utrecht (Supplementary Table S2).
Genetic Analysis
The GISTIC 2.0 algorithm28 was used to analyze Agilent CGH array data corresponding to 538 glioblastoma samples obtained from The Cancer Genome Atlas (TCGA) using the following dataset: (http://gdac.broadinstitute.org/runs/analyses__2012_02_17/data/GBM/20120217/gdac.broadinstitute.org_GBM.CopyNumber_Gistic2.Level_4.2012021700.0.0.tar.gz). Both pre-threshold and threshold copy number (CN) values were used to perform the correlations with mRNA expression data. Copy number analysis was also run using the Broad Institute version of the GISTIC 2.0 algorithm on 3 independent datasets, including 74 primary glioblastomas analyzed on an Affymetrix 100K array,28 97 glioblastomas analyzed on an Affymetrix 100K array from the Repository of Molecular Brain Neoplasia Data (REMBRANDT; http://rembrandt.nci.nih.gov), and 42 primary glioblastomas from the University Hospital of Utrecht, analyzed with Affymetrix 250 and SNP 6.0 arrays. Both the Genepattern platform of the Broad institute and the Partek suite were used to generate these analyses.
RNA-Seq sequencing data from 158 tumors deposited in TCGA of GBM were downloaded from The Cancer Genomics Hub (https://cghub.ucsc.edu/). Variant call format files were created with samtools and bcftools (http://samtools.sourceforge.net/mpileup.shtml), and mutation analysis was performed using SnpEff for the effect of the variations on gene gap junction beta-6 (GJB6) ---> NON_SYNONYMOUS_CODING, SYNONYMOUS_CODING polymorphisms in the GJB6 sequence.
Messenger RNA levels for transcripts carrying identified polymorphisms were determined on Agilent G4502A_07 TCGA data. The mRNA expression levels of the GJB6 gene of 42 primary glioblastomas from the University Hospital of Utrecht were analyzed with the Affymetrix GeneChip Human Genome U133 Plus 2.0 array. The U133 Plus 2.0 array of 97 glioblastomas from REMBRANDT were also examined for transcript levels.
The human methylation_27 array of 284 glioblastoma samples from TCGA were used without further processing to determine the methylation status of the chr13:19703656–0,cg03473518 and chr13:19703679–0,cg22377389 loci of the GJB6 promoter.
Compounds
We obtained 17-allylamino-17-demethoxygeldanamycin (17-AAG) from Sigma-Aldrich, in the form of lyophilized powder; it was stored in the dark at −20°C and reconstituted in dimethyl sulfoxide before use. Cells were treated with 17-AAG as described previously.37
Transient Transfections
U87 cells and GM1 cells were plated at low confluence for 24 h in a 100-mm culture dish or in T75 culture flasks (Corning). Appropriate quantities of pCMV6-XL5 plasmid (Origene Technologies) encoding or not for Cx30 DNA (Clongen) and dissolved in TansIT LT1 transfection reagent (Mirus Bio) were added to the cells according to the manufacturer's instructions. Cells were then incubated for 24–48 h prior to initiating further experiments, as described in the text.
Western Blot Analysis
Whole-cell protein extracts (20–50 µg) obtained using the radioimmunoprecipitation assay (RIPA) buffer extraction kit (Santa Cruz Biotechnology) or the mitochondrial extraction kit for cultured cells (Pierce) were separated on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel, using a Tris-glycine–sodium dodecyl sulfate buffer solution. After transfer using a Tris-glycine-methanol transfer buffer solution to a nylon transfer membrane (Immobilon-P, Merck Millipore) and blocking with Tris-buffered saline Tween 20 plus 5% dry milk powder, the membranes were incubated overnight at 4°C with a primary antibody. We used antibodies directed against Cx30 (rabbit polyclonal, 1: 500; Life Technologies), α-tubulin (mouse monoclonal, 1: 30 000; Sigma-Aldrich), extracellular signal-regulated kinase (ERK)1/2 (rabbit polyclonal, 1:5000; Sigma-Aldrich), diphosphorylated ERK1/2 (mouse monoclonal, 1:3000; Sigma-Aldrich), p21 (rabbit polyclonal, 1:1000; Santa Cruz), p27 (mouse monoclonal, 1:2000; BD Transduction Laboratories), CD1 (mouse monoclonal, 1:2000; Cell Signaling Technology), cMyc (rabbit polyclonal, 1:1000; Cell Signaling), survivin (rabbit polyclonal, 1:1000; Novus), cytochrome C (mouse monoclonal, 1:1000; BD Pharmigen), cytochrome C oxidase IV (rabbit monoclonal, 1:1000; Cell Signaling), phosphoAkt (T308) (rabbit monoclonal antibody, 1:1000; Cell Signaling), and Akt (mouse monoclonal antibody, 1:1000; Cell Signaling). The membranes were washed and finally incubated for 1 h with secondary peroxidase-conjugated anti-rabbit or anti-mouse antibodies (Dako) at a 1:2000 final dilution. The reaction was revealed by the enhanced chemoluminescence detection method (ECL Western Blotting Substrate, Pierce Biotechnology; or ECL Plus Western Blotting Detection Reagents, GE Healthcare) according to the manufacturer's instructions.
Quantitative Reverse Transcription Polymerase Chain Reaction
Cx30 mRNA levels were determined with reverse transcription (RT) PCR. Total RNA was extracted from U87 cells using the RNeasy Kit (Qiagen) according to the manufacturer's instructions. Recovered RNA was quantified by spectometry, and 1 μg RNA was subjected to RT using the Transcriptor First Strand cDNA Synthesis kit system for RT-PCR (Roche). The cDNA was amplified by TaqMan quantitative PCR according to the manufacturer's instruction and using SYBR Green and commercially available specific primers (Tebu-Bio reference PPH06117A24). For the rat Cx30, primers were as follows: right, GACATCAAACGGCAGAAGGT; left, TCTCTGGGCTGTGTTCTCT (Eurogentec).
Luciferase Reporter Assay
U87 cells (5 × 103 cells/well) were seeded in 24-well cell culture plates (Corning) and transfected with a Cx30-expressing vector or a control vector as described in the text. After 24 h, cells were further transfected with a test reporter (firefly luciferase), as described by Robe and colleagues83 for nuclear factor (NF)–kappaB and according to the TOPflash (Merck Millipore) transcription factor (TCF) reporter gene instructions for beta-catenin. These assays were normalized by cotransfecting cells with a Renilla luciferase reporter driven by a constitutive promoter. After lysis, firefly and Renilla luciferase activities of cell lysates were measured according to the manufacturer's instructions for the Dual Luciferase Reporter Assay System (Promega) in a luminometer. The relative firefly luciferase activity was calculated by normalizing transfection efficiency to Renilla luciferase activity.
Proliferation Assays
Cells (5 × 103) were seeded in 24-well culture plates (Corning). Cells were trypsinized and counted on a hemocytometer by the trypan blue exclusion method every day for 5 days. For some experiments, proliferation assays based on MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide) were used on cells grown in 96-well plates, as described previously.83
Kinase Assay
Cells were lysed using the RIPA buffer extraction kit (Santa Cruz), and lysates (300 μg) were incubated for 4 h at 4°C with anti–casein kinase 2 (CK2) antibody (clone 1AD9, Millipore). We added 25 μL GammaBind G Sepharose (GE Healthcare) to the extract and incubated it overnight. In vitro kinase assays were performed with immunoprecipitated proteins according to the manufacturer's instruction with the Casein Kinase 2 Assay Kit (Merck Millipore).
Intracranial Implantation of Tumor Cells in Immunodeficient Mice
All animal care and experiments were performed in agreement with the the European and Belgian regulations for animal care and under authorization from the Animal Ethics Committee of the University of Liège. Mice were anesthetized with an intraperitoneal injection of a 1:1 (volume/volume) solution of Rompum (sedativum 2%, Bayer) and Ketalar (ketamine 50 mg/mL, Pfizer). A suspension of 50 000 glioblastoma cells in 2 μL phosphate buffered saline (PBS) solution was stereotactically implanted by intracranial injection into the right striatum of the athymic nude mice. Mice were sacrificed at 28 days after injection.
Processing of Tissue Sections and Cell Cultures
Mice were anesthetized with pentobarbital (Nembutal, Ceva Sante Animal) before an intracardiac perfusion with NaCl 0.9% (VWR International, Prolabo), followed by paraformaldehyde 4% at 4°C. Brains were removed and postfixed, then cryoprotected for 48 h in PBS containing 20% sucrose before being frozen at −80°C in a 2- methylbutane solution (Sigma). Fourteen-micrometer-thick coronal and sagittal sections were cut on a cryostat and stored at −80°C.
Immunohistofluorescence and Histology
Permeabilization and blocking of nonspecific binding sites were obtained by a 30-min incubation at room temperature in PBS containing 10% donkey serum and 0.3% Triton X-100 in PBS. Brain sections were incubated with the primary antibodies (1:250, Zymed rabbit antibody to Cx30 and mouse monoclonal IgG1 to human mitochondria [Millipore], diluted in a carrier solution containing 0.2% donkey serum and 0.1% Triton X-100 in PBS), washed, and incubated with the relevant fluorescent-dye coupled secondary antibodies (Jackson Immunoresearch Laboratories). Tissue sections were mounted in an assembly Vectashield solution (Vector). For histology, some slides were stained with a hematoxylin/eosin solution, and the area of the maximal tumor section was calculated using the formula: (small diameter × large diameter/2). Sections were imaged and examined using a laser-scanning confocal microscope equipped with a krypton/argon gas layer (Olympus FluoView 1000). Figures were composed using ImageJ (National Institutes of Health) and Gimp software (GNOME Foundation). Cultures and injections were at least performed in triplicate.
Clonogenic Assays
Exponentially growing cells were seeded at low density (104/T75 flask; Corning) for 6 h to allow their attachment and then irradiated using a Gammacell 40 Exactor irradiator at a dose of 0, 4 Gy, or 10 Gy. Cells were then allowed to grow for 7–14 days in culture medium. Media were then removed and replaced with a solution of trypan blue. Live colonies of more than 20 cells were then counted in 3 low-power (10×) random microscopic fields. Results represent the mean of 3 experiments.
Comet Assays
About 10 000 cells were mixed with 75 of 1% agarose solution at 37°C. After gentle mixing, the cell suspension was rapidly spread on the CometSlide and maintained at 4°C for 10 min. Each slide was immersed in freshly prepared lysis solution (Comet Assay Kit, Trevigen) at 4°C for 1 h to remove the cell membrane and soluble contents of the cytoplasm and nucleoplasm, except for nuclear material. The slides were then treated with fresh alkaline solution (10 mM NaOH, 1 mM EDTA) at room temperature for 30 min to cleave the alkali-labile sites. The slides were treated with freshly prepared Tris-EDTA buffer (10 mM TrisCl, pH 7.5, 1 mM EDTA) for 5 min and then placed horizontally on an electrophoresis tray, which was filled with the alkaline solution (300 mM NaOH, 1 mM EDTA). Electrophoresis was then conducted at room temperature with an electrical field of 25V and a current of 300 mA for 40 min. After electrophoresis, the slides were dehydrated in 70% ethanol and stained with SYBR Green (Comet Assay Kit, Trevigen). The slides were examined using a fluorescence microscope (Zeiss Axioskop 40) attached to a video camera (Moticam 2000).
DNA Damage Studies
The incidence of DNA double-strand breaks (DSBs) was assessed using phosphoH2AX immunocytological detection. We used the anti-phosphoH2AX antibody (mouse monoclonal, 1:250; Upstate, Millipore) and the relevant fluorescent-dye coupled secondary antibodies (Jackson Immunoresearch) in blocking solution. At various times following irradiation, cells were fixed with paraformaldehyde 4%, were permeabilized with 0.1% Triton X-100 in PBS and blocking of nonspecific binding sites was obtained by a 30-min incubation at room temperature in PBS containing 4% donkey serum. Cells were then processed for immunohistochemistry and analyzed using a laser-scanning confocal microscope equipped with a krypton/argon gas layer (Olympus FluoView 1000). Results are representative of 3 independent experiments.
Senescence Assays—Beta-galactosidase Staining
Cellular senescence assays were performed using the Cellular Senescence Assay Kit (Chemicon) according to the manufacturer's instructions.
ATP Tests
ATP levels were measured using the ATP Assay Kit (Calbiochem) according to the manufacturer's instructions. Briefly, cells (5 × 103/well) were seeded in a 24-well cell culture plate (Corning) and grown for 24 h. They were then irradiated with a Gammacell 40 Exactor irradiator (4 Gy). After a given period of time (1–3 h), culture medium was removed and cells were treated with 120 μL nuclear releasing reagent for 5 min at room temperature with gentle shaking. Then, 1 L of ATP monitoring enzyme was added to the cell lysate, and samples were read in 1 min in a luminometer. Appropriate standard curves were obtained to verify the linearity of the reaction in the assessed range of measures. Experiments were reproduced 3 times independently.
Proximity Ligation Assays
Formalin-fixed paraffin embedded (FFPE) sections were deparafffinized in xylene and rehydrated through graded alcohols. Endogenous peroxidase was blocked with a 10-min incubation in 0.3% hydrogen peroxide in de-ionized water. Sections were then incubated in citrate buffer (pH 6) for 12 min at 126°C for antigen retrieval. After 3 washes in Tris-buffered saline, sections were stained with antibodies against Cx30 (rabbit polyclonal, 1:500; Life Technologies) and human mitochondria (mouse monoclonal, 1:250; Millipore) for 1 h at room temperature and proximity ligation assays were performed using Duolink immunoassay reagents, according to the manufacturer's instructions.
Immunofluorescence Microscopy
Immunofluorescence analysis was performed according to standard protocols. Briefly, cells were fixed with paraformaldehyde 4%, then rinsed with PBS and successively treated with 0.1% Triton X-100 for 5 min and rinsed in PBS for 15 min; blocking of nonspecific binding sites was obtained using a 4% donkey serum solution prepared in PBS. Cells were then incubated overnight at 4°C with antibodies against Cx30 (rabbit polyclonal, 1:500; Life Technologies), human mitochondria (mouse monoclonal, 1:250; Millipore), and beta-catenin (immunofluorescence preferred; mouse monoclonal, 1:1000; Cell Signaling) in blocking solution. After being rinsed with PBS, cells were incubated for 1 h at room temperature using fluorescein isothiocyanate or rhodamine-conjugated secondary antibodies (Jackson Immunoresearch) in blocking solution. Antibodies were used at a dilution of 1:2000. Cells were then incubated for 5 min with a PBS solution containing 4′,6′-diamidino-2-phenylindole (Sigma-Aldrich) to stain cell nuclei and then rinsed 3 times with PBS before examination with a laser-scanning confocal microscope equipped with a krypton/argon gas layer (Olympus FluoView 1000).
Statistical Analysis
Fisher's exact contingency test, ANOVA, 2-way ANOVA, and t-tests were performed using GraphPad Prism software. Kaplan–Meier survival curves were generated with this software, while Weibull multiple regression survival models were generated with Statview software (SAS).
Results
Connexin 30 Gene Deletion, mRNA Expression, and Pattern of Protein Expression in Malignant Glioma Patients
A GJB6 gene loss at 13q12 was found in 127 cases (23.6%), and amplification was found in 10 tumors (1.8%) of 538 glioblastomas in TCGA. These results were confirmed on 3 independent datasets: (i) a series of 74 primary glioblastomas from the Broad Institute,28 yielding 30 deletions (40.5%) versus 2 amplifications (2.7%); (ii) 97 glioblastomas from REMBRANDT, finding GJB6 deletions in 30 tumors (30.9%) and amplifications in 2 (2%); and (iii) a proprietary series of 42 primary glioblastomas operated on at the University Hospital of Utrecht, yielding 7 deletions (16.7%) and no amplification. Thus, on a total of 751 analyzed tumors, these analyses demonstrate a deletion of the GJB6 gene in 194 tumors (25.8%) and amplification in 14 (1.8%).
The exon sequencing data of 158 of the tumors of TCGA were then analyzed for GJB6 mutations, revealing frameshift mutations in 23 of these tumors (14.5%, 22 samples with 13_20796938 * +T and 1 with 13_20796938 * +TT) as well as 2 synonymous coding mutations (13_20797443 T→C and 13_20797590 G→A). These mutations were found in a similar percentage of tumors that displayed normal and reduced CN for GJB6 (17% and 19.3%, respectively, nonsignificant [NS], Fisher's exact test).
The methylation level of 2 loci (chr13:19703656-0,cg03473518 and chr13:19703679-0,cg22377389) of the GJB6 promoter was analyzed on 284 of the samples from TCGA. At a calculated beta-value cutoff of 0.7, corresponding to 95% of cytosine–phosphate–guanine island hypermethylation sites in glioblastomas,29 these sites were concomitantly hypermethylated in 38.2% of the tumors.
Neither GJB6 CN nor promoter methylation correlated with the mRNA expression of the TCGA tumors (NS, linear correlation and Student's t-test between GISTIC 2.0 CN unthresholded values and expression log2 values, n = 220). Similarly, we did not find any correlation between gene dosage and mRNA expression in our own samples and in the REMBRANDT dataset. On the other hand, however, glioblastoma samples that presented either of the 2 frameshift mutations of the GJB6 gene expressed significantly more GJB6 mRNA than GJB6 wild type–expressing tumors (P < .0001, Student's t-test, TCGA dataset).
The FFPE samples corresponding to our fresh frozen specimens were then analyzed for their expression of Cx30 by immunohistology. The expression of the protein was absent in 11 of 42 of these tumors (26.2%). The FFPE surgical specimen of 2 additional proprietary cohorts of glioblastomas were analyzed by immunohistochemistry for their expression of Cx30. Cohort 1 consisted of 50 patients operated on between 1999 and 2001 and treated first-line with radiation therapy alone. Cohort 2 consisted of 95 patients treated first-line with surgery followed by radiation therapy and concomitant plus adjuvant temozolomide chemotherapy. Seventeen patients in cohort 1 (34%) and 25 patients in cohort 2 (26.3%) did not display any immunoreactivity for Cx30.
Growth Inhibitory Effects of Connexin 30
Two cell lines (LN18 and U87) and 2 primary cultures (GM1 and GM2) of malignant gliomas were analyzed using Affymetrix 250K SNP arrays for their GJB6 CN, using a circular binary algorithm, and for Cx30 protein expression. All 4 cell types were found to lack Cx30 protein expression, both on confocal microscopy after immunostaining and on western blots. U87 cells additionally presented a deletion of the GJB6 gene. Using quantitative RT-PCR, the human U87 glioma cell line and GBM_GM1 human glioblastoma primary culture cells were found to strongly underexpress the Cx30 mRNA with respect to surgical samples of epileptic, nontumoral human cortex (Supplementary Fig. S1).
The in vitro transfection of U87 and GM1 primary cell cultures with plasmids encoding human Cx30 mRNA reproducibly and significantly inhibited their growth with respect to mock-transfected cells, as assessed by proliferating cell nuclear antigen immunolabeling, sequential hemocytometer counts, and clonogenic assays (Supplementary Fig. S2A–C and data not shown).
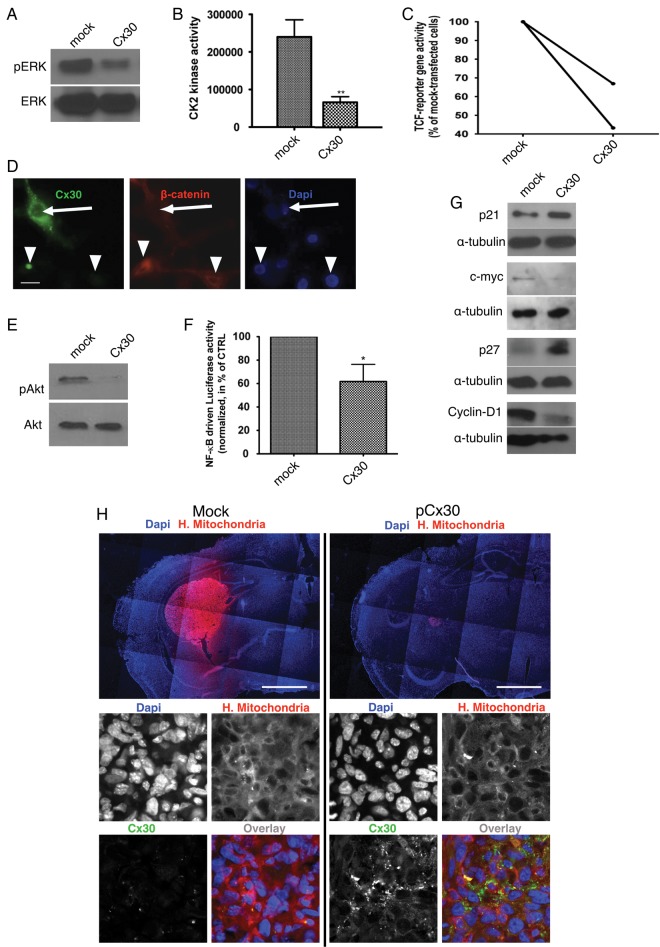
The restoration of Cx30 expression with a plasmid encoding GJB6 also reduced ERK/mitogen-activated protein kinase phosphorylation in U87 cells (Fig. 1A). It was recently reported that ERK could regulate the activity of CK2 in glioblastoma cells and hence beta-catenin/TCF-dependent transcription.30 Using specific kinase assays, we likewise observed that Cx30 expression inhibited the activity of CK2 in U87 cells (3.32 folds in C6 and 3.6 folds in U87 cells, P < .01, Student's t-test; Fig. 1B and data not shown). Using a specific transcription reporter plasmid, transient expression of Cx30 in U87 cells reproducibly halved the beta-catenin/TCF driven luciferase activity with respect to the mock-transfected control cells (Fig. 1C). Finally, immunofluorescence analysis of Cx30+ and Cx30− U87 cells also showed that beta-catenin was trapped in the cytoplasm of connexin-expressing cells (Fig. 1D). Likewise, forced expression of Cx30 in those cells reproducibly decreased the ratio of pAkt/Akt in these cells (Fig. 1E), as well as NF-kappaB transcriptional activity. Using canonical reporter plasmids, this latter was reduced by 38.25 ± 14.66% in U87 cells expressing Cx30 compared with mock-transfected cells (P < .05, paired Student's t-test; Fig. 1F). Downstream of these various signaling pathways, the levels of p21Cip and p27Kip on the one hand and cyclin D1 and cMyc on the other hand were respectively increased and decreased in Cx30-expressing U87 cells (Fig. 1G). Densitometric analysis for the western blots shown in Fig. 2A is provided in Supplementary Fig. S3.
Fig. 1.
Mechanisms of Cx30 effects on cell growth and proliferation in glioma cells. (A) Western blot analysis of phosphoERK and ERK expression in Cx30-expressing U87 cells compared with mock-transfected cells. This is representative of 3 independent experiments. (B) CK2 kinase activity is decreased in U87 cells following restoration of Cx30 expression. Kinase activities are shown as the mean ± SD of 3 independent experiments (**P < .01, Student's t-test, see text for details). (C) Forced expression of Cx30 reproducibly decreased beta-catenin TCF reporter gene activity in U87 cells (the results of 2 independent experiments are shown). (D) Immunofluorescence analysis showing a mainly cytoplasmic localization of beta-catenin (red) in Cx30-expressing cells (green) and a perinuclear localization in nontransfected cells (arrows). Nuclei are stained with 4′,6′-diamidino-2-phenylindole (DAPI) (blue). Scale bar = 20 μm. (E) Western blot analysis of phosphoAkt and Akt expression showed a decreased phosphorylation of this kinase in Cx30-expressing U87 cells compared with mock-transfected cells. This is representative of 3 independent experiments. (F) NF-kappaB driven luciferase activity is decreased in Cx30-expressing U87 cells compared with mock-transfected cells (n = 3, *P < .05, paired Student's t-test). (G) Forced expression of Cx30 in U87 cells respectively enhanced the expression of p21Cip and p27Kip and inhibited that of cyclin D1 and cMyc. These western blots are representative of 3 independent experiments. (H) In vivo effect of Cx30 expression on human glioma tumor growth. Intracranial implantation of a suspension of U87 cells transfected with a Cx30 expression vector or a control vector in the striatum of immunodeficient mice. Brain slices were stained using a monoclonal antibody to human mitochondria and red-fluorescent dye-coupled secondary antibody. After 4 weeks, the tumor mass was significantly smaller in Cx30-expressing xenografts in comparison with the mock-transfected ones (see text for details). Inserts: Immunofluorescence staining for Cx30 (green), human mitochondria (red), and human nuclei (blue). Scale bars: 1100 μm (upper panel) and 30 μm (lower panel).
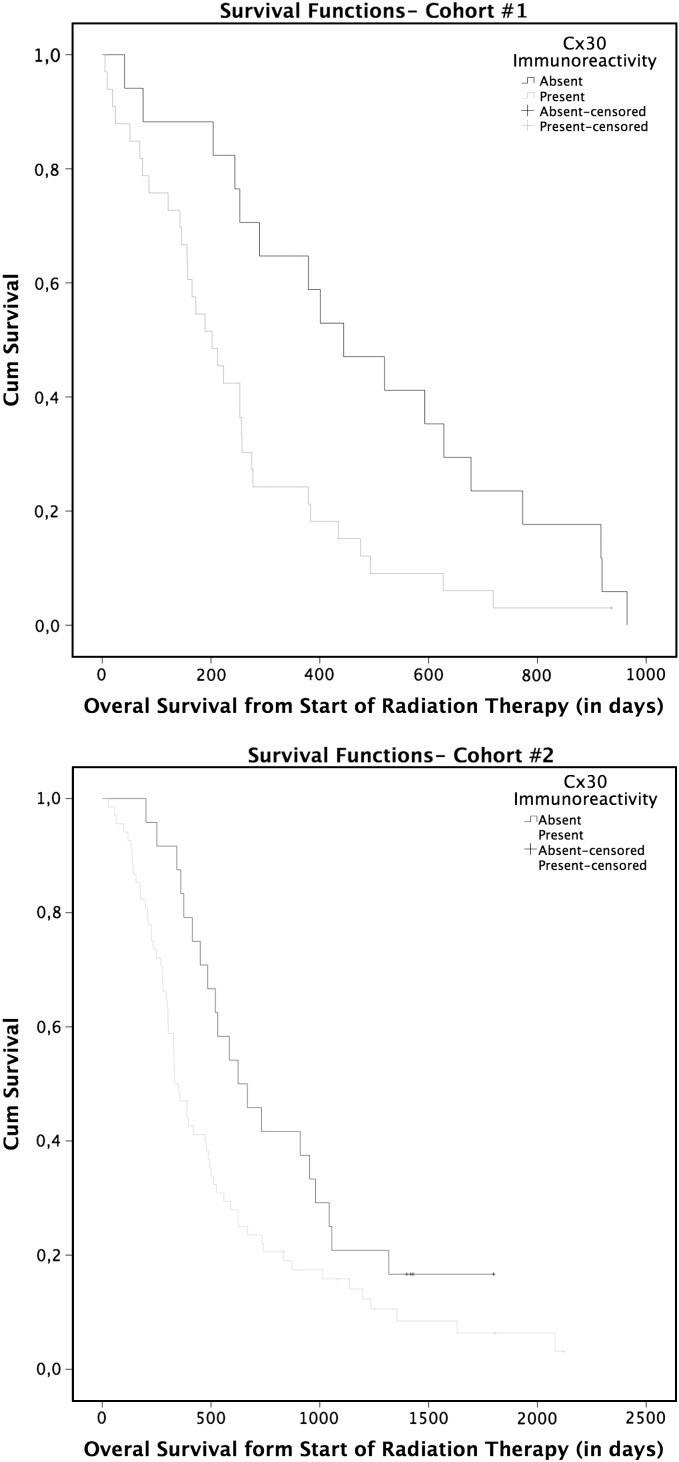
Fig. 2.
Patients survival. Kaplan–Meier curves Cohort 1 and Cohort 2, showing patients survival in comparison with Cx30 immunoreactivity in their GBM. Cx30 associates with an earlier mortality rate in patients expressing this protein (see text for details).
To corroborate our findings in vitro, we analyzed p27Kip nuclear expression by immunochemistry on the 95 FFPE samples of our second cohort of GBM patients. We observed a significant increase in p27Kip expression in Cx30+ patients compared with Cx30− tumors (independent samples t-test, P = .034).
We then proceeded to confirm these results in vivo using xenografts of mock-transfected and Cx30-expressing U87 cells in the striatum of immunodeficient mice. Mock-transfected U87 transplant animals developed significantly much larger tumors than the Cx30-expressing xenografts, with mean maximal section areas of 453000 ± 231100 μm2 and 20190 ± 9315 μm2, respectively (mean ± SEM, P < .05, n = 10 for each condition, Student's t-test). Immunofluorescence staining for Cx30 confirmed that the cells of these smaller tumors still expressed this connexin at the time of sacrifice (Fig. 1H).
Connexin 30 Expression in Malignant Glioma Adversely Affects Patient Survival
At the genetic level, overall patient survival did not correlate with the presence of GJB6 deletions in 517 analyzable tumors from TCGA and 67 tumors from REMBRANDT (log rank: P > .05, data not shown). Likewise, patient survival did not correlate with GJB6 mRNA expression in patients from TCGA (Cox proportional hazards model, NS).
We then analyzed the relationship between Cx30 protein immunostaining and survival in 2 independent series of patients. Cohort 1 consisted of 50 patients accrued between 1999 and 2001 when the standard of care consisted of radiation therapy alone following surgery or biopsy, while cohort 2 included patients treated with surgery or biopsy followed by radiation therapy in combination with temozolomide4 between 2005 and 2008. In the first cohort, Cx30 expression adversely influenced survival both in univariate analysis (log-rank: P < .05) and in a multivariate analysis using a Cox proportional hazards model and taking into consideration Cx30 (P < .001), KPS (P < .005), age (NS), and the type of surgery (biopsy vs resection, NS). In cohort 2 as well, Cx30 immunoreactivity also adversely influenced survival in univariate analysis (log rank: P < .05) and in multivariate analysis (P < .05) using a similar Cox model (age: P < .001, KPS: P < .05, and type of surgery: P < .05) (Fig. 2 and Supplementary Table S1A and B).
Connexin 30 Reduces Radiation Sensitivity In vitro
As the deleterious influence of Cx30 on glioblastoma patient survival contrasted with its growth suppressive properties, we investigated whether Cx30 modulated the radiation resistance of these tumors.
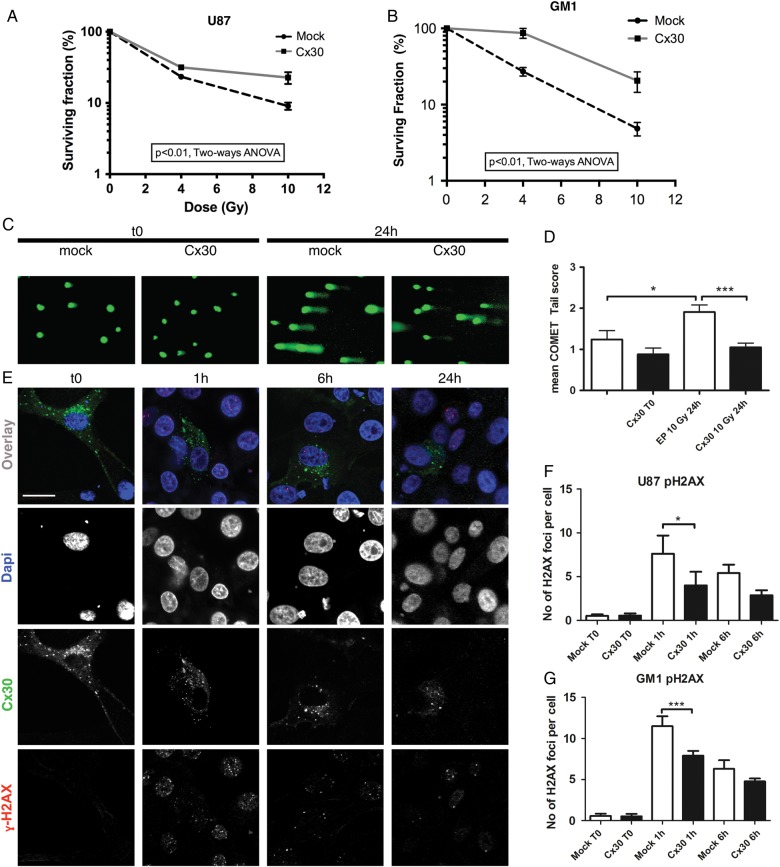
First, the clonogenic survival of Cx30-expressing U87 and GM1 cells was measured after 0, 4 Gy, and 10 Gy of gamma-irradiation and compared with that of mock-transfected cells. Forced Cx30 expression resulted in reduced radiation sensitivity in both cell types (Fig. 3A and B).
Fig. 3.
Clonogenic assays and DNA damage studies. Clonogenic survival curves assessed at baseline and 10 days after gamma irradiation (4 and 10 Gy) in (A) U87 and (B) GM1 cells expressing Cx30 compared with mock-transfected cells (P < .01 for both cell types, 2-way ANOVA). (C) Illustration of the DNA comet tails observed at baseline and 24 h after gamma irradiation (10 Gy) in Cx30-expressing U87 cells compared with mock-transfected cells. (D) Visual scoring analysis (mean ± SEM, 1-way ANOVA, mock nonirradiated vs mock irradiated cells P < .05, Cx30+ nonirradiated vs Cx30+ irradiated cells P < .001). (E) Confocal immunofluorescence illustration of the gamma H2AX foci at baseline and following gamma radiation treatment in Cx30-expressing cells. Dapi, 4′,6′-diamidino-2-phenylindole. Scale bars = 20 μm. (F) Average number of gamma H2AX foci per cell in U87 cells; at least 100 cells were counted per condition in 3 independent experiments (mean ± SD, 1-way ANOVA, P < .0001, Bonferroni multiple comparison test) and in (G) GM1 cells (at least 150 cells were counted per condition in 3 independent experiments) at baseline and 1 h and 6 h following gamma irradiation (mean ± SD, 1-way ANOVA, P < .0001, Bonferroni multiple comparison test).
Second, the influence of Cx30 expression on DNA DSBs, a hallmark of radiation-induced cytotoxicity,31 was explored on U87 cells using single cell gel electrophoresis (comet assay). Cx30-expressing U87 cells showed significantly smaller DNA tails than mock-transfected cells 24 h after a radiation treatment of 10 Gy, suggesting either reduced radiosensitivity or enhanced DNA repair in Cx30-expressing cells (Fig. 3C and D). The phosphorylation of histone H2AX (γH2AX), another marker of radiation-induced DSBs, was then assessed by immunochemistry at 1 and 6 h following a radiation exposure of 10 Gy gamma32 and was found to be consistently less intense in Cx30-expressing cells than mock-transfected cells (Fig. 3E–G and Supplementary Fig. S4).
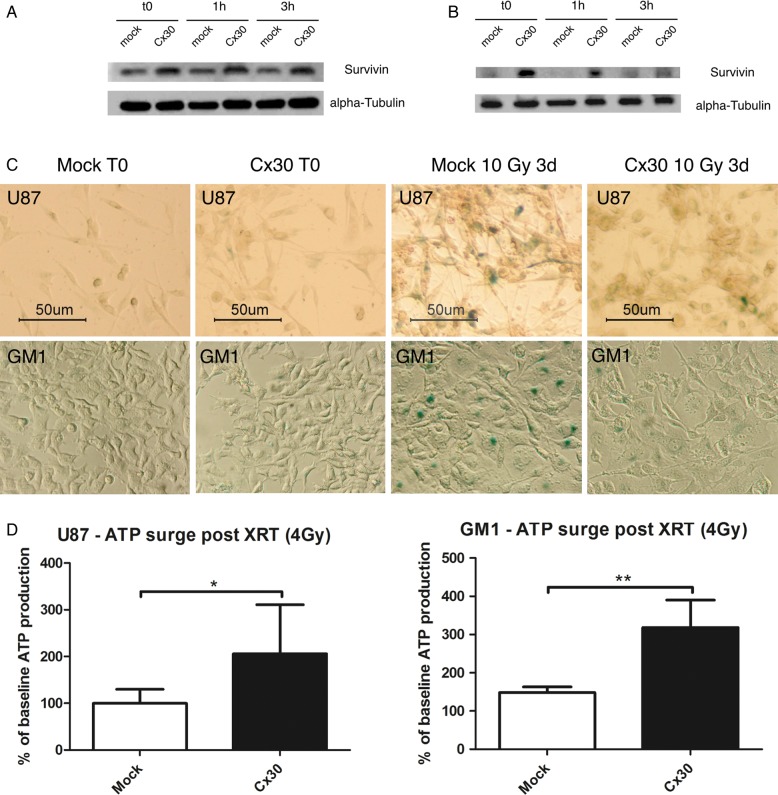
The expression levels of survivin, an anti-apoptotic protein previously associated with radioresistance in malignant glioma cells,33 were then observed. Western blots demonstrated a higher baseline and a further induction of the expression of this protein 1 h and 3 h following radiation treatment (4 Gy) in Cx30-expressing U87 and GM1 cells with respect to control cells (Fig. 4A and B and Supplementary Fig. S5). We did not observe any Cx30-mediated alteration of apoptotic cell death in our U87 cell cultures following irradiation (up to 10 Gy), based on caspase-9, -7, and -3 expression and activation western blots, apoptosis inducing factor nuclear translocation assays, and experiments by terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling (data not shown). Mitotic catastrophes also remained scarce irrespective of the Cx30 status of the cells at these doses of radiation, and we did not observe any autophagic cell death in our cultures using beclin-1 western blots, microtubule-associated protein/light chain 3 immunolabeling, and monodansylcadaverine incorporation (data not shown). In contrast, however, beta-galactosidase assays evidenced a significant radiation-induced senescence 3 days after radiation treatment (10 Gy) in Cx30− U87 and GM1 cells, but not in Cx30+ cells (Fig. 4C).
Fig. 4.
(A) Western blot analysis showed increased survivin expression levels in Cx30-expressing U87 cells and in (B) GM1 cells, at baseline and 1 h and 3 h after radiation treatment, compared with mock-transfected cells. This is representative of 3 independent experiments. (C) Beta-galactosidase staining showed a decreased radiation-induced senescence (10 Gy) in Cx30-expressing U87 and GM1 cells compared with mock-transfected cells, 3 days after gamma irradiation. Scale bars = 50 μm. (D) Analysis of the ATP content at baseline and 1 h following gamma irradiation in U87 and GM1 cells expressing Cx30 compared with mock-transfected cells (mean ± SD, unpaired Student's t-test, P < .05 for U87 [99.89 ± 10.62 vs 205.8 ± 37.19]; P < .01 for GM1 [148.2 ± 14.93 vs 317.9 ± 72.12]). XRT, external beam radiotherapy.
Radiation-Induced, Heat Shock Protein 90–Dependent Connexin 30 Mitochondrial Translocation and Energy Production
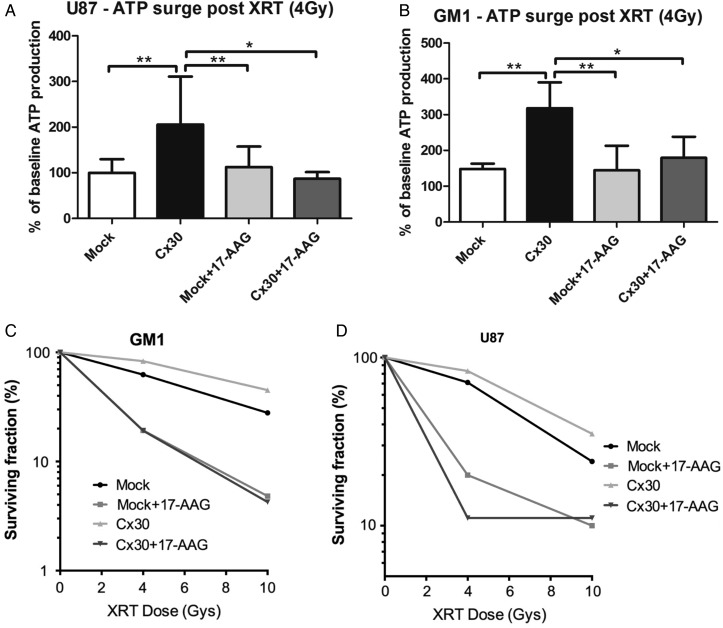
Since we previously showed that radioresistant glioma cells show increased ATP production following irradiation,33 we examined the effect of Cx30 expression on the cellular energy metabolism. The intracellular ATP levels increased in U87 and GM1 cells within 1 h following a radiation treatment of 4 Gy, but this increase was significantly stronger in Cx30-expressing cells (Fig. 4D).
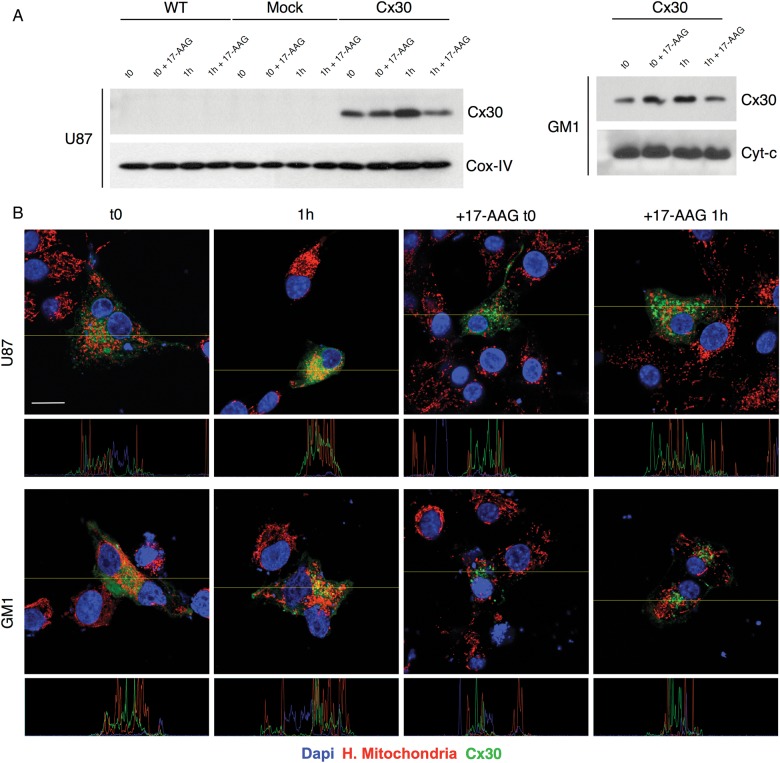
Another member of the connexin family of transmembrane proteins, Cx43, has been shown to also localize in the mitochondria of cardiac myocytes and neurons and to further translocate into these organelles in response to cellular stresses such as ischemia and in a translocase of the outer membrane– and ATP-driven HSP90 molecular chaperone–dependent fashion.34–36 Based on this evidence, we investigated whether Cx30 was localized in the mitochondria of human malignant glioma cells in our biopsies. Proximity ligation assays were performed on the second cohort of tumors using specific antibodies to Cx30 and to human mitochondria and showed a mitochondrial localization of Cx30 in a total of 77.6% of the Cx30+ tumors (Supplementary Fig. S6). Western blots performed on isolated mitochondrial extracts and double immunofluorescence experiments using specific antibodies to human mitochondria showed that in U87 and GM1 glioma cells engineered to express Cx30, Cx30 would also further translocate toward the mitochondria within 1 h after a radiation exposure of 4 Gy. Pretreatment with 17-AAG, an inhibitor of HSP-90,37 reproducibly countered this phenomenon, suggesting that HSP90 is involved in the radiation-induced translocation of Cx30 into the mitochondria of these cells (Fig. 5A and B and Supplementary Fig. S5). In parallel, 17-AAG pretreatment also blunted the ATP production observed in Cx30-expressing U87 and GM1 glioma cells compared with mock-transfected cells (one-sided Student's t-test, GM1 P < .01, U87 P < .05; Fig. 6A and B) and restored the sensitivity of Cx30-expressing U87 and GM1 cells to radiation (4 Gy and 10 Gy), as assessed after 3 weeks using clonogenic assays (Fig. 6C and D).
Fig. 5.
(A) Detection of the presence of Cx30 in mitochondria 1 h following gamma irradiation, in U87 and GM1 cells, by western blot analysis and (B) by confocal immunofluorescence, with or without pretreatment with 17-AAG. Scale bar = 20 μm. Dapi, 4′,6′-diamidino-2-phenylindole.
Fig. 6.
(A) Analysis of the ATP content 1 h following gamma irradiation and related to the baseline values, in U87 (mean ± SD, 1-way ANOVA P < .01, Bonferroni multiple comparison posttest) and (B) GM1 cells expressing Cx30 compared with mock-transfected cells, with or without pretreatment with 17-AAG (mean ± SD, 1-way ANOVA P < .01, Bonferroni multiple comparison posttest). XRT, external beam radiotherapy. Experiment representative of a minimum of 3 independent experiments for each cell type. (C) Clonogenic survival curves following gamma irradiation (4 Gy and 10 Gy) and related to the baseline values, in U87 and (D) GM1 cells expressing Cx30 compared with mock-transfected cells, with or without 17-AAG pretreatment (2-way ANOVA for both cells).
Discussion
While radiation therapy is a pillar of the therapeutic approach of glioblastoma, it extends patient survival by only a few months,4 while molecular knowledge of the radiation resistance of GBM remains limited.5 Connexins are known to regulate cell proliferation and are frequently underexpressed in tumors.38–41 In addition to their growth suppressive effects, however, Cx43 and Cx32 have been reported to protect rat glioma cells against a wide range of cellular injuries, including UV radiation and oxidative stress,42 and a Cx32-mediated radiation resistance of thyroid follicular cells has been hypothesized.43
Astrocytes normally express Cx43 and Cx30.44 Human malignant gliomas frequently lack Cx43 expression,11 and we have shown rodent gliomas to lack Cx30 as well.14 We have here observed that the GJB6 gene is deleted in over 25% of 751 human glioblastomas and mutated in 15% and that its encoded protein, Cx30, cannot be evidenced in almost 30% of the assessed tumors. These GJB6 CN alterations were similarly distributed in tumors of the different expression classification subtypes in the study by Verhaak and colleagues45 and did not associate with the methylator phenotype46 (data not shown).
Genomic rearrangements encompassing the GJB6 locus (13q12.11) are common in glioblastomas.47–50 This region is close to that of the tumor suppressor retinoblastoma 1 (13q14.2) and both sites are codeleted in 96% of cases (data not shown), raising the question as to whether GJB6 deletion could be a passenger event during gliomagenesis. The fact that this proportion is similar (94.5%) in 220 low-grade tumors from TCGA (data not shown) also supports this view but does not exclude that GJB6 exerts a proper tumor suppressive role, as do other connexins. For instance, deletions of the GJA1 gene encoding Cx43 and situated on 6q22.31 occur in only 11.3% of GBM51 and yet are thought to contribute to gliomagenesis.13,52–54 In support of a proper oncosuppressive role of GJB6 as well, we found a significant number of GJB6 frameshift focal deletions (15%) and promoter methylation (38%) in tumors, indicating that multiple mechanisms contribute to the loss of the expression of Cx30 in GBM. Moreover, microRNAs were recently shown to alter the expression of other connexins (Cx43)—in the myocardium, for instance.55
The frequency and various mechanisms of alterations of GJB6 led us to assess the growth suppressive effects of Cx30 in human gliomas. As in the case of rodent gliomas,14 the restoration of the expression of this protein strongly inhibited the growth and in situ tumorigenicity of human glial tumors. To the best of our knowledge, this is the first evidence of the growth suppressor role of Cx30 in human gliomas. We could, however, not directly assess the role of Cx30 in the control of cell proliferation in human gliomas, as all our primary or established tumor cell lines (>20 primary cultures tested, data not shown) had lost Cx30 expression in culture, which did not allow any knockdown experiment. We therefore chose to restore Cx30 expression in cell cultures. This expression did not enhance the gap junction intercellular communication (GJIC) capacities in our cells, as assessed by fluorescence activated cell sorting analysis of calcein dye transfer assays (data not shown). GJIC is, however, thought to play only a partial role in the tumor suppressive properties of other connexins, such as Cx43 and Cx32.15–17,39,56–58 In our experiments, the restoration of Cx30 expression inhibited ERK, CK2, and phosphatidylinositol-3 kinase–dependent signaling in our U87 glioma cells. It also repressed the nuclear translocation and transcriptional activity of beta-catenin and blunted p50/p65 NF-kappaB transcriptional activity, which might result from the observed inhibition of Akt59,60 and/or CK2 activity.61,62 While connexin-related ERK and beta-catenin/TCF inhibitions have previously been reported for Cx43,21,56,63,64 our work demonstrates for the first time an association between connexin expression and the activity of CK2, NF-kappaB, and Akt. All these pathways are major modulators of cell proliferation and survival65–69 and we indeed found that Cx30 reexpression upregulated the p21 and p27 cell cycle inhibitors and the repressed cell cycle enhancers cMyc and cyclin D1.
In spite of all this evidence for the growth suppressive role of Cx30, we found that the expression of this protein adversely correlated with survival in 2 unrelated retrospective cohorts of glioblastoma patients treated with surgery and either radiation therapy alone (cohort 1) or radiation therapy and adjuvant temozolomide chemotherapy (cohort 2, Stupp protocol).4 This effect of Cx30 on patient survival remained significant in multivariate analysis taking age, KPS, and type of surgery (biopsy vs resection) into account. This suggests a prognostic value of Cx30 expression as a marker of radiosensitivity in glioblastomas, an hypothesis that deserves confirmatory prospective clinical studies. As both series received radiation therapy, we wondered whether Cx30 could have dual antiproliferative and radioprotective effects, as described for other potential tumor suppressors.70,71 Forced Cx30 expression indeed reproducibly improved the clonogenic survival of U87 and GM1 cells after exposure to gamma radiation. Several mechanisms may contribute to this finding. First, Cx30 upregulated p21 and p27 and inhibited cMyc and cyclin D1 expression, preventing the progression of the cell cycle to the radiosensitive G2 and M phases of the cell cycle and slowing cell divisions, 2 long-known determinants of radiosensitivity.72–74 Second, Cx30+ cells expressed more survivin, a known factor of radioresistance,33 than Cx30− at baseline as well as following irradiation. Since Cx30 expression inhibited TCF and NF-kappaB, 2 transcriptional activators of survivin,75,76 it is likely that Cx30 also alters some other, uncharacterized, cell signaling pathways. Aside from its anti-apoptotic role, survivin can modulate the progression of the cell cycle at the G2/M checkpoint and thereby reduce radiosensitivity77 and can prevent cell senescence upon genotoxic stress,78 in agreement with our postradiation beta-galactosidase assays. Third, Cx30+ cells exhibited significantly less DSB damage than Cx30− cells 1 h after treatment, suggesting that Cx30 expression also improves the efficiency of DNA repair mechanisms.
We observed an HSP90-dependent, radiation-induced Cx30 mitochondrial translocation that correlated with ATP production and cell survival. These mitochondrial and metabolic responses to cellular stress are similar to those of Cx43 in neurons and cardiomyocytes in response to ischemic stress34,35 and might contribute to the radioresistance of the cells.33,79,80 Previous reports highlighted the radiosensitizing effect of HSP90 inhibitors in glioma cells,37 and these drugs are currently being evaluated in cancer clinical trials.81 Our data provide an additional mechanism to explain the activity of such agents in some Cx30-expressing cancers when administered in combination with radiation therapy. In addition, we also observed Cx30 expression in the mitochondria of surgical glioblastoma samples using proximity ligation assays, suggesting that Cx30 might have a role in mitochondrial homeostasis in these tumors beyond resistance to radiation therapy, for instance in response to other cellular stresses associated with cancerogenesis.
In conclusion, Cx30 exerts both growth suppressive and radioprotective effects in human glioblastomas, and its expression significantly—but adversely—correlates with patient survival following irradiation. These results pave the way for clinical studies aimed at confirming the value of Cx30 as a prognostic biomarker and as a target for sequential therapeutic intervention against glioblastoma.
Supplementary Material
Funding
This study was supported by a grant PNC 29-006 of the Belgian Ministry of Health to V.B. and P.A.R., grant 1.5.162.10 of the National Research Fund of Belgium to P.A.R., a grant from the Belgian Foundation against Cancer to V.B. and P.A.R., a grant from the Centre Anticancéreux of the University of Liege to V.B. and the T&P Bohnenn Fund for Neuro-Oncology Research to PR and TS and Televie grants to M.A and L.S.
Supplementary Material
Acknowledgments
We wish to thank Dr Sandra Ormenese for her help with flow cytometry, as well as Miguel Lopez y Cadenas, Patricia Ernst Gengoux, Olivier Hougrand, and Catherine Waltener for their expert technical assistance.
Conflict of interest statement. None declared.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Noda S-E, El-Jawahri A, Patel D, et al. Molecular advances of brain tumors in radiation oncology. Sem Radiat Oncol. 2009;19(3):171–178. doi: 10.1016/j.semradonc.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen PA. Molecular cloning, functional expression, and tissue distribution of a novel human gap junction-forming protein, connexin-31.9. interaction with zona occludens protein-1. J Biol Chem. 2002;277(41):38272–38283. doi: 10.1074/jbc.M205348200. [DOI] [PubMed] [Google Scholar]
- 7.Yamasaki H. Gap junctional communication and carcinogenesis. Carcinogenesis. 1990;11(7):1051–1058. doi: 10.1093/carcin/11.7.1051. [DOI] [PubMed] [Google Scholar]
- 8.Nagy JI, Rash JE. Connexins and gap junctions of astrocytes and oligodendrocytes in the CNS. Brain Res Rev. 2000;32(1):29–44. doi: 10.1016/s0165-0173(99)00066-1. [DOI] [PubMed] [Google Scholar]
- 9.Nagy J, Li X, Rempel J. Connexin26 in adult rodent central nervous system: demonstration at astrocytic gap junctions and colocalization with connexin30 and connexin43. J Comp Neurol. 2001;441(4):302–323. doi: 10.1002/cne.1414. [DOI] [PubMed] [Google Scholar]
- 10.Nagy JI, Dudek FE, Rash JE. Update on connexins and gap junctions in neurons and glia in the mammalian nervous system. Brain Res Rev. 2004;47(1–3):191–215. doi: 10.1016/j.brainresrev.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Shinoura N, Chen L, Wani MA, et al. Protein and messenger RNA expression of connexin43 in astrocytomas: implications in brain tumor gene therapy. J Neurosurg. 1996;84(5):839–845. doi: 10.3171/jns.1996.84.5.0839. discussion 846. [DOI] [PubMed] [Google Scholar]
- 12.Huang RP, Fan Y, Hossain MZ, et al. Reversion of the neoplastic phenotype of human glioblastoma cells by connexin 43 (cx43) Cancer Res. 1998;58(22):5089–5096. [PubMed] [Google Scholar]
- 13.Naus CCG, Elisevich K, Zhu D, et al. In vivo growth of C6 glioma cells transfected with connexin43 cDNA. Cancer Res. 1992;52(15):4208–4213. [PubMed] [Google Scholar]
- 14.Princen F, Robe P, Gros D, et al. Rat gap junction connexin-30 inhibits proliferation of glioma cell lines. Carcinogenesis. 2001;22(3):507–513. doi: 10.1093/carcin/22.3.507. [DOI] [PubMed] [Google Scholar]
- 15.Zhang YW. The gap junction-independent tumor-suppressing effect of connexin 43. J Biol Chem. 2003;278(45):44852–44856. doi: 10.1074/jbc.M305072200. [DOI] [PubMed] [Google Scholar]
- 16.Olbina G, Eckhart W. Mutations in the second extracellular region of connexin 43 prevent localization to the plasma membrane, but do not affect its ability to suppress cell growth. Mol Cancer Res. 2003;1(9):690–700. [PubMed] [Google Scholar]
- 17.Giepmans B. Gap junctions and connexin-interacting proteins. Cardiovasc Res. 2004;62(2):233–245. doi: 10.1016/j.cardiores.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Giepmans BN, Moolenaar WH. The gap junction protein connexin43 interacts with the second PDZ domain of the zona occludens-1 protein. Curr Biol. 1998;8(16):931–934. doi: 10.1016/s0960-9822(07)00375-2. [DOI] [PubMed] [Google Scholar]
- 19.Penes MC, Li X, Nagy JI. Expression of zonula occludens-1 (ZO-1) and the transcription factor ZO-1-associated nucleic acid-binding protein (ZONAB)-MsY3 in glial cells and colocalization at oligodendrocyte and astrocyte gap junctions in mouse brain. Eur J Neurosci. 2005;22(2):404–418. doi: 10.1111/j.1460-9568.2005.04225.x. [DOI] [PubMed] [Google Scholar]
- 20.Talhouk RS, Mroue R, Mokalled M, et al. Heterocellular interaction enhances recruitment of α and β-catenins and ZO-2 into functional gap-junction complexes and induces gap junction-dependant differentiation of mammary epithelial cells. Exp Cell Res. 2008;314(18):3275–3291. doi: 10.1016/j.yexcr.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 21.Ai Z, Fischer A, Spray DC, et al. Wnt-1 regulation of connexin43 in cardiac myocytes. J Clin Invest. 2000;105(2):161–171. doi: 10.1172/JCI7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koffler L, Roshong S, Kyu Park I, et al. Growth inhibition in G(1) and altered expression of cyclin D1 and p27(kip-1)after forced connexin expression in lung and liver carcinoma cells. J Cell Biochem. 2000;79(3):347–354. doi: 10.1002/1097-4644(20001201)79:3<347::aid-jcb10>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Tabernero A, Sánchez-Alvarez R, Medina JM. Increased levels of cyclins D1 and D3 after inhibition of gap junctional communication in astrocytes. J Neurochem. 2006;96(4):973–982. doi: 10.1111/j.1471-4159.2005.03623.x. [DOI] [PubMed] [Google Scholar]
- 24.Ruiz-Meana M, Rodriguez-Sinovas A, Cabestrero A, et al. Mitochondrial connexin43 as a new player in the pathophysiology of myocardial ischaemia-reperfusion injury. Cardiovasc Res. 2008;77(2):325–333. doi: 10.1093/cvr/cvm062. [DOI] [PubMed] [Google Scholar]
- 25.de Feijter AW, Matesic DF, Ruch RJ, et al. Localization and function of the connexin 43 gap-junction protein in normal and various oncogene-expressing rat liver epithelial cells. Mol Carcinog. 1996;16(4):203–212. doi: 10.1002/(SICI)1098-2744(199608)16:4<203::AID-MC4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 26.Dang X, Doble BW, Kardami E. The carboxy-tail of connexin-43 localizes to the nucleus and inhibits cell growth. Mol Cell Biochem. 2003;242(1–2):35–38. [PubMed] [Google Scholar]
- 27.Mennecier G, Derangeon M, Coronas V, et al. Aberrant expression and localization of connexin43 and connexin30 in a rat glioma cell line. Mol Carcinog. 2008;47(5):391–401. doi: 10.1002/mc.20393. [DOI] [PubMed] [Google Scholar]
- 28.Beroukhim R, Getz G, Nghiemphu L, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104(50):20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Ji Y, Qiu P. Identification of thresholds for dichotomizing DNA methylation data. EURASIP J Bioinform Syst Biol. 2013;2013(1):8. doi: 10.1186/1687-4153-2013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji H, Wang J, Nika H, et al. EGF-induced erk activation promotes CK2-mediated disassociation of α-catenin from β-catenin and transactivation of β-catenin. Mol Cell. 2009;36(4):547–559. doi: 10.1016/j.molcel.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332(2):237–248. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Mah L-J, El-Osta A, Karagiannis TC. GammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24(4):679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 33.Chakravarti A, Zhai GG, Zhang M, et al. Survivin enhances radiation resistance in primary human glioblastoma cells via caspase-independent mechanisms. Oncogene. 2004;23(45):7494–7506. doi: 10.1038/sj.onc.1208049. [DOI] [PubMed] [Google Scholar]
- 34.Azarashvili T, Baburina Y, Grachev D, et al. Calcium-induced permeability transition in rat brain mitochondria is promoted by carbenoxolone through targeting connexin43. Am J Physiol Cell Physiol. 2011;300(3):C707–C720. doi: 10.1152/ajpcell.00061.2010. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Sinovas A, Boengler K, Cabestrero A, et al. Translocation of connexin 43 to the inner mitochondrial membrane of cardiomyocytes through the heat shock protein 90-dependent TOM pathway and its importance for cardioprotection. Circ Res. 2006;99(1):93–101. doi: 10.1161/01.RES.0000230315.56904.de. [DOI] [PubMed] [Google Scholar]
- 36.Ellis RJ. Molecular chaperones: Plugging the transport gap. Nature. 2003;421(1):801–802. doi: 10.1038/421801a. [DOI] [PubMed] [Google Scholar]
- 37.Sauvageot CME, Weatherbee JL, Kesari S, et al. Efficacy of the HSP90 inhibitor 17-AAG in human glioma cell lines and tumorigenic glioma stem cells. Neuro Onc. 2009;11(2):109–121. doi: 10.1215/15228517-2008-060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamasaki H. Commentary: role of connexin genes in growth control. Carcinogenesis. 1996;11(6):1199–1213. doi: 10.1093/carcin/17.6.1199. [DOI] [PubMed] [Google Scholar]
- 39.King TJ, Gurley KE, Prunty J, et al. Deficiency in the gap junction protein connexin32 alters p27Kip1 tumor suppression and MAPK activation in a tissue-specific manner. Oncogene. 2005;24(10):1718–1726. doi: 10.1038/sj.onc.1208355. [DOI] [PubMed] [Google Scholar]
- 40.Cottin S, Gould PV, Cantin L, et al. Gap junctions in human glioblastomas: implications for suicide gene therapy. Cancer Gene Ther. 2011;18(9):674–681. doi: 10.1038/cgt.2011.38. [DOI] [PubMed] [Google Scholar]
- 41.Mesnil M, Crespin S, Avanzo J, et al. Defective gap junctional intercellular communication in the carcinogenic process. Biochim Biophys Acta. 2005;1719(1–2):125–145. doi: 10.1016/j.bbamem.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 42.Lin JH-C, Yang J, Liu S, et al. Connexin mediates gap junction-independent resistance to cellular injury. J Neurosci. 2003;23(2):430–441. doi: 10.1523/JNEUROSCI.23-02-00430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green LM, Tran DT, Murray DK, et al. Response of thyroid follicular cells to gamma irradiation compared to proton irradiation: II. The role of connexin 32. Radiat Res. 2002;158(4):475–485. doi: 10.1667/0033-7587(2002)158[0475:rotfct]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 44.Nagy JI, Patel D, Ochalski PA, et al. Connexin30 in rodent, cat and human brain: selective expression in gray matter astrocytes, co-localization with connexin43 at gap junctions and late developmental appearance. Neuroscience. 1999;88(2):447–468. doi: 10.1016/s0306-4522(98)00191-2. [DOI] [PubMed] [Google Scholar]
- 45.Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bredel M, Scholtens DM, Harsh GR, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302(3):261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J, Jiang C, Ng H-K, et al. Genome-wide allelotype study of primary glioblastoma multiforme. Chin Med J. 2003;116(4):577–583. [PubMed] [Google Scholar]
- 49.Kros JM, van Run PR, Alers JC, et al. Spatial variability of genomic aberrations in a large glioblastoma resection specimen. Acta Neuropathol. 2001;102(1):103–109. doi: 10.1007/s004010000327. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura M, Yang F, Fujisawa H, et al. Loss of heterozygosity on chromosome 19 in secondary glioblastomas. J Neuropathol Exp Neurol. 2000;59(6):539–543. doi: 10.1093/jnen/59.6.539. [DOI] [PubMed] [Google Scholar]
- 51.Sin WC, Crespin S, Mesnil M. Opposing roles of connexin43 in glioma progression. Biochim Biophys Acta. 2012;1818(8):2058–2067. doi: 10.1016/j.bbamem.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 52.Zhu D, Caveney S, Kidder GM, et al. Transfection of C6 glioma cells with connexin 43 cDNA: analysis of expression, intercellular coupling, and cell proliferation. Proc Natl Acad Sci U S A. 1991;88(5):1883–1887. doi: 10.1073/pnas.88.5.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu CT. CCN3 (NOV) Interacts with connexin43 in c6 glioma cells: possible mechanism of connexin-mediated growth suppression. J Biol Chem. 2004;279(35):36943–36950. doi: 10.1074/jbc.M403952200. [DOI] [PubMed] [Google Scholar]
- 54.Pu P, Xia Z, Yu S, et al. Altered expression of Cx43 in astrocytic tumors. Clin Neurol Neurosurg. 2004;107(1):49–54. doi: 10.1016/j.clineuro.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 55.Danielson LS, Park DS, Rotllan N, et al. Cardiovascular dysregulation of miR-17–92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 2013;27(4):1460–1467. doi: 10.1096/fj.12-221994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stains JP, Civitelli R. Gap junctions regulate extracellular signal-regulated kinase signaling to affect gene transcription. Mol Biol Cell. 2005;16(1):64–72. doi: 10.1091/mbc.E04-04-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sin WC, Bechberger JF, Rushlow WJ, et al. Dose-dependent differential upregulation of CCN1/Cyr61 and CCN3/NOV by the gap junction protein connexin43 in glioma cells. J Cell Biochem. 2008;103(6):1772–1782. doi: 10.1002/jcb.21571. [DOI] [PubMed] [Google Scholar]
- 58.King TJ, Lampe PD. Mice deficient for the gap junction protein Connexin32 exhibit increased radiation-induced tumorigenesis associated with elevated mitogen-activated protein kinase (p44/Erk1, p42/Erk2) activation. Carcinogenesis. 2004;25(5):669–680. doi: 10.1093/carcin/bgh071. [DOI] [PubMed] [Google Scholar]
- 59.Romashkova JA, Makarov SS. NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature. 1999;401(6748):86–90. doi: 10.1038/43474. [DOI] [PubMed] [Google Scholar]
- 60.Dan HC, Cooper MJ, Cogswell PC, et al. Akt-dependent regulation of NF-kappaB is controlled by mTOR and Raptor in association with IKK. Genes Dev. 2008;22(11):1490–1500. doi: 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kato T, Delhase M, Hoffmann A, et al. CK2 Is a C-Terminal IkappaB kinase responsible for NF-kappaB activation during the UV response? Mol Cell. 2003;12(4):829–839. doi: 10.1016/s1097-2765(03)00358-7. [DOI] [PubMed] [Google Scholar]
- 62.Yu M. Protein kinase casein kinase 2 mediates inhibitor-kappab kinase and aberrant nuclear factor-kappab activation by serum factor(s) in head and neck squamous carcinoma cells. Cancer Res. 2006;66(13):6722–6731. doi: 10.1158/0008-5472.CAN-05-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamei J, Toyofuku T, Hori M. Negative regulation of p21 by beta-catenin/TCF signaling: a novel mechanism by which cell adhesion molecules regulate cell proliferation. Biochem Biophys Res Comm. 2003;312(2):380–387. doi: 10.1016/j.bbrc.2003.10.129. [DOI] [PubMed] [Google Scholar]
- 64.Sirnes S, Bruun J, Kolberg M, et al. Connexin43 acts as a colorectal cancer tumor suppressor and predicts disease outcome. Int J Cancer. 2011;131(3):570–581. doi: 10.1002/ijc.26392. [DOI] [PubMed] [Google Scholar]
- 65.Olivier S, Robe P, Bours V. Can NF-kappaB be a target for novel and efficient anti-cancer agents? Biochem Pharmacol. 2006;72(9):1054–1068. doi: 10.1016/j.bcp.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 66.Bredel M, Bredel C, Juric D, et al. Tumor necrosis factor-alpha-induced protein 3 as a putative regulator of nuclear factor-kappaB-mediated resistance to O6-alkylating agents in human glioblastomas. J Clin Oncol. 2006;24(2):274–287. doi: 10.1200/JCO.2005.02.9405. [DOI] [PubMed] [Google Scholar]
- 67.Naugler WE, Karin M. NF-κB and cancer—identifying targets and mechanisms. Curr Opin Genet Dev. 2008;18(1):19–26. doi: 10.1016/j.gde.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411(6835):355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 69.Lopez-Ramos M, Prudent R, Moucadel V, et al. New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights. FASEB J. 2010;24(9):3171–3185. doi: 10.1096/fj.09-143743. [DOI] [PubMed] [Google Scholar]
- 70.Han Y, Huang H, Xiao Z, et al. Integrated analysis of gene expression profiles associated with response of platinum/paclitaxel-based treatment in epithelial ovarian cancer. PLoS ONE. 2012;7(12):e52745. doi: 10.1371/journal.pone.0052745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hötte GJ, Linam-Lennon N, Reynolds JV, et al. Radiation sensitivity of esophageal adenocarcinoma: the contribution of the RNA-binding protein RNPC1 and p21-mediated cell cycle arrest to radioresistance. Radiat Res. 2012;177(3):272–279. doi: 10.1667/rr2776.1. [DOI] [PubMed] [Google Scholar]
- 72.Packard C. The relation between division rate and the radiosensitivity of cells. Cancer Res. 1930;14(3):359–369. [Google Scholar]
- 73.Slowinski J, Mazurek U, Bierzynska-Macyszyn G, et al. Cell proliferative activity estimated by histone H2B mRNA level correlates with cytogenetic damage induced by radiation in human glioblastoma cell lines. J Neurooncol. 2005;71(3):237–243. doi: 10.1007/s11060-004-2030-6. [DOI] [PubMed] [Google Scholar]
- 74.Moeller BJ, Dreher MR, Rabbani ZN, et al. Pleiotropic effects of HIF-1 blockade on tumor radiosensitivity. Cancer Cell. 2005;8(2):99–110. doi: 10.1016/j.ccr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 75.Papanikolaou V, Iliopoulos D, Dimou I, et al. Survivin regulation by HER2 through NF-κB and c-myc in irradiated breast cancer cells. J Cell Mol Med. 2011;15(7):1542–1550. doi: 10.1111/j.1582-4934.2010.01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tapia JC, Torres VA, Rodriguez DA, et al. Casein kinase 2 (CK2) increases survivin expression via enhanced beta-catenin-T cell factor/lymphoid enhancer binding factor-dependent transcription. Proc Natl Acad Sci U S A. 2006;103(41):15079–15084. doi: 10.1073/pnas.0606845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rödel F, Hoffmann J, Distel L, et al. Survivin as a radioresistance factor, and prognostic and therapeutic target for radiotherapy in rectal cancer. Cancer Res. 2005;65(11):4881–4887. doi: 10.1158/0008-5472.CAN-04-3028. [DOI] [PubMed] [Google Scholar]
- 78.Wang Q, Wu PC, Roberson RS, et al. Survivin and escaping in therapy-induced cellular senescence. Int J Cancer. 2011;128(7):1546–1558. doi: 10.1002/ijc.25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kulkarni R, Reither A, Thomas RA, et al. Mitochondrial mutant cells are hypersensitive to ionizing radiation, phleomycin and mitomycin C. Mutat Res. 2009;663(1-2):46–51. doi: 10.1016/j.mrfmmm.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 80.Liu J, Hou M, Yuan T, et al. Enhanced cytotoxic effect of low doses of metformin combined with ionizing radiation on hepatoma cells via ATP deprivation and inhibition of DNA repair. Oncol Rep. 2012;28(4):1406–1412. doi: 10.3892/or.2012.1932. [DOI] [PubMed] [Google Scholar]
- 81.Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012;18(1):64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robe PA. In vitro and in vivo activity of the nuclear factor-kappab inhibitor sulfasalazine in human glioblastomas. Clin Cancer Res. 2004;10(16):5595–5603. doi: 10.1158/1078-0432.CCR-03-0392. [DOI] [PubMed] [Google Scholar]
- 83.Robe PA, Jolois O, N'Guyen M, et al. Modulation of the HSV-TK/ganciclovir bystander effect by n-butyrate in glioblastoma: correlation with gap-junction intercellular communication. Int J Oncol. 2004;25(1):187–192. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.