Abstract
Background
Regulatory T cells (Tregs) are potentially prognostic indicators in patients with glioblastoma. If differences in frequency of Tregs in tumor or blood account for substantial variation in patient survival, then reliably measuring Tregs may enhance treatment selection and improve outcomes.
Methods
We measured Tregs and CD3+ T cells in tumors and blood from 25 patients with newly diagnosed glioblastoma. Tumor-infiltrating Tregs and CD3+ T cells, measured by quantitative DNA demethylation analysis (epigenetic qPCR) and by immunohistochemistry, and peripheral blood Treg proportions measured by flow cytometry were correlated with patient survival. Additionally, we analyzed data from The Cancer Genome Atlas (TCGA) to correlate the expression of Treg markers with patient survival and glioblastoma subtypes.
Results
Tregs, as measured in tumor tissue and peripheral blood, did not correlate with patient survival. Although there was a correlation between tumor-infiltrating Tregs expression by epigenetic qPCR and immunohistochemistry, epigenetic qPCR was more sensitive and specific. Using data from TCGA, mRNA expression of Forkhead box protein 3 (FoxP3) and Helios and FoxP3 methylation level did not predict survival. While the classical glioblastoma subtype corresponded to lower expression of Treg markers, these markers did not predict survival in any of the glioblastoma subtypes.
Conclusions
Although immunosuppression is a hallmark of glioblastoma, Tregs as measured in tissue by gene expression, immunohistochemistry, or demethylation and Tregs in peripheral blood measured by flow cytometry do not predict survival of patients. Quantitative DNA demethylation analysis provides an objective, sensitive, and specific way of identifying Tregs and CD3+ T cells in glioblastoma.
Keywords: epigenetic qPCR, glioblastoma, immunotherapy, regulatory T cell, Tregs
Immunotherapy is emerging as an effective new treatment for many cancers,1 but it has yet to translate into a proven clinical benefit for patients with glioblastoma.2 A purported limitation of applying immunotherapy to gliomas is the tumor-induced impairment of the immune response, especially within the tumor microenvironment.3 Regulatory T cells (Tregs), a subpopulation of CD4+ T cells, characterized by the expression of the unique transcription factor Forkhead box protein 3 (FoxP3), depress immune function. Expression of Helios, a member of the Ikaros transcription factor family, is a marker for thymic or natural Tregs (nTregs).4 Although Tregs share functionality and suppression mechanisms across cancers, an increasing number of reports indicate tumor-specific variability in the proportion of Tregs in peripheral blood and in the tumor microenvironment. There are even reports where elevated numbers of Tregs inversely correlate with patient survival and/or response to immune therapies.5–7
The association between glioblastoma and immunosuppression suggests that accurately assessing immune competency could help to predict survival and identify patients most likely to benefit from immunotherapy. Severe CD4+ lymphopenia early in treatment correlates with shorter survival,8 while extensive lymphocytic infiltration of glioblastoma improves survival.9,10 In patients with glioblastoma, the percentage of Tregs in peripheral blood has been reported to be increased compared with normal controls.11,12 In tumor tissue, the number of Tregs correlates with persistent tumor burden and poor immune antitumor response.13
Several recent studies examining the prognostic implications of Tregs in patients with glioblastoma have reported inconsistent results. Heimberger et al14 and Lohr et al9 found no association between Tregs and prognosis in patients with glioblastoma, while Jacobs et al15 reported a moderate, although not statistically significant, inverse association between tumor Tregs and survival. In contrast, Yue et al16 found a significant association between the density of Tregs infiltrating glioblastoma and poor prognosis. Comparing measurements of Tregs from immunohistochemistry (IHC) and flow cytometry across studies is challenging because there are variations in surface and intracellular markers measured and differences in the parent population used to quantify Treg proportions. In this prospective study, we used epigenetic quantitative (q)PCR, a novel methodology to more accurately measure lymphocyte populations, to determine the association between Tregs measured in tumor tissue and peripheral blood and overall survival (OS) in patients with newly diagnosed glioblastoma.
Methods
Patients and Tissue Acquisition
We collected tissue specimens from 25 consecutive patients with glioblastoma surgically resected between January 1, 2005 and January 1, 2009 at Dartmouth Hitchcock Medical Center. Peripheral blood mononuclear cells (PBMCs) were available from all patients, and 23 glioblastoma tissue specimens were available for analysis. Twenty-two patients were deceased at the time of evaluation. At the time of surgery, all 25 patients had signed consent forms approved by the institutional Committee for the Protection of Human Subjects for use of brain tumor tissue for research. The 3 patients alive at the time of this study signed an additional “Authorization for Use/Disclosure of Protected Health Information” form consenting to their medical information being used. Normal brain tissue was obtained from the frontal lobes of autopsy tissue determined to be without abnormality by pathologic review. This study was conducted in accordance with recognized international scientific and ethical standards, including the principles of the Declaration of Helsinki.
The pathologic diagnosis of glioblastoma was confirmed by a certified neuropathologist. The O-methyl-guanine-methyltransferase (MGMT) methylation status of the tumor and the mutation status of isocitrate dehydrogenase (IDH)1 and IDH2 were determined when enough tissue was available, as described below. The clinical characteristics recorded for all cases included age at diagnosis, sex, Karnofsky Performance Scale (KPS) score, disease-free interval (defined as the interval between surgery and diagnosis of recurrent disease or death), date of diagnosis of recurrent disease, date of documented recurrence by MRI, and date of death. The treatment history was reviewed to obtain information about extent and date of surgical excision, chemotherapy administered, and radiation and targeted or investigational therapies delivered. The objective of this study was to correlate Treg frequency in surgically resected glioblastoma and OS, defined as the time from surgical resection until death or last follow-up for patients who were alive.
Lymphocyte Profiling by Epigenetic Quantitative PCR Analysis
Genomic DNA from formalin-fixed, paraffin-embedded (FFPE) (for glioblastoma tissue) or fresh frozen tissue (for normal brain) was isolated using the QIAamp DNA FFPE Tissue Kit and DNeasy Blood and Tissue Kit, respectively (Qiagen), according to the manufacturer's recommendations. As starting material, 6 to 8 tissue slices 10 µm thick were used for FFPE samples, and 25 mg of tissue for fresh frozen biopsies. All recovered DNA from FFPE samples and 5 µg DNA from fresh frozen tissue samples were bisulfite converted using the EpiTect Bisulfite kit (Qiagen). Epigenetic FoxP3 and CD3 qPCR analysis was performed by Epiontis GmbH as published previously.17,18 Measured cell subset values were expressed as percentages of Tregs or CD3+ T cells in the total cell population of the tissue sample, which included tumor cells, endothelial cells, and other immune infiltrating cells.
Immunohistochemical Analysis
IHC was performed to identify selected populations of tumor-infiltrating cells in FFPE tissue specimens. Deparaffinization, antigen retrieval, and staining were performed on the Leica Bond Max automated tissue staining instrument (Leica Microsystems). Glioblastoma tissues were sectioned (4 µm thick), air dried, then loaded onto the Leica Bond Max and baked for 30 min at 60°C. After deparaffinization, antigen retrieval was performed by heating at 100°C for 20 to 30 min (antibody specific) at pH 8.9, with automated Bond Epitope Retrieval 2. Staining was done with the Leica Bond Polymer Refine kit. Sections were incubated for 15 min with the optimized dilution of primary antibody against either CD3 (undiluted; Leica PA0553) or FoxP3 (1:100 dilution; Biolegend 623801). Slides were then incubated for 8 min with rabbit anti-mouse IgG, 8 min with anti-rabbit IgG–horseradish peroxidase, 8 min in polymer, then 5 min in 3% hydrogen peroxide to block endogenous peroxidase activity. Antibody was visualized with diaminobenzidine chromogen and hematoxylin counterstain (all steps as per the Leica Bond Polymer Refine kit).
Tregs in the tissue were evaluated by examining the slide images from a quantitative microscope using at least 3 different high-power fields (40×). Each of the specimens was examined microscopically by 2 independent blinded observers. Due to the low numbers of FoxP3-positive cells per high-power field, FoxP3 staining was rated as present or absent after review of multiple high-power fields. The number of CD3 cells that stained positively within an established area of the slide (3 high-power fields) was counted. Interrater reliability was determined using a correlation analysis (R = 0.991). Discrepancies between the recorded numbers of CD3+ cells prompted recounting of the cells in the specimens, with final arbitration conducted by the study neuropathologist.
Flow Cytometry
PBMCs were isolated from whole blood by Ficoll density centrifugation, washed, frozen in 90% pooled Human Serum AB (Gemini Bio-Products) plus 10% dimethyl sulfoxide (Sigma) freeze media, and stored at −140°C until use. Blood was drawn from patients after surgery and after completing concomitant radiation and temozolomide therapy. PBMCs for both timepoints from each patient were thawed, washed, and stained concurrently with combinations of the following fluorochrome-conjugated monoclonal antibodies for the surface staining of the cells: CD3 PerCPCy5.5 (BD Biosciences), CD4 APC Alexa 750, and CD25 PE-Cy7 (Biolegend). Intranuclear FoxP3 staining was performed using the Biolegend FoxP3-phycoerythrin kit in accordance with the manufacturer's instructions. The proportion and absolute number of peripheral blood Tregs were defined as the percent or number of CD3+ cells that were CD4+CD25+FoxP3+. Examining the proportion of Tregs in the T-cell population (CD3+) rather than as a proportion of the CD4+ fraction incorporates the interpatient range of values for the circulating CD4+ cell subset into the Treg analyses and facilitates comparison with epigenetic qPCR results.
MGMT Methylation Assay
Genomic DNA was isolated from FFPE using the Gentra PureGene Blood Kit Plus (Qiagen). Fifty nanograms of patient DNA, CpGenome Universal Unmethylated DNA control, and CpGenome Universal Methylated DNA control (Chemicon International) were treated with bisulfite using the MethylEdge Bisulfite Conversion System (Promega). MGMT methylation status was assessed using MethyLight, a quantitative assay that uses fluorescence-based real-time PCR (TaqMan) technology. Real-time PCR was performed using primers and TaqMan probes for the methylated MGMT gene and for the β-actin gene (ACTB), which was used as reference. A standard curve for MGMT and ACTB was generated using the methylated control DNA to obtain the relative amounts of methylated MGMT and ACTB amplicons in each sample. To evaluate the relative methylation level, the percentage of methylated reference (PMR) was calculated by dividing the MGMT sample/methylated control ratio by the ACTB sample/methylated control. A PMR cutoff of 4 was previously validated (data not shown). Samples showing a PMR <4 were defined as unmethylated, and samples with a PMR ≥4 were defined as methylated.
Isocitrate Dehydrogenase 1/2 Assessment
Screening for mutations in exon 4 of IDH1 and IDH2 genes was performed using a SNaPshot genotyping method. Multiplexed amplification of DNA targets was achieved by PCR with unlabeled oligonucleotide primers. Multiplexed single-base primer extension was performed with fluorescently labeled dideoxynucleotide triphosphate, and analysis of labeled primer-extension products was performed using capillary electrophoresis. All samples and a positive control were amplified with unlabeled primers and subjected to a multiplexed extension primer reaction using the SNaPshot Ready Reaction Mix (Life Technologies). Capillary electrophoresis of PCR products was performed using the ABI 3500 Genetic Analyzer with POP-7 polymer and 50 cm capillary. The genotyping results were analyzed using Applied Biosystems GeneMapper 4.1 software.
Analysis of The Cancer Genome Atlas Data
The Cancer Genome Atlas (TCGA) database was mined as previously described.19 Briefly, data from TCGA were obtained using Cbioportal (www.cbioportal.org) and TCGA data portal (http://cancergenome.nih.gov/). All glioblastoma samples available from the 2008 TCGA study20 were included in our analysis (n = 206) except where methylation was studied and those analyses included all patients with available HM27K methylation data (n = 285). It has been shown that nTregs are the predominant population of lymphocytes infiltrating human glioblastoma.4 We examined FoxP3 and Helios (expressed by nTregs)4 gene expression, as well as FoxP3 demethylation, and correlated them with survival in the tissue sample dataset available in TCGA. While Helios alone cannot identify Tregs, FoxP3 and Helios together can identify Tregs more accurately.21 Using patient data from the 2008 TCGA study,20 we further examined whether glioblastoma subtypes have different expression of Treg markers. We evaluated the mRNA expression of FoxP3 and Helios in the 4 subtypes and then grouped patients based on subtype to look for correlations between the expression of Treg markers and OS.
Statistics
The null hypothesis was that Treg frequencies in the glioblastoma specimens did not correlate with survival. The primary alternative hypothesis was that Treg frequencies in the glioblastoma specimens did correlate with survival. The primary analysis was to compare 2 groups as defined by Treg frequency. Patients were dichotomized into a high Treg frequency group and a low Treg frequency group based on median Treg' frequencies. All statistical tests were 2-sided, and the overall type I error was 0.05. We used a Kaplan–Meier plot to demonstrate the differences in survival probability for high Treg patients versus low Treg patients. A log-rank test (Mantel–Cox) was used to determine if the 2 survival curves were significantly different from each other. A Cox proportional hazards model including age and MGMT status as confounding variables was applied to test if there was a variable of interest that was associated with survival after the adjustment. The adjusted P-value was calculated using a Wald test. Power analysis was performed using R statistical software. Briefly, the power function in R was used to compute the statistical power based on sample size, variance, significance level (.05), and null hypothesis value.
A log transformation was performed for variables that were not normally distributed, including cell count (IHC) and ratios. A simple linear regression method was applied to test for correlation of 2 continuous measurements, while a Pearson correlation test was used for comparison of a continuous and noncontinuous variable. To further characterize the association between the 2 methods used to identify tissue Tregs, the dichotomous outcome measure (IHC), and the continuous measure (epigenetic qPCR), we applied a receiver operating characteristic (ROC) curve. A 1-way multiple-comparison ANOVA (Tukey post hoc test) was performed on the 2008 TCGA dataset for glioblastoma to test for differential expression of Treg markers in subtypes of glioblastoma.
Results
Patient Characteristics
Patient demographic characteristics are summarized in Table 1. All patients had a KPS score of at least 70. The median OS for the 25 patients was 18 months, with 2 patients still alive more than 5 years after diagnosis as previously published.22 All patients had at least a partial resection (mean resected tumor volume 73.9% ± 17.9% by MRI volumetric analysis using ITKsnap).23 Corticosteroids were tapered after surgery to a minimum effective dose to control cerebral edema. Tumor tissue was available from 18 patients for IDH1 and IDH2 analysis. One tumor was found to have an IDH1 mutation, and there were no IDH2 mutations (n = 17).
Table 1.
Patient characteristics
| Demographic | Statistics |
|---|---|
| Patients | N = 25 |
| Age, y, mean ± SD | 68.4 ± 11.2 |
| OS, mo, median/mean ± SD | 18/25.8 ± 18.4 |
| MGMT hypermethylation | N = 8/23 (34.8%) |
| Percent resection, mean ± SD | 73.9% ± 17.9% |
Briefly, as reported in our earlier paper,22 based on multicolor flow cytometry cell subset analysis, treatment (radiation plus temozolomide) increased the proportion of functional Tregs in peripheral blood, but their absolute number remained stable. Ten of the patients reported on in the current cohort were treated on a protocol using a dendritic cell vaccine.22 At time of progression, 13 patients were treated with bevacizumab plus irinotecan, 3 were treated with bevacizumab plus temozolomide, 2 were treated with temozolomide plus irinotecan, and 1 received temozolomide alone. Two patients received other chemotherapy at time of progression (gimatecan and enzastaurin, respectively). The remaining 3 patients received supportive care without further chemotherapy.
Infiltrating T Cells and Tregs, Circulating Tregs, and Patient Survival
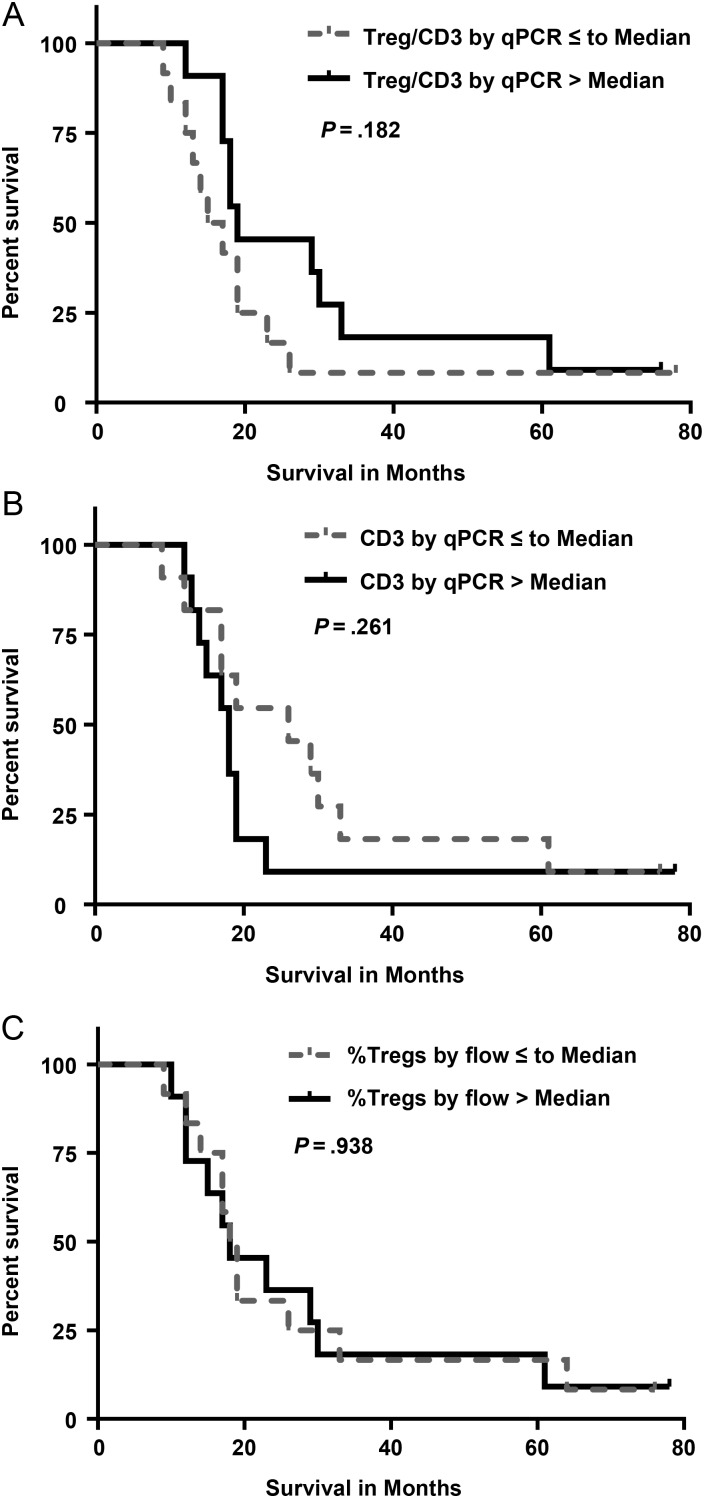
Based on epigenetic qPCR of glioblastoma tissue, the median percent of total cells that were Tregs was 0.3% (range, 0%–3%) and that of CD3+ T cells was 3.7% (range, 0.6%–12.9%). The average ratio of Tregs to CD3+ T cells was 0.13 ± 0.13 (range, 0–0.64). The extent of tumor infiltration by Tregs (Tregs:CD3 ratio; Fig. 1A) and CD3+ T cells (Fig. 1B) did not correlate with OS (P = .18 and P = .26, respectively), even after adjusting for age and MGMT status (P = .55 and P = .29, respectively). In addition, we examined the relationship between OS and the proportion of Tregs in the T-cell population in peripheral blood by flow cytometry. Due to limitations in the number of PBMCs available, we were unable to additionally assess peripheral blood Tregs using the epigenetic qPCR method. We found no correlation between the pretreatment (postsurgery) percentage of Tregs in the peripheral blood T-cell population and survival for this patient cohort (P = .94; Fig. 1C).
Fig. 1.
(A) Kaplan–Meier survival curves for patients with a high ratio (above median value) versus low ratio (below or equal to median value) of tumor-infiltrating CD3+ cells that are Tregs as determined by epigenetic qPCR method, P = .182. (B) Kaplan–Meier survival curves for patients based on the proportion of tumor-infiltrating CD3+ T cells as determined by epigenetic qPCR (P = .261). Patients were divided into those with a percentage of CD3+ T cells above the median value and those with a percentage of CD3+ T cells below or equal to the median value. (C) Kaplan–Meier survival curves for the pretreatment (postsurgery) percentage of Tregs in the peripheral blood T-cell population determined by flow cytometry (P = .938, percent of CD3+ cells that are CD4+CD25+FoxP3+). Patients were divided into those with a high percentage of Tregs (above median value) and a low percentage of Tregs (below or equal to median value).
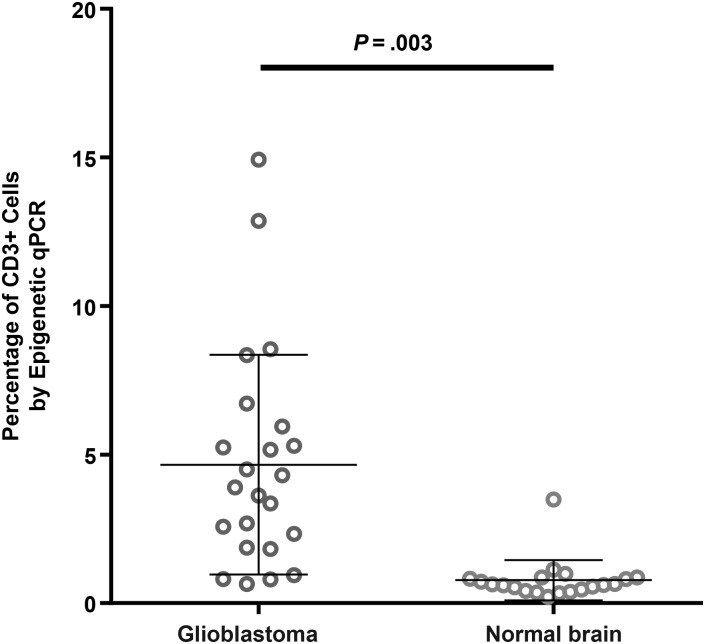
To further examine the sensitivity of the novel method of epigenetic qPCR for quantifying Tregs, we also analyzed 20 normal brain samples from necropsy material of the frontal lobe and found that Tregs ranged from 0% to 0.1% of total cells, close to the detection limit for the quantity of DNA available, and CD3+ cells were 0.62% of total cells. Compared with healthy brain tissue, glioblastoma tissue had significantly greater percentages of infiltrating CD3+ cells and a more heterogeneous range of values (P = .003; Fig. 2).
Fig. 2.
Results of epigenetic qPCR determination of percent of CD3 frequency in brain tumor tissue from patients with glioblastoma (n = 23) and normal brain tissue from necropsy specimens (n = 20) showed a significant difference in infiltrating CD3 cells (P = .003).
By IHC analysis, FoxP3+ cells were observed in 47% of the glioblastoma samples. By IHC counts, a median of 38.9 CD3+ T cells/mm2 were found in glioma tissue (range, 1–307 cells/mm2). We found no relationship between adjusted OS and the presence or absence of detected Tregs (P = .53) or the number of tumor-infiltrating CD3+ cells (P = .58).
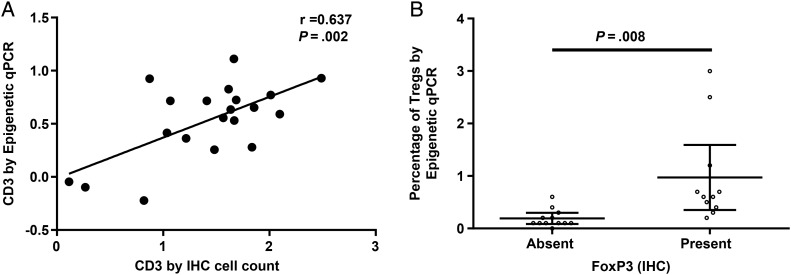
We compared the results from the IHC and epigenetic qPCR methods of quantifying Tregs and T cells in glioblastoma tissue. The 2 methodologies correlated for determination of CD3+ tumor-infiltrating T cells (correlation coefficient 0.64, P = .002; Fig. 3A). A 2-sample t-test demonstrated a significant relationship between the dichotomous (IHC) and continuous (epigenetic qPCR) Treg measures (P = .008; Fig. 3B), which was further supported by an area under the ROC curve of 0.92.
Fig. 3.
(A) Correlation between log percents of CD3+ tumor-infiltrating T cells determined by epigenetic qPCR and log-transformed absolute numbers of CD3+ tumor-infiltrating T cells determined by IHC analysis, P = .002. (B) Relationship between tumor-infiltrating Tregs as determined by IHC and epigenetic qPCR assay, P = .008. Tregs were divided into present or absent based on IHC, and the corresponding percent of Tregs as determined by epigenetic qPCR (%FoxP3) for each patient was plotted. Error bars represent 95% confidence intervals.
Tregs and Patient Survival in TCGA Database
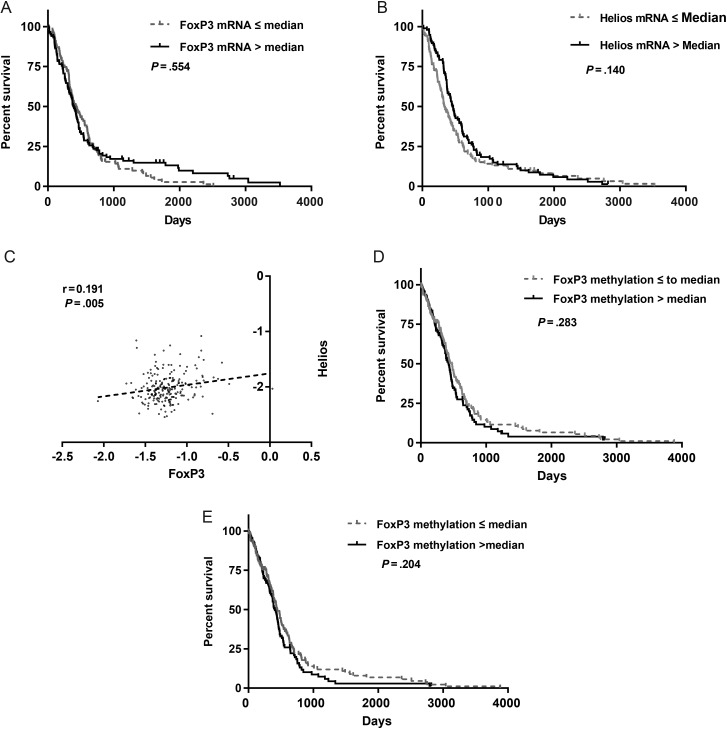
To corroborate results for our institution's cohort of patients, we analyzed whether markers of Tregs could predict survival in a larger glioblastoma patient cohort (n = 206) from TCGA.20 There were no tumors with IDH1 or IDH2 mutations in this cohort. We dichotomized patients with glioblastoma from TCGA according to expression or methylation of Treg markers in tumors relative to the median value for the specific gene. We found that patients with glioblastoma who had low FoxP3 and Helios expression (Z ≤ 0) did not have a survival advantage compared with the patients with high tumor expression (Z > 0) (Fig. 4A and B). Based on a Pearson correlation test, direct correlation between FoxP3 and Helios mRNA expression was observed (r = 0.191, P = .005; Fig. 4C). Further, we analyzed whether methylation of FoxP3 could predict survival for patients with glioblastoma. For this analysis we used the cohort of TCGA with available HM27K methylation data (n = 285). Our results show that patients with high methylation (defined as greater than the median) of FoxP3 in their tumors did not have a survival advantage compared with patients with low methylation of FoxP3 (P = .28; Fig. 4D). When we excluded the 9 patients in the cohort with IDH1 mutations, we still did not find a correlation between survival and methylation of FoxP3 (P = .20; Fig. 4E).
Fig. 4.
(A) Kaplan–Meier survival analysis of patients with glioblastoma from TCGA analysis whose tumors expressed variable levels of FoxP3 and (B) Helios mRNA. (C) Dot plot demonstrating direct correlation between FoxP3 and Helios mRNA levels in glioblastoma tissue. Slope of linear regression is 0.208 ± 0.075. The r-value is 0.191 and the Pearson P-value was significant and equal to .005. (D) Kaplan–Meier survival curves of patients from TCGA analysis with tumor levels of FoxP3 DNA methylation above and below the median value (P = .28). (E) Kaplan–Meier survival curves for patients from TCGA analysis, excluding the 9 patients with IDH1 mutations. Patients were divided identically to (D), and there was still no significant difference in OS (P = .204).
Glioblastoma Subtype Analysis
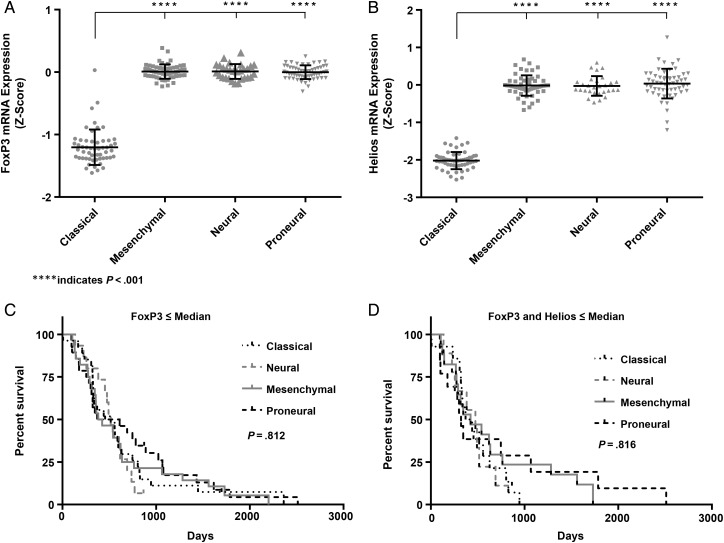
Transcriptome analysis of glioblastoma has revealed 4 molecular subtypes: proneural, neural, classical, and mesenchymal.24 In our analysis of the 2008 TCGA database (n = 206), the classical subtype of glioblastoma had a significantly reduced expression of Tregs, FoxP3, and Helios markers (Fig. 5A and B). Neither FoxP3 nor Helios expression predicted survival in any of the glioblastoma subtypes. Even when expression of FoxP3 and of Helios were combined, there was no correlation between survival and glioblastoma subtypes (Fig. 5C and D).
Fig. 5.
(A) mRNA expression analysis of FoxP3 and (B) Helios in tumors of patients with different subtypes of glioblastoma. Multiple-comparison 1-way ANOVA indicated significant difference in the expression of Treg markers between the classical group and other subtypes. (C) Glioblastoma patients with low FoxP3 mRNA or (D) low FoxP3 and Helios mRNA were analyzed for OS. The cohort analyzed included 54 classical, 56 proneural, 56 mesenchymal, and 29 neural glioblastoma cases. No difference was found between groups for either FoxP3 (P = .812) or FoxP3 and Helios combined (P = .816).
Discussion
We hypothesized that the frequency of Tregs in tissue or peripheral blood would correlate with survival of patients with glioblastoma. We detected Treg infiltration in tumor tissue by IHC in approximately half of the patient samples, consistent with results in the literature.9,14 Epigenetic qPCR was more sensitive and identified infiltrating Tregs in all but one of the glioblastoma specimens examined. Methodologically, we found a strong correlation between IHC and epigenetic qPCR measures of Tregs in tissue. We found no correlation between survival and Tregs in the tumor microenvironment or blood, even when adjusting for prognostic factors including age, MGMT status, and KPS score. Further, using the glioblastoma TCGA database, we interrogated markers of Tregs, including FoxP3 and Helios gene expression and methylation of FoxP3 DNA, none of which predicted patient survival.
It is now well established from transcriptome analysis that glioblastoma has 4 distinct molecular subtypes: proneural, classical, neural, and mesenchymal.24 Although the classical glioblastoma subtype had a significantly lower expression level of Treg markers, little if any differences in marker expression could be found between the other subtypes. Further, we found no differences in OS that could be predicted by these markers.
Despite suggestions that the presence of Tregs in the glioblastoma microenvironment correlates with poorer overall patient survival,12 several studies have failed to show such an association.9,14,15 We found that 47% of the glioblastomas in our sample had measurable Tregs, similar to another study that found 48% of the glioblastomas had measurable Treg infiltration by IHC tissue array, and the survival of the group with Treg infiltration was not significantly different than survival of the group without Tregs.14 On the other hand, a recent publication demonstrated a correlation in progression-free survival and OS with tumor-infiltrating Tregs as identified by FoxP3+ IHC in 62 patients with glioblastoma.16 While our study and others14 dichotomized Tregs based on presence or absence of FoxP3+ T cells by IHC, these authors reported FoxP3 density, defined as number of cells per square millimeter in 5 randomly selected fields.16 This finding, taken in context with ours and others, highlights the technical issues associated with evaluating tumor-infiltrating lymphocytes.
Reports that tumor-infiltrating Tregs inversely correlate with patient survival in other types of tumors25–27 raise the question of why this is not the case in glioblastoma. A plausible explanation is that Tregs are just one of multiple mechanisms of immune suppression and may be an epiphenomenon of the tumor secretion of cytokines that induce preferential migration, proliferation, and survival of Tregs.13 In glioblastoma, the number of tumor-infiltrating Tregs is low compared with other tumors in which presence of Tregs correlates with outcome.18 By IHC, the median FoxP3+ cell density in glioblastoma tissue has been reported as 5.6 cells/mm2,16 while in colorectal cancer, in which Tregs correlate with improved survival, the density is reported as 116 cells/mm2.28
Studies using IHC and flow cytometry are limited in that not all cells that express FoxP3 are Tregs, and there are many variations of staining marker combinations and gating strategies for quantifying Tregs. In our study, we applied a novel assay method that targets the demethylation of the FoxP3 gene, a specific epigenetic marker observed in only functionally active stable Tregs and not in any other leukocyte cell type, including other T cells that may show transient FoxP3 expression.17,29,30 Recent studies suggest that thymic nTregs are the most prevalent type in the glioblastoma microenvironment.13,31,32 Our study provides additional evidence that glioblastoma tissue FoxP3+ cells are primarily stable functional Tregs, based on the high correlation between the IHC and the epigenetic qPCR results.
To our knowledge, only one other study has attempted to identify Tregs by quantitative DNA demethylation assay in patients with brain tumors.33 In that study, analysis of 120 patients with gliomas representing many different histological subtypes—ependymomas, oligodendrogliomas, and astrocytomas of World Health Organization (WHO) grades I–IV—demonstrated that tumor-infiltrating T cells correlated positively with tumor grade. While the number of infiltrating Tregs was inversely correlated with survival, survival was not adjusted for tumor grade, thus the survival disadvantage may reflect the more aggressive biology and natural history of glioblastoma. All 25 patients analyzed in our study, as well as the 206 TCGA patients, had glioblastoma (WHO grade IV). Thus, we believe that our study provides more specific insight into the correlation of Tregs with survival of patients with glioblastoma. Our report supports the use of an epigenetic qPCR methylation method that more consistently measures tumor-infiltrating lymphocytes across studies. Using this methodology, confirmed by analysis of a larger sample of TCGA data, in combination with several prior observations,9,14,15 we demonstrate that glioblastoma-infiltrating Tregs do not correlate with patient OS.
Our study has limitations that may have influenced the results—first, our small sample size, although our results were corroborated by analysis of the glioblastoma cases in TCGA. Second, our cohort was not perfectly uniform. All patients underwent partial resection, radiation, and temozolomide chemotherapy, but 10 patients received vaccine therapy in addition to the aforementioned standard of care. In a separate analysis of this subgroup, we found no difference in tumor-infiltrating or peripheral Tregs in patients treated with or without vaccine (data not shown). Finally, various methodological details may have added variability to our results. The DNA used for epigenetic qPCR was obtained from a representative area of the tumor, but the sample may not reflect the heterogeneity of the whole tumor. There was an interval of several weeks between the time of surgery, when the tissue was procured, and the time the peripheral blood for Treg isolation was obtained. Excision of the tumor may have changed the peripheral lymphocyte subpopulation distribution, and we did not have samples to determine whether the peripheral Treg frequency before surgical removal correlated with survival. The staining and analysis variability inherent in IHC and flow cytometry may explain some of the discordance across reported Treg populations in tissue and blood within the glioblastoma literature as well as across other tumor types. In the future, we believe that the use of methods like epigenetic qPCR will provide a more consistent quantitation of Tregs.
The experience with immunotherapy for glioblastoma suggests that there is an immune system “threshold” that must be exceeded if a cancer patient is to respond positively to therapy. It is likely that a combination of immune measures including but not limited to tumor-infiltrating Tregs and T cells will help to define this threshold. Epigenetic qPCR is a sensitive and specific methodology that can improve consistency across studies and aid in future efforts to quantify immune measures and increase understanding of glioblastoma.
Funding
This work was supported in part by grants CCSG P30, CA023108, 5P20RR024475-02, 8 P20 GM103534-02, and RO1-HL074175, Medical Oncology Immunotherapy, and the Neuro-Oncology Programs at Dartmouth Hitchcock Medical Center.
Acknowledgments
We would like to thank Mary Robinson for her role in preparing the manuscript for submission and Christopher Dant for critical review of the writing. Flow cytometry was carried out in the DartLab: Immunoassay and Flow Cytometry Shared Resource at the Geisel School of Medicine at Dartmouth.
Conflict of interest statement. None declared.
References
- 1.Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342(6165):1432–1433. [DOI] [PubMed] [Google Scholar]
- 2.Rolle CE, Sengupta S, Lesniak MS. Challenges in clinical design of immunotherapy trials for malignant glioma. Neurosurg Clin N Am. 2010;21(A):201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomez GG, Kruse CA. Mechanisms of malignant glioma immune resistance and sources of immunosuppression. Gene Ther Mol Biol. 2006;10(1):133–146. [PMC free article] [PubMed] [Google Scholar]
- 4.Adeegbe DO, Nishikawa H. Natural and induced T regulatory cells in cancer. Front Immunol. 2013;4(190):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilke CM, Wu K, Zhao E, et al. Prognostic significance of regulatory T cells in tumor. Int J Cancer. 2010;127(4):748–758. [DOI] [PubMed] [Google Scholar]
- 6.de Leeuw RJ, Kost SE, Kakal JA, et al. The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin Cancer Res. 2012;18(11):3022–3029. [DOI] [PubMed] [Google Scholar]
- 7.Schwarzer A, Wolf B, Fisher JL, et al. Regulatory T-cells and associated pathways in metastatic renal cell carcinoma (mRCC) patients undergoing DC-vaccination and cytokine-therapy. PloS One. 2012;7(10):e46600 epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossman SA, Ye X, Lesser G, et al. Immunosuppression in patients with high-grade gliomas treated with radiation and temozolomide. Clin Cancer Res. 2011;17(16):5473–5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lohr J, Ratliff T, Huppertz A, et al. Effector T-cell infiltration positively impacts survival of glioblastoma patients and is impaired by tumor-derived TGF-beta. Clin Cancer Res. 2011;17(13):4296–4308. [DOI] [PubMed] [Google Scholar]
- 10.Palma L, Di Lorenzo N, Guidetti B. Lymphocytic infiltrates in primary glioblastomas and recidivous gliomas. Incidence, fate, and relevance to prognosis in 228 operated cases. J Neurosurg. 1978;49(6):854–861. [DOI] [PubMed] [Google Scholar]
- 11.El Andaloussi A, Lesniak MS. An increase in CD4+CD25+FOXP3+ regulatory T cells in tumor-infiltrating lymphocytes of human glioblastoma multiforme. Neuro Oncol. 2006;8(3):234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fecci PE, Mitchell DA, Whitesides JF, et al. Increased regulatory T-cell fraction amidst a diminished CD4 compartment explains cellular immune defects in patients with malignant glioma. Cancer Res. 2006;66(6):3294–3302. [DOI] [PubMed] [Google Scholar]
- 13.Crane CA, Ahn BJ, Han SJ, et al. Soluble factors secreted by glioblastoma cell lines facilitate recruitment, survival, and expansion of regulatory T cells: implications for immunotherapy. Neuro Oncol. 2012;14(5):584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heimberger AB, Abou-Ghazal M, Reina-Ortiz C, et al. Incidence and prognostic impact of FoxP3+ regulatory T cells in human gliomas. Clin Cancer Res. 2008;14(16):5166–5172. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs JF, Idema AJ, Bol KF, et al. Prognostic significance and mechanism of Treg infiltration in human brain tumors. J Neuroimmunol. 2010;225(1–2):195–199. [DOI] [PubMed] [Google Scholar]
- 16.Yue Q, Zhang X, Ye HX, et al. The prognostic value of Foxp3+ tumor-infiltrating lymphocytes in patients with glioblastoma. J Neurooncol. 2014;116(2):251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wieczorek G, Asemissen A, Model F, et al. Quantitative DNA methylation analysis of FOXP3 as a new method for counting regulatory T cells in peripheral blood and solid tissue. Cancer Res. 2009;69(2):599–608. [DOI] [PubMed] [Google Scholar]
- 18.Sehouli J, Loddenkemper C, Cornu T, et al. Epigenetic quantification of tumor-infiltrating T-lymphocytes. Epigenetics. 2011;6(2):236–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahme GJ, Israel MA. Id4 suppresses MMP2-mediated invasion of glioblastoma-derived cells by direct inactivation of Twist1 function. Oncogene. 2015;34(1):53–62. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Himmel ME, MacDonald KG, Garcia RV, et al. Helios+ and Helios‒ cells coexist within the natural FOXP3+ T regulatory cell subset in humans. J Immunol. 2013;190(5):2001–2008. [DOI] [PubMed] [Google Scholar]
- 22.Fadul CE, Fisher JL, Gui J, et al. Immune modulation effects of concomitant temozolomide and radiation therapy on peripheral blood mononuclear cells in patients with glioblastoma multiforme. Neuro Oncol. 2011;13(4):393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31(3):1116–1128. [DOI] [PubMed] [Google Scholar]
- 24.Verhaak RG, Hoadley KA, Purdom E, et al. An integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Wang FM, Wang T, et al. Tumor-infiltrating FoxP3+ Tregs and CD8+ T cells affect the prognosis of hepatocellular carcinoma patients. Digestion. 2012;86(4):329–337. [DOI] [PubMed] [Google Scholar]
- 26.Kinoshita T, Ishii G, Hiraoka N, et al. Forkhead box P3 regulatory T cells coexisting with cancer associated fibroblasts are correlated with a poor outcome in lung adenocarcinoma. Cancer Sci. 2013;104(4):409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang JM, Chung HG, Oh SJ, et al. Increased expression of intracellular HLA-DM but not on the surface of blood monocyte-derived dendritic cells during maturation. Yonsei Med J. 2003;44(2):293–298. [DOI] [PubMed] [Google Scholar]
- 28.Salama P, Phillips M, Grieu F, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009;27(2):186–192. [DOI] [PubMed] [Google Scholar]
- 29.Baron U, Floess S, Wieczorek G, et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol. 2007;37(9):2378–2389. [DOI] [PubMed] [Google Scholar]
- 30.Floess S, Freyer J, Siewert C, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5(2):e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wainwright DA, Dey M, Chang A, et al. Targeting Tregs in malignant brain cancer: overcoming IDO. Front Immunol. 2013;4:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wainwright DA, Sengupta S, Han Y, et al. Thymus-derived rather than tumor-induced regulatory T cells predominate in brain tumors. Neuro Oncol. 2011;13(12):1308–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiencke JK, Accomando WP, Zheng S, et al. Epigenetic biomarkers of T-cells in human glioma. Epigenetics. 2012;7(12):1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]