Abstract
Background
In resource-limited settings, HIV infection is often diagnosed using two rapid tests. If the results are discordant, a third tie-breaker test is often used to determine HIV status. This study characterized samples with discordant rapid tests and compared different testing strategies for determining HIV status in these cases.
Methods
Samples were previously collected from 173 African adults in a population-based survey who had discordant rapid test results. Samples were classified as HIV positive or HIV negative using a rigorous testing algorithm that included two fourth-generation tests, a discriminatory test, and two HIV RNA tests. Tie-breaker tests were evaluated, including: rapid tests (one performed in-country), a third-generation enzyme immunoassay (EIA), and two fourth-generation tests. Selected samples were further characterized using additional assays.
Results
Twenty-nine (16.8%) samples were classified as HIV positive; 24 (82.8%) of those samples had undetectable HIV RNA. Antiretroviral drugs were detected in one sample. Sensitivity was 8.3%–43% for the rapid tests; 24.1% for the third-generation EIA; 95.8% and 96.6% for the fourth-generation tests. Specificity was lower for the fourth-generation tests than the other tests. Accuracy ranged from 79.5–91.3%.
Conclusions
In this population-based survey, most HIV-infected adults with discordant rapid tests were virally suppressed without antiretroviral drugs. Use of individual assays as tie-breaker tests was not a reliable method for determining HIV status in these individuals. More extensive testing algorithms that use a fourth-generation screening test with a discriminatory test and HIV RNA test are preferable for determining HIV status in these cases.
Keywords: HIV, rapid test, discordant, Africa
INTRODUCTION
The United States Centers for Disease Control and Prevention (US CDC) previously recommended using an immunoassay with a confirmatory Western blot or indirect immunofluorescence assay for HIV diagnosis.1 In 2014, an updated testing algorithm was recommended that increases sensitivity for detecting acute HIV infections and differentiates between HIV-1 and HIV-2 infection.2 In the current algorithm, samples are first tested using an assay that detects both HIV antigen and antibody (fourth-generation assay). If that assay is reactive, an HIV-1/HIV-2 discriminatory assay is performed. If the discriminatory assay is non-reactive or indeterminate, a nucleic acid test is performed.2
In resource-limited settings, the World Health Organization (WHO) recommends using two HIV rapid tests for diagnosis.3,4 These tests can be performed at the point of care, facilitating linkage to counseling services and HIV care.4 If the results of the two rapid tests are discordant (one reactive, one non-reactive), a tie-breaker test is recommended3 (e.g., a third rapid test or an enzyme immunoassay [EIA]5,6). Despite the widespread use of a tie-breaker approach for HIV diagnosis, and the availability of performance data for individual HIV assays, there are limited data comparing the performance of different testing strategies for determining HIV status in individuals with discordant rapid test results. Previous studies have evaluated individuals with discordant HIV rapid tests in populations at increased risk of HIV infection (e.g., sexual partners of HIV-infected individuals,7 women attending prenatal or antenatal clinics,8,9 adults attending clinics for sexually transmitted diseases,10,11 and adults in HIV screening programs with very high HIV prevalence12). In those studies, the frequency of discordant rapid test results ranged from 0.7–2.3%, and many participants had acute HIV infection. However, because the window period for acute HIV infection is short,13 individuals with acute HIV infection may comprise a smaller portion of those with discordant rapid tests in cohorts at lower risk of HIV acquisition.
In this study, we analyzed samples from participants in a population-based survey in Tanzania and South Africa. The goals of this study were to compare the performance of different testing strategies for determining HIV status in individuals with discordant HIV rapid test results, and to investigate factors associated with discordant rapid tests in HIV-infected individuals.
Methods
Study population and samples used for analysis
Samples were obtained from the National Institute of Mental Health (NIMH) Project Accept trial (HIV Prevention Trials Network [HPTN] 043) (NCT00203749). HPTN 043 was a large, community-randomized clinical trial in Africa and Thailand that assessed the impact of integrated behavioral interventions on HIV incidence.14 HIV incidence was assessed at the end of the trial in a single cross-sectional survey.15 The survey included over 50,000 participants aged 18–32 years, randomly sampled from 48 communities at five study sites. All eligible individuals in each household could participate in the survey.
This report describes analysis of samples collected at three HPTN 043 sites (Kisarawe, Tanzania; Soweto and Vulindlela, South Africa). In HPTN 043, one 10 mL EDTA-anticoagulated blood sample was collected from each participant. The sample was used for in-country HIV testing and then used to prepare plasma that was frozen at −80°C and shipped to the HPTN Laboratory Center at Johns Hopkins University, Baltimore, MD, USA for further testing; a 4 mL sample was also collected for in-country CD4 cell count testing. Laboratories at the study sites participated in external quality assurance programs; external quality assurance assessments were also performed by the HPTN Laboratory Center. In HPTN 043, 255 (0.7%) of 34,813 samples from Tanzania and South Africa had discordant HIV rapid test results (187 [2.1%] of 9,041 samples from Tanzania; 68 [0.3%] of 25,772 samples from South Africa, Table 1).
Table 1.
Samples used for analysis.
| Prevalence | Incidence | Participants tested |
Participants with discordant test resultsa |
Samples evaluatedb |
Samples classified as HIV positivec |
|
|---|---|---|---|---|---|---|
| Tanzania | 5.9% | 0.78% | 9,041 | 187 (2.1%) | 127 (67.9%) | 24 (18.9%) |
| South Africa | 21.8% | 2.3% | 25,772 | 68 (0.3%) | 46 (67.6%) | 5 (10.9%) |
| Soweto | 14.1% | 1.18% | 13,929 | 22 (0.2%) | 16 (72.7%) | 3 (18.8%) |
| Vulindlela | 30.8% | 3.90% | 11,843 | 46 (0.4%) | 30 (65.2%) | 2 (6.7%) |
| Total | 17.7% | 1.85% | 34,813 | 255 (0.7%) | 173 (67.8%) | 29 (16.8%) |
Samples were tested in-country using two HIV rapid tests. The number and percentage of samples that had discordant HIV rapid test results (one reactive result and one non-reactive result) are shown.
Samples from 173 study participants were evaluated in this study. Samples from the remaining 82 participants who had discordant HIV rapid test results were not evaluated for the following reasons: insufficient plasma available (N=80), in-country sample contamination (N=1), and in-country sample mix-up (N=1).
Samples were classified as HIV positive or HIV negative using the testing algorithm shown in Figure 1. The number and percentage of samples evaluated in this study that were classified as HIV positive are shown.
In-country HIV rapid testing
Samples were initially tested in-country at local laboratories using two HIV rapid tests performed in parallel on fresh samples. The following rapid tests were used for this initial assessment: Determine HIV-1/2 (Inverness Medical Innovations, Phetchabun, Japan), SD Bioline HIV 1/2 v3 (Youngin-Si, South Korea), or Uni-Gold HIV Test (Trinity Biotech, Bray, Ireland; Soweto only). Samples with discordant rapid test results were analyzed in-country using a tie-breaker test. The site in Tanzania used a third HIV rapid test as the tie-breaker test (Uni-Gold HIV Test). The two sites in South Africa used a fourth-generation chemiluminescent immunoassay (CMIA) as the tie-breaker test (the ARCHITECT HIV Ag/Ab Combo assay, Abbott Laboratories, Wiesbaden, Germany; referred to below as the Abbott Combo assay). All other testing described in this report was performed at the HPTN Laboratory Center.
Classification of samples with discordant rapid test results as HIV positive or HIV negative
Samples were classified as HIV positive or HIV negative using the testing algorithm shown in Figure 1. This algorithm is similar to the current US CDC testing algorithm2, but was modified to include two fourth-generation assays and two HIV RNA assays to maximize sensitivity for detecting HIV infection. The two fourth-generation assays were performed in parallel (Abbott Combo assay; and GS HIV Combo Ag/Ab EIA, Bio-Rad Laboratories, Redmond, WA; referred to below as the Bio-Rad Combo assay). If one or both assay was reactive, samples were tested using the Multispot HIV-1/HIV-2 Rapid Test (Bio-Rad Laboratories, Redmond, WA; referred to below as the discriminatory assay). If the discriminatory assay was negative or indeterminate, samples were tested using two HIV RNA assays (APTIMA HIV-1 RNA Qualitative Assay, Hologic Gen-Probe Inc., San Diego, CA, referred to below as the Aptima RNA assay; and Roche COBAS AMPLICOR HIV-1 MONITOR test, v1.5, Roche Diagnostics, Branchburg, NJ; referred to below as the Roche viral load assay). Samples were classified as HIV positive if they had any of the following test results: positive discriminatory test, positive Aptima RNA assay, or positive Roche viral load assay (Figure 1).
Figure 1. Testing algorithm used to classify samples with discordant HIV rapid tests as HIV positive or HIV negative.
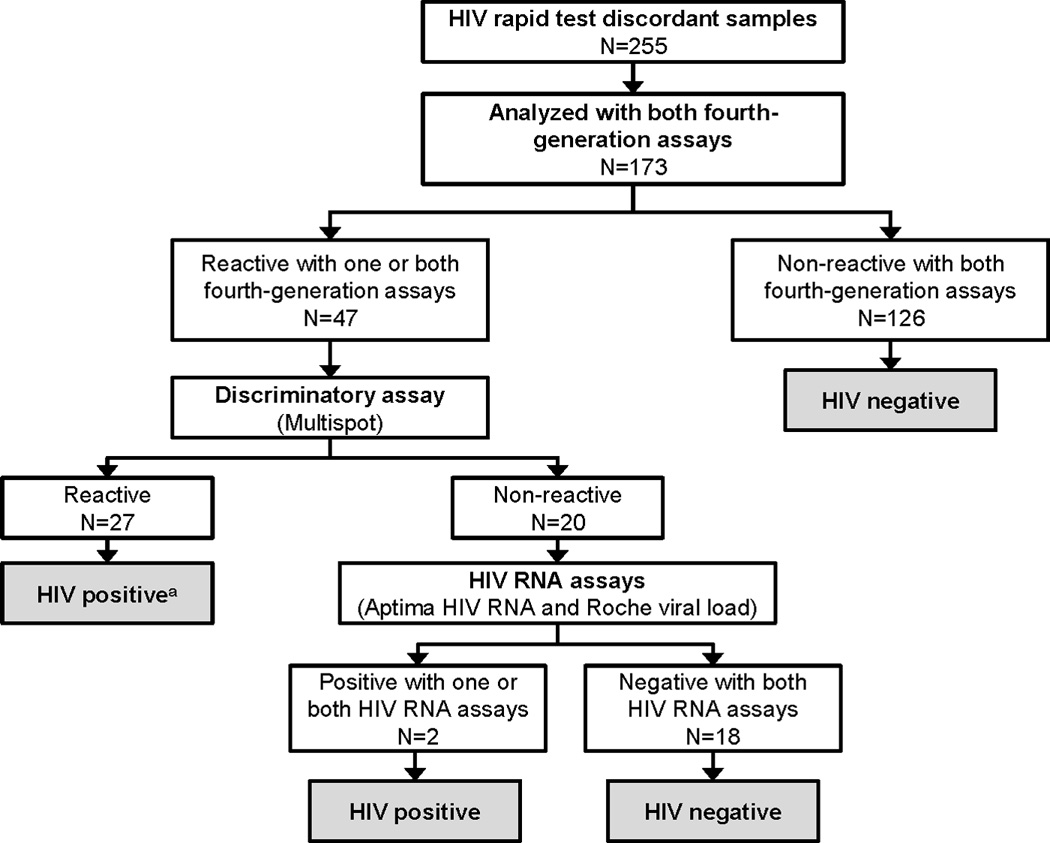
Assays used for testing were: Fourth-generation assays: the GS HIV Combo Ag/Ab EIA (BioRad Combo), the ARCHITECT HIV Ag/Ab Combo assay (Abbott Combo); Discriminatory assay: Multispot HIV-1/HIV-2 Rapid Test (Multispot); HIV RNA assays: the APTIMA HIV-1 RNA Qualitative Assay (Aptima HIV RNA) and the Roche COBAS AMPLICOR HIV-1 MONITOR test, v1.5 (Roche viral load).
a Three (11.1%) of the 27 samples that had reactive results with the discriminatory assay were positive with one or both of the HIV RNA assays (Aptima HIV RNA, Roche viral load). The remaining 24 samples had undetectable HIV RNA.
Comparison of the performance of different tie-breaker assays
Samples with discordant rapid tests were tested with four tie-breaker assays: (1) a third HIV rapid test (Uni-Gold HIV Test; performed in-country, Tanzania only); (2) a third-generation EIA (VITROS Anti-HIV 1+2 assay, Ortho Clinical Diagnostics, Rochester, NY, referred to below as the Vitros EIA), (3) the Bio-Rad Combo assay, and (4) the Abbott Combo assay.
Further characterization of samples with discordant rapid test results
Samples classified as HIV positive were further characterized with two additional HIV rapid tests (Uni-Gold Recombigen HIV-1/2; Trinity Biotech, Bray, Ireland; and OraQuick Advance Rapid HIV-1/2 Antibody Test; OraSure Technologies, Inc, Bethlehem, PA), a Western blot test, and a qualitative multi-drug screening that detects 15 antiretroviral drugs, including protease inhibitors, nucleoside/nucleotide reverse transcriptase inhibitors, and non-nucleoside reverse transcriptase inhibitors.16,17 Samples classified as HIV negative that had at least one reactive (false-positive) serologic assay were also further characterized with a Western blot test and antiretroviral drug screening.
Ethical Approval
The work described was carried out in accordance with the Declaration of Helsinki. The HPTN 043 trial was conducted in close partnership with established community advisory boards and local government departments. Consent was obtained (written consent in South Africa; oral consent in Tanzania) at the community level for trial participation. To approach household members to participate in the post-intervention survey, investigators needed permission from the head of the household. Oral consent was obtained from each participant for collection and testing of blood samples.14 The study was approved by ethics committees for each site and by all participating academic institutions.
RESULTS
Classification of HIV discordant samples as HIV positive or HIV negative
This study evaluated 173 (67.8%) of the 255 HIV discordant samples identified in the HPTN 043 study (Table 1). Eighty-two samples were excluded (80 had insufficient plasma available, one was contaminated in-country, and one was involved in an in-country sample mix-up). The 173 samples were first classified as HIV positive or HIV negative using the HIV testing algorithm shown in Figure 1. This classification was used as a gold standard for the evaluation of simpler, tie-breaker testing approaches. Twenty-nine (16.8%) of the 173 samples were classified as HIV positive (24/127 from Tanzania; 5/46 samples from South Africa, P=0.3, Fisher’s exact test); 144 samples were classified as HIV negative.
Comparison of tie-breaker approaches for determining the HIV status of samples with discordant HIV test results
We next evaluated four tie-breaker approaches for determining the HIV status of individuals with discordant HIV rapid test results (Table 2): (1) a third-generation rapid test (the Uni-Gold HIV Test, performed in-country, Tanzania only), (2) a third-generation EIA (the Vitros EIA), (3) a fourth-generation EIA (the Bio-Rad Combo assay), and (4) a fourth-generation CMIA (the Abbott Combo assay). The two third generation tests had low sensitivity (8.3% and 24.1%) with specificities of 96.1% and 98.6%. The fourth-generation assays were more sensitive (93.1% and 96.6%), but had lower specificity (88.2% and 90.3%). The accuracy of the four tests ranged from 79.5% to 91.3% (Table 2).
Table 2.
Comparison of different testing approaches for evaluating samples with discordant HIV rapid test results.*
| Samples evaluated N=173 HIV positive N=29 (Tanzania N=24, South Africa N=5) HIV negative N=144 (Tanzania N=103, South Africa N=41) |
|||||||
|---|---|---|---|---|---|---|---|
| Testing approach | True Positive | False positive | True Negative | False negative | Sensitivity | Specificity | Accuracy |
| Rapid testa | |||||||
| Tanzania | 2 | 4 | 99 | 22 | 8.3% | 96.1% | 79.5% |
| 3rd-gen EIA | |||||||
| Tanzania | 3 | 1 | 102 | 21 | 12.5% | 99.0% | |
| South Africa | 4 | 1 | 40 | 1 | 80% | 97.6% | |
| Total | 7 | 2 | 142 | 22 | 24.1% | 98.6% | 86.1% |
| Bio-Rad 4th-gen | |||||||
| Tanzania | 23 | 16 | 87 | 1 | 95.8% | 84.5% | |
| South Africa | 4 | 1 | 40 | 1 | 80% | 97.6% | |
| Total | 27 | 17 | 127 | 2 | 93.1% | 88.2% | 89.0% |
| Abbott 4th-gen | |||||||
| Tanzania | 24 | 13 | 90 | 0 | 100% | 87.4% | |
| South Africa | 4 | 1 | 40 | 1 | 80% | 97.6% | |
| Total | 28 | 14 | 130 | 1 | 96.6% | 90.3% | 91.3% |
Abbreviations: 3rd-gen: third-generation; 4th-gen: fourth-generation; EIA: enzyme immunoassay.
The assays used for testing (See Methods) included: Rapid test: Uni-Gold HIV Test; 3rd-gen EIA: VITROS Anti-HIV 1+2 assay; Bio-Rad 4th-gen: GS HIV Combo Ag/Ab EIA; Abbott 4th-gen: ARCHITECT HIV Ag/Ab Combo assay.
The HIV rapid test (third test, tie-breaker) was performed in Tanzania only; all other testing was performed at the HPTN Laboratory Center in Baltimore, MD, USA.
Further evaluation of samples classified as HIV positive
Further testing was performed to characterize the 29 HIV positive samples (Table 3). The mean CD4 cell count of 29 participants was 1050 cells/µL (range: 222–1826); two had a CD4 cell count <350 cells/µL (case 1: 252; case 4: 222). As noted above, the HIV rapid test used as a tie-breaker in Tanzania had a sensitivity of 8.3% (Uni-Gold HIV test). For comparison, the HIV positive samples were tested using two HIV rapid tests approved by the US Food and Drug Administration (FDA, the Uni-Gold Recombigen HIV-1/2 Test and the OraQuick Advance HIV-1/2 Antibody Test). This testing was performed at the HPTN Laboratory Center and included samples from both Tanzania and South Africa (Table 3; note that some samples were depleted in previous testing). The Uni-Gold Recombigen HIV-1/2 Test was reactive for 4/16 samples tested (sensitivity: 25.0%). The OraQuick Advance HIV-1/2 Antibody Test was reactive for 11/26 samples tested (sensitivity: 42.3%). In several cases where these tests were reactive, the test line was faint and could easily have been missed (one of four samples that was reactive with the Uni-Gold Recombigen HIV-1/2 Test had an extremely faint T line; nine of 11 samples that were reactive with the OraQuick Advance HIV-1/2 Antibody Test had a faint or extremely faint T-line). Sensitivity was even lower when a Western blot test was used to confirm HIV infection (Table 3, range: 4.2% to 12.5%).
Table 3.
Further evaluation of samples classified as HIV positive using the testing algorithm shown in Figure 1*.
| ID# | Rapid test 1b |
Rapid test 2c |
Rapid test 3d |
3rd-gen EIA | WB | WB Bands | BioRad 4th-gen |
BioRad S/COe |
Abbott 4th-gen |
Abbott S/COe |
Discrim assay |
Aptima testf |
Viral load (c/mL) |
ARV drugs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | R | R | R | R | Pos | -- | R | Max | R | 318.06 | R | n/a | 9,052 | No |
| 2 | R | NR | NR | NR | Ind | 31+/− | R | 9.93 | R | 6.06 | R | NR | <400 | No |
| 3a | n/a | NR | R | R | Pos | -- | R | Max | R | 111.27 | R | QNS | <400 | No |
| 4a | n/a | QNS | QNS | R | Pos | -- | R | Max | R | 970.75 | R | n/a | 1,280 | 3TC, NVP |
| 5 | NR | NR | R | R | Pos | -- | R | 6.54 | R | 9.06 | NR | n/a | 2,651 | No |
| 6a | n/a | R | NR | R | Pos | -- | R | 10.75 | R | 3.88 | R | NR | <400 | No |
| 7 | NR | NR | NR | R | Neg | -- | R | 9.41 | R | 5.54 | NR | n/a | 459,246 | No |
| 8a | n/a | QNS | NR | R | Neg | -- | R | 1.37 | NR | 0.17 | R | NR | <400 | No |
| 9 | NR | NR | R | NR | Ind | 24+/− | NR | 0.06 | R | 1.19 | R | NR | <400 | No |
| 10a | n/a | NR | NR | NR | Neg | -- | NR | 0.67 | R | 3.11 | R | NR | <400 | No |
| 11 | NR | QNS | NR | NR | Ind | 160+ | R | Max | R | 458.32 | R | R | <400 | No |
| 12 | NR | QNS | NR | NR | Ind | 160+/− | R | 11.53 | R | 2.58 | R | NR | <400 | No |
| 13 | NR | NR | R | NR | Ind | 18+/−, 31+/−, 160+ | R | Max | R | 11.53 | R | NR | <400 | No |
| 14 | NR | QNS | NR | NR | Ind | 160+/− | R | 8.61 | R | 3.00 | R | NR | <400 | No |
| 15 | NR | QNS | R | NR | Ind | 160+/− | R | 12.09 | R | 28.11 | R | NR | <400 | No |
| 16 | NR | QNS | QNS | NR | Ind | NA | R | 11.84 | R | 158.13 | R | NR | <400 | QNS |
| 17 | NR | QNS | NR | NR | Ind | 160+/− | R | 10.97 | R | 28.29 | R | NR | QNS | QNS |
| 18 | NR | QNS | NR | NR | Ind | 18+/−, 160+/− | R | Max | R | 44.53 | R | NR | <400 | No |
| 19 | NR | QNS | QNS | NR | Ind | NA | R | 13.67 | R | 56.00 | R | NR | <400 | No |
| 20 | NR | R | R | NR | Ind | 160+ | R | Max | R | 137.92 | R | NR | <400 | QNS |
| 21 | NR | QNS | R | NR | Ind | 160+ | R | 13.28 | R | 6.78 | R | NR | QNS | QNS |
| 22 | NR | R | NR | NR | Ind | 160+ | R | Max | R | 133.60 | R | NR | <400 | No |
| 23 | NR | QNS | NR | NR | Ind | 18+, 160+/− | R | Max | R | 71.17 | R | NR | <400 | No |
| 24 | NR | NR | NR | NR | Ind | 160+/− | R | Max | R | 55.89 | R | NR | <400 | No |
| 25 | NR | NR | R | NR | Ind | 160+ | R | 10.13 | R | 10.15 | R | NR | <400 | No |
| 26 | NR | QNS | NR | NR | Ind | 160+/− | R | 5.75 | R | 2.13 | R | NR | <400 | No |
| 27 | NR | NR | R | NR | Ind | 160+ | R | 13.19 | R | 4.02 | R | NR | <400 | No |
| 28 | NR | NR | R | NR | Ind | 160+, DB+/− | R | Max | R | 18.97 | R | NR | <400 | No |
| 29 | NR | NR | NR | NR | Neg | -- | R | 4.32 | R | 3.91 | R | NR | <400 | No |
The assays used for testing (see Methods) included: Rapid tests: Uni-Gold HIV Test; Uni-Gold Recombigen HIV Test; Oraquick 3rd-gen EIA: VITROS Anti-HIV 1+2 assay; WB: Genetics System HIV-1 Western Blot; Bio-Rad 4th-gen: GS HIV Combo Ag/Ab EIA; Abbott 4th-gen: ARCHITECT HIV Ag/Ab Combo assay; Discrim assay: Multispot HIV-1/HIV-2 Rapid Test; Aptima test: APTIMA HIV-1 RNA Qualitative Assay; Viral load: Roche COBAS AMPLICOR HIV-1 MONITOR test, v1.5 (validated modified assay for low-volume samples); ARV assay: qualitative multi-drug screening assay that detects 15 ARV drugs.
Abbreviations: ID#: sample identification number; 3rd-gen: third generation; EIA: enzyme immunoassay; WB: Western blot; 4th-gen: fourth-generation; Discrim: discriminatory; c/mL: copies per milliliter; ARV: antiretroviral drug; R: reactive; NR: non-reactive; Pos: positive; Neg: negative; Ind: indeterminate; n/a: not applicable/not performed; QNS: quantity not sufficient (insufficient plasma available for testing); 3TC: lamivudine; NVP: nevirapine.
Cases 3, 4, 6, 8, and 10 are from South Africa; the rest of the cases are from Tanzania.
Rapid test 1: Uni-Gold HIV test; performed in Tanzania only.
Rapid test 2: Uni-Gold Recombigen HIV-1/2 Test (FDA-cleared); 13 samples did not have sufficient plasma for testing.
Rapid test 3: OraQuick ADVANCE® Rapid HIV-1/2 Antibody Test (FDA-cleared); 3 samples did not have sufficient plasma for testing.
The signal/cutoff ratio (S/CO) is considered non-reactive if the value is <1. For the GS HIV Combo Ag/Ab EIA assay, a reactive sample with an absorbance over the plate reader is reported as having a maximal S/CO (Max).
Samples were not tested with the Aptima HIV RNA assay if the viral load was >400 copies/mL.
Twenty-four (82.8%) of the 29 samples were reactive with both fourth-generation assays and the discriminatory test. Three of the remaining five samples were reactive with only one of the two fourth-generation assays (one was reactive with the Bio-Rad Combo assay only; two were reactive with the Abbott Combo assay only); all three of these samples were positive using the discriminatory assay. One or two of these three samples would have been missed using the testing algorithm recommended by the US CDC,2 depending on which one of two fourth-generation assays was used for screening. The remaining two samples were reactive using both fourth-generation assays and the third-generation EIA, but had non-reactive results using the discriminatory assay.
Samples were next tested using the Roche viral load assay (performed using a validated method for low-volume samples, limit of detection: 400 copies/mL; two samples did not have sufficient plasma available for testing). Samples with viral loads <400 copies/mL were tested using the Aptima HIV RNA assay (limit of detection: 30 copies/mL; one sample did not have sufficient plasma for testing). Four (14.8%) of 27 samples tested had viral loads >400 copies/mL (viral load: 1,280–459,246 copies/mL); one sample with <400 copies/mL HIV RNA had a reactive Aptima HIV RNA assay. The remaining 24 samples did not have detectable HIV RNA.
As a final step, samples were tested using a qualitative assay that detects 15 antiretroviral drugs (four samples did not have sufficient plasma for testing). Only one sample had a positive result: a sample from a pregnant women in South Africa with a viral load of 1,280 copies/mL; lamivudine and nevirapine were detected (case 4). These results indicate that the majority of the HIV-infected samples with discordant HIV rapid test results (21/25=84.0%) were from individuals who were virally suppressed in the absence of antiretroviral treatment (ART).
Further evaluation of samples classified as HIV negative
Samples from 144 study participants were classified as HIV negative using the testing algorithm shown in Figure 1. These individuals had a mean CD4 cell count of 986 cells/µL (range: 481–5,833; one value was missing). In 24 (16.7%) of those cases, one or more serologic assay was reactive (in addition to one HIV rapid test initially performed at study sites, Table 4). In 11 of the 24 cases, only one of the assays had a reactive tie-breaker test (four had a reactive rapid test, two had a reactive third-generation EIA, four had a reactive Bio-Rad Combo assay, one had a reactive Abbott Combo assay). A Western blot test was performed for the seven samples that had a reactive third-generation EIA or fourth-generation assay; one Western blot was indeterminate and six were negative. In the remaining 13 cases (cases 12–24), both fourth-generation assays were reactive; a Western blot was indeterminate in 9 cases, negative in three cases, and positive in one case. In all of the cases, the discriminatory assay and the Aptima HIV RNA assay were negative. As a final step, the samples were tested for antiretroviral drugs (12 samples did not have sufficient plasma for testing); none of the samples had antiretroviral drugs detected. Since longitudinal samples were not collected in the HPTN 043 trial and cellular DNA was not stored for testing (e.g., whole blood samples), it was not possible to determine conclusively whether or not these individuals were HIV infected.
Table 4.
Evaluation of a subset of samples classified as HIV negative using the testing algorithm shown in Figure 1.*
| ID | Rapid testb |
3rd-gen EIA |
WB | WB Bands | BioRad 4th-gen |
BioRad S/COc |
Abbott 4th-gen |
Abbott S/COc |
Discrim assayd |
Aptima HIV RNA |
ARV drugs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | R | NR | n/a | -- | NR | 0.26 | NR | 0.15 | n/a | NR | None |
| 2 | R | NR | n/a | -- | NR | 0.25 | NR | 0.18 | n/a | NR | QNS |
| 3 | R | NR | n/a | -- | NR | 0.03 | NR | 0.10 | n/a | NR | None |
| 4 | R | NR | n/a | -- | NR | 0.20 | NR | 0.13 | n/a | NR | None |
| 5 | NR | R | Neg | -- | NR | 0.63 | NR | 0.14 | NR | NR | None |
| 6a | n/a | R | Neg | -- | NR | 0.21 | NR | 0.29 | NR | NR | None |
| 7 | NR | NR | Ind | n/a | R | 4.06 | NR | 0.67 | NR | NR | QNS |
| 8 | NR | NR | Neg | -- | R | Max | NR | 0.12 | NR | NR | None |
| 9 | NR | NR | Neg | -- | R | 6.91 | NR | 0.17 | NR | NR | QNS |
| 10a | n/a | NR | Neg | -- | R | 1.32 | NR | 0.75 | NR | NR | QNS |
| 11a | n/a | NR | Neg | -- | NR | 0.27 | R | 1.12 | NR | NR | QNS |
| 12 | NR | NR | Pos | -- | R | 1.39 | R | 57.52 | NR | NR | None |
| 13 | NR | NR | Ind | n/a | R | Max | R | 14.09 | NR | NR | QNS |
| 14 | NR | NR | Ind | 18+/− | R | 12.71 | R | 14.30 | NR | NR | QNS |
| 15 | NR | NR | Ind | 160+/− | R | 11.54 | R | 13.07 | NR | NR | QNS |
| 16 | NR | NR | Ind | 160+/− | R | 11.15 | R | 2.62 | NR | NR | None |
| 17 | NR | NR | Ind | 18+/−, 160+/− | R | 13.39 | R | 33.29 | NR | NR | None |
| 18 | NR | NR | Ind | 18+, 160+/− | R | 7.85 | R | 2.15 | NR | NR | None |
| 19 | NR | NR | Ind | 160+/− | R | 6.18 | R | 2.72 | NR | NR | None |
| 20 | NR | NR | Ind | 160+/− | R | 3.50 | R | 17.66 | NR | NR | QNS |
| 21 | NR | NR | Ind | 160+/− | R | 9.94 | R | 1.84 | NR | NR | QNS |
| 22 | NR | NR | Neg | -- | R | 7.12 | R | 3.45 | NR | NR | None |
| 23 | NR | NR | Neg | -- | R | 3.43 | R | 1.40 | NR | NR | QNS |
| 24 | NR | NR | Neg | -- | R | 10.53 | R | 5.37 | NR | NR | QNS |
Samples included in this table had at least one reactive serologic assay (in addition to the one of the two HIV rapid tests initially performed at study sites). The assays used for testing (see Methods) included: Rapid test: Uni-Gold HIV Test; 3rd-gen EIA: VITROS Anti-HIV 1+2 assay; WB: Genetics System HIV-1 Western Blot; Bio-Rad 4th-gen: GS HIV Combo Ag/Ab EIA; Abbott 4th-gen: ARCHITECT HIV Ag/Ab Combo assay; Discrim assay: Multispot HIV-1/HIV-2 Rapid Test; RNA assay: APTIMA HIV-1 RNA Qualitative Assay; ARV assay: qualitative multi-drug screening assay that detects 15 ARV drugs.
Abbreviations: ID#: sample identification number; 3rd-gen: third generation; EIA: enzyme immunoassay; WB: Western blot; 4th-gen: fourth-generation; S/CO: signal-to-cutoff ratio; Discrim: discriminatory; ARV: antiretroviral drug; R: reactive; NR: non-reactive; n/a: not applicable/not performed; Neg: negative; Ind: indeterminate; QNS: quantity not sufficient (insufficient plasma available for testing).
Cases 6, 10 and 11 are from South Africa; the rest of the cases are from Tanzania.
The HIV rapid test (third test, tie-breaker) was performed in Tanzania only; all other testing was performed at the HPTN Laboratory Center in Baltimore, MD, USA.
The signal/cutoff ratio (S/CO) is considered non-reactive if the value is <1. For the GS HIV Combo Ag/Ab EIA assay, a reactive sample with an absorbance over the plate reader is reported as having a maximal S/CO (Max).
The discriminatory assay was not performed if in-country HIV rapid tests were the only tests with reactive/positive test results.
DISCUSSION
At the three HPTN 043 sites included in this study, HIV prevalence ranged from 5.9% to 30.8%, and HIV incidence ranged from 0.78% to 3.90%. Overall, 255 (0.7%) of the 34,813 participants had discordant HIV rapid test results (0.3% in South Africa, 2.1% in Tanzania). This report describes analysis of plasma samples from 173 of the 255 participants; 29 (16.8%) of the samples were classified as HIV positive using a rigorous testing algorithm. Because the number of HIV positive samples from South Africa was small (N=5), we did not compare the performance of testing approaches in the two countries.
We compared the sensitivity and specificity of four tie-breaker assays for determining HIV status in these discordant cases. The sensitivity of third-generation assays was low (HIV rapid test performed in Tanzania: 8.3%; third-generation EIA: 24.1%); specificity was 96.1% and 98.6%, respectively. A higher sensitivity was obtained when samples were tested in the US using FDA-cleared HIV rapid tests (25.0% and 42.3%); however, many of the reactive test results were very weak and could easily have been missed, especially in an environment with sub-optimal lighting. Use of a Western blot to confirm a reactive third-generation assay (HIV rapid test or EIA) further reduced the sensitivity for detecting HIV infection. The sensitivity of fourth-generation assays was considerably higher (Bio-Rad Combo assay: 93.1%; Abbott Combo assay: 96.6%); however, specificity was only 88.2%, 90.3%, respectively. Accuracy was highest for the Abbott Combo assay (91.3%). These results indicate that more rigorous testing approaches (beyond a single tie-breaker assay or a third-generation assay with confirmatory Western blot), should be considered when resolving HIV status in individuals with discordant rapid test results.
Previous studies found that some individuals with discordant rapid tests had acute HIV infection.10–12 Those studies were performed in cohorts at increased risk of HIV acquisition (those attending clinics for sexually transmitted infections, or participating in a screening program with very high HIV prevalence).10–12 It is notable that those studies considered individuals with undetectable HIV RNA to be HIV uninfected. In this study, which was performed in the context of a general population survey, almost all of the HIV-infected individuals with discordant HIV rapid tests had undetectable HIV RNA using two assays validated for different HIV subtypes.18,19 Since most of these individuals were virally suppressed in the absence of ART, they are likely to have been elite controllers. Criteria used to identify elite controllers usually include documentation of viremic control for at least one year.20 Unfortunately, longitudinal samples were not collected in the HPTN 043 trial. DNA PCR testing may also be helpful for confirming a diagnosis of HIV infection in elite controllers; however, some elite controllers have negative DNA PCR tests.21,22 Samples suitable for HIV DNA testing were not collected in the HPTN 043 trial. It was not possible to determine the proportion of elite controllers in the entire HPTN 043 cohort, since viral load testing was only performed for a small proportion of the HIV-infected individuals. Other reports suggest that elite controllers represent <1% of HIV-infected individuals;20,23 however, higher prevalence of elite controllers has been reported in some studies.24
Viral suppression, whether natural or from ART, has been associated with false negative HIV screening tests.25,26 Down-regulation of anti-HIV antibodies in virally-suppressed individuals has been documented using serologic assays developed for HIV incidence estimation,27–30 The low levels of circulating virus (and viral antigen) in virally-suppressed individuals may also increase the likelihood of obtaining a false-negative test result using a fourth-generation assay.
Failure to detect HIV infection in virally-suppressed individuals has practical implications. While most individuals on ART are not likely to be tested for HIV infection (since their HIV status is known), some may chose not to disclose knowledge of their HIV status to care providers.31 Individuals on ART may also be tested for HIV infection in population-based surveys and clinical trials (e.g., if self-reported data on HIV status is not collected, or if individuals chose not to disclose their HIV status to study staff32). Elite controllers may not be aware of their HIV status and may be incorrectly classified as HIV-uninfected because of false-negative HIV tests.33 Viral suppression may also complicate identification of newly-infected individuals. We recently described a cohort in the US in which 21% of HIV seroconverters had low or undetectable HIV RNA in the absence of antiretroviral drug use.17 In the same study, HIV infection was missed at multiple visits in two virally-suppressed participants using an HIV rapid test; in one case, HIV infection was also missed using a fourth-generation assay.33 Missed HIV infection may become more common in resource-limited settings that rely on third-generation HIV screening assays and simple HIV testing algorithms, particularly as ART is scaled up for HIV treatment and prevention.34–36
Another factor that may potentially impact the performance of different HIV testing algorithms is HIV-1 subtype. Subtype D HIV infection is associated with lower levels of anti-HIV antibodies and lower avidity of anti-HIV antibodies for target antigens.37,38 In HPTN 043, HIV prevalence and incidence were higher in South Africa than in Tanzania. However, the percentage of individuals with discordant HIV rapid test results and the proportion of the discordant samples that were from HIV-infected individuals were both higher in Tanzania. This may reflect differences in the prevalent HIV subtypes in these two countries. In South Africa, the overwhelming majority of HIV infections are subtype C, while various viral subtypes are found in Tanzania.39 Previous analysis of samples from a subset of HPTN 043 participants in Tanzania revealed a mixture of HIV subtypes (44% Subtype A, 22% subtype C, 24% intersubtype recombinant, 10% subtype D15). In this study, we were not able to assess the contribution of HIV subtype to HIV test outcomes, since the majority of the HIV-infected individuals with discordant HIV rapid tests were virally suppressed; in the three cases where HIV subtype was determined, one individual had subtype A1 infection and two had subtype C infection.
In this study, individuals classified as HIV positive were likely to have been HIV infected, since they had positive/reactive tests with numerous HIV assays (one third-generation rapid test, at least one fourth-generation HIV assay, and the discriminatory assay or an RNA assay). We do note, however, that the HIV testing algorithm currently recommended by the US CDC has not been fully evaluated for use in Africa. Further testing is needed to evaluate performance of this algorithm in different settings. In this study, some false positive test results were obtained using fourth-generation assays as tie-breaker tests. A variety of factors have been associated with false positive HIV test results, including the presence of cross-reacting antibodies to other infectious agents.40–43 The high frequency of other infectious diseases in Africa could contribute to a higher rate of false-positive test results due to immune stimulation and/or formation of antibodies that cross-react with target antigens in HIV assays. Interestingly, the extended sample testing performed in this study identified 13 study participants who were classified as HIV negative using a rigorous testing algorithm, but had reactive results using both the Bio-Rad and Abbott Combo fourth-generation assays. These participants do not meet the criteria for HIV infection using the testing algorithm currently recommended by the US CDC, since both the differentiation assay and HIV RNA tests were negative. In HPTN 043, samples were not stored for HIV DNA testing, which may have been helpful in resolving the HIV status in these cases.
Taken together, the findings from this report suggest that while simple testing algorithms are practical for determining HIV status in the majority of individuals in resource-limited settings, extensive, laboratory-based testing may be needed to resolve the HIV status in individuals with discordant HIV rapid tests.
ACKNOWLEDGMENTS
The authors thank the HPTN 043 study team and study participants for providing samples for this study, and thank the laboratory staff at the study sites and at the HPTN Laboratory Center with assistance for sample management and testing. The authors thank Laura Robins-Morris for assistance with data management in the HPTN 043 study.
Sources of Funding
Abbott Laboratories has provided reagents and performed testing for some collaborative studies. Dr. Eshleman received an honorarium in 2009 for a presentation at a symposium sponsored by Abbott Laboratories.
This project was supported by the following awards: the HIV Prevention Trials Network, sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), the National Institute of Mental Health (NMH), and the National Institute of Drug Abuse (NIDA), Office of AIDS Research, of the National Institutes of Health [NIH, grants U01-AI068613/UM1-AI068613 (Eshleman); U01-AI068617/UM1-AI068617 (Donnell); and U01-AI068619/UM1-AI068619 (Vermund). Additional support for NIMH Project Accept (HPTN 043) was provided by the NIMH (U01-MH066687, U01-MH066688, U01-MH066701, and U01-MH066702). The funders had no role in study design; collection, analysis or interpretation of data; writing the report; or deciding to submit the manuscript for publication.
Footnotes
Conflicts of Interest
None of the authors has a conflict of interest or potential conflict of interest, with the following exception: Dr. Eshleman has collaborated on research studies with investigators from Abbott Laboratories.
AUTHORS CONTRIBUTIONS:
| Jessica M. Fogel | Designed the study; coordinated testing; assisted with data analysis and interpretation; drafted the manuscript |
| Estelle Piwowar-Manning | HPTN Laboratory Center Representative for Project Accept (HPTN 043); designed the study; coordinated testing |
| Kelsey Donohue | Coordinated and performed HIV testing; analysed test results |
| Vanessa Cummings | Coordinated and performed HIV testing; analysed test results |
| Mark A. Marzinke | Responsible for antiretroviral drug testing and interpretation of test results |
| William Clarke | Responsible for antiretroviral drug testing and interpretation of test results |
| Autumn Breaud | Coordinated and performed antiretroviral drug testing; analysed test results |
| Agnès Fiamma | Study Coordinator for NIMH Project Accept (HPTN 043); provided data for the study |
| Deborah Donnell | Statistician for NIMH Project Accept (HPTN 043); provided data for the study |
| Michal Kulich | Statistician for NIMH Project Accept (HPTN 043); provided data for the study |
| Jessie K. K. Mbwambo | Site PI for NIMH Project Accept (HPTN 043); provided samples and data for the study |
| Linda Richter | Site PI for NIMH Project Accept (HPTN 043); provided samples and data for the study |
| Glenda Gray | Site PI for NIMH Project Accept (HPTN 043); provided samples and data for the study |
| Michael Sweat | Investigator for NIMH Project Accept (HPTN 043); provided samples and data for the study |
| Thomas J. Coates | PI of NIMH Project Accept (HPTN 043); provided samples and data for the study |
| Susan H. Eshleman | PI of the HPTN Laboratory Center; designed the study; responsible for data analysis and interpretation; drafted the manuscript |
Contributor Information
Jessica M. Fogel, Email: jfogel@jhmi.edu, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, USA.
Estelle Piwowar-Manning, Email: epiwowa@jhmi.edu, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, USA.
Kelsey Donohue, Email: kdonohu8@jhmi.edu, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, USA.
Vanessa Cummings, Email: Vcummin1@jhmi.edu, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, USA.
Mark A. Marzinke, Email: mmarzin1@jhmi.edu, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, USA.
William Clarke, Email: wclarke@jhmi.edu, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, USA.
Autumn Breaud, Email: abreaud1@jhmi.edu, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, USA.
Agnès Fiamma, Email: afiamma@gmail.com, Program in Global Health, University of California at Los Angeles, Los Angeles, CA, USA.
Deborah Donnell, Email: Deborah@scharp.org, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; Dept. of Global Health, Univ. of Washington, Seattle, WA, USA.
Michal Kulich, Email: kulich@karlin.mff.cuni.cz, Dept. of Probability and Statistics, Faculty of Mathematics and Physics, Charles Univ., Prague, Czech Republic.
Jessie K. K. Mbwambo, Email: jmbwambo@gmail.com, Muhimbili Univ. of Health and Allied Sciences, Muhimbili Univ. Teaching Hospital, Dares Salaam, Tanzania.
Linda Richter, Email: lrichter@hsrc.ac.za, DST-NRF Centre of Excellence in Human Development, Universities of the Witwatersrand and KwaZulu-Natal; Human Sciences Research Council, Durban, South Africa.
Glenda Gray, Email: Glenda.Gray@mrc.ac.za, Perinatal HIV Research Unit, Chris Hani Baragwanath Hospital, Univ. of the Witwatersrand, Johannesburg, South Africa; South African Medical Research Council, Cape Town, South Africa.
Michael Sweat, Email: sweatm@musc.edu, Dept. of Psychiatry and Behavioral Sciences, the Medical Univ. of South Carolina, Charleston, SC, USA.
Thomas J. Coates, Email: tcoates@mednet.ucla.edu, Center for World Health, David Geffen School of Medicine and UCLA Health, Los Angeles, CA, USA.
Susan H. Eshleman, Email: seshlem@jhmi.edu, Dept. of Pathology, Johns Hopkins Univ. School of Medicine, Baltimore, MD, USA.
References
- 1.Centers for Disease Control and Prevention. Detection of acute HIV infection in two evaluations of a new HIV diagnostic testing algorithm - United States, 2011–2013. MMWR Morb Mortal Wkly Rep. 2013;62(24):489–494. [PMC free article] [PubMed] [Google Scholar]
- 2.Branson BM, Owen SM, Wesolowski LG, et al. Laboratory testing for the diagnosis of HIV infections. [Accessed June 30, 2014];2014 Available at: http://www.cdc.gov/hiv/pdf/HIVtestingAlgorithmRecommendation-Final.pdf.
- 3.World Health Organization (WHO) Service delivery approaches to HIV testing and counseling (HTC): a strategic HTC policy framework. [Accessed Feb 12, 2014];2012 Available at: http://apps.who.int/iris/bitstream/10665/75206/1/9789241593877_eng.pdf.
- 4.World Health Organization (WHO) Rapid HIV Tests: Guidelines for use in HIV testing and counseling services in resource-limited settings. [Accessed: Mar 4, 2014];2004 Available at: http://applications.emro.who.int/aiecf/web28.pdf.
- 5.Ministry of Health and Social Welfare. Tanzania HIV Rapid Test Algorithm. [Accessed: Mar 4 2014];2007 Available at: http://www.jica.go.jp/project/tanzania/001/materials/pdf/vct_07pdf.
- 6.South Africa Minister of Health. HIV counseling and testing (HCT) policy guidelines. [Accessed: March 4 2014];2010 Available at: http://www.sanac.org.za/resources/cat_view/1-resources.
- 7.Boeras DI, Luisi N, Karita E, et al. Indeterminate and discrepant rapid HIV test results in couples' HIV testing and counselling centres in Africa. J Int AIDS Soc. 2011;14:18. doi: 10.1186/1758-2652-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granade TC, Parekh BS, Tih PM, et al. Evaluation of rapid prenatal human immunodeficiency virus testing in rural Cameroon. Clin Diagn Lab Immunol. 2005;12(7):855–860. doi: 10.1128/CDLI.12.7.855-860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koblavi-Deme S, Maurice C, Yavo D, et al. Sensitivity and specificity of human immunodeficiency virus rapid serologic assays and testing algorithms in an antenatal clinic in Abidjan, Ivory Coast. J Clin Microbiol. 2001;39(5):1808–1812. doi: 10.1128/JCM.39.5.1808-1812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powers KA, Miller WC, Pilcher CD, et al. Improved detection of acute HIV-1 infection in sub-Saharan Africa: development of a risk score algorithm. AIDS. 2007;21(16):2237–2242. doi: 10.1097/QAD.0b013e3282f08b4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiscus SA, Pilcher CD, Miller WC, et al. Rapid, real-time detection of acute HIV infection in patients in Africa. J Infect Dis. 2007;195(3):416–424. doi: 10.1086/510755. [DOI] [PubMed] [Google Scholar]
- 12.Bassett IV, Chetty S, Giddy J, et al. Screening for acute HIV infection in South Africa: finding acute and chronic disease. HIV Med. 2011;12(1):46–53. doi: 10.1111/j.1468-1293.2010.00850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17(13):1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 14.Coates TJ, Kulich M, Zelaya CE, et al. Outcomes from NIMH Project Accept (HPTN 043): A cluster-randomized trial of community mobilization, mobile HIV testing, post-test support services, and real-time performance feedback. Lancet Global Health. 2014;2:e267–e277. doi: 10.1016/S2214-109X(14)70032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laeyendecker O, Piwowar-Manning E, Fiamma A, et al. Estimation of HIV Incidence in a large, community-based, randomized clinical trial: NIMH Project Accept (HIV Prevention Trials Network 043) PLoS One. 2013;8(7):e68349. doi: 10.1371/journal.pone.0068349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marzinke MA, Breaud A, Parsons TL, et al. The development and validation of a full scan-high resolution accurate mass spectrometric (HRMS) screening method for the qualitative monitoring of antiretroviral agents in human blood. Clin Chim Acta. 2014;433:157–168. doi: 10.1016/j.cca.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen I, Cummings V, Fogel JM, et al. Low-level viremia early in HIV infection. J Acquir Immune Defic Syndr. 2014;67(4):405–408. doi: 10.1097/QAI.0000000000000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gen-Probe. Product package insert. APTIMA HIV-1 RNA Qualitative Assay. [Accessed Oct 20, 2014]; Available at: http://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/UCM149927.pdf.
- 19.Michael NL, Herman SA, Kwok S, et al. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J Clin Microbiol. 1999;37(8):2557–2563. doi: 10.1128/jcm.37.8.2557-2563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol. 2013;13(7):487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- 21.Lambotte O, Boufassa F, Madec Y, et al. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin Infect Dis. 2005 Oct 1;41(7):1053–1056. doi: 10.1086/433188. [DOI] [PubMed] [Google Scholar]
- 22.Hatano H, Delwart EL, Norris PJ, et al. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J Virol. 2009 Jan;83(1):329–335. doi: 10.1128/JVI.01763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okulicz JF, Marconi VC, Landrum ML, et al. Clinical outcomes of elite controllers, viremic controllers, and long-term nonprogressors in the US Department of Defense HIV natural history study. J Infect Dis. 2009;200(11):1714–1723. doi: 10.1086/646609. [DOI] [PubMed] [Google Scholar]
- 24.Goujard C, Chaix ML, Lambotte O, et al. Spontaneous control of viral replication during primary HIV infection: when is "HIV controller" status established? Clin Infect Dis. 2009;49(6):982–986. doi: 10.1086/605504. [DOI] [PubMed] [Google Scholar]
- 25.Merchant M, Wright M, Kabat W, et al. Long-term highly suppressed HIV-infected children and adolescents with negative rapid HIV tests due to significant antibody loss. J Clin Virol. 2014;59(3):172–176. doi: 10.1016/j.jcv.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Claassen M, van Zyl GU, Korsman SN, et al. Pitfalls with rapid HIV antibody testing in HIV-infected children in the Western Cape, South Africa. J Clin Virol. 2006;37(1):68–71. doi: 10.1016/j.jcv.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Wendel SK, Mullis CE, Eshleman SH, et al. Effect of natural and ARV-induced viral suppression and viral breakthrough on anti-HIV antibody proportion and avidity in patients with HIV-1 subtype B infection. PLoS One. 2013;8(2):e55525. doi: 10.1371/journal.pone.0055525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laeyendecker O, Rothman RE, Henson C, et al. The effect of viral suppression on cross-sectional incidence testing in the Johns Hopkins hospital emergency department. J Acquir Immune Defic Syndr. 2008;48(2):211–215. doi: 10.1097/QAI.0b013e3181743980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marinda ET, Hargrove J, Preiser W, et al. Significantly diminished long-term specificity of the BED capture enzyme immunoassay among patients with HIV-1 with very low CD4 counts and those on antiretroviral therapy. J Acquir Immune Defic Syndr. 2010;53(4):496–499. doi: 10.1097/qai.0b013e3181b61938. [DOI] [PubMed] [Google Scholar]
- 30.Longosz AF, Mehta SH, Kirk GD, et al. Incorrect identification of recent HIV infection in adults in the United States using a limiting-antigen avidity assay. AIDS. 2014;28(8):1227–1232. doi: 10.1097/QAD.0000000000000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan AK, Savage EJ, Lowndes CM, et al. Non-disclosure of HIV status in UK sexual health clinics--a pilot study to identify non-disclosure within a national unlinked anonymous seroprevalence survey. Sex Transm Infect. 2013;89(2):120–121. doi: 10.1136/sextrans-2012-050801. [DOI] [PubMed] [Google Scholar]
- 32.Marzinke MA, Clarke W, Wang L, et al. Non-disclosure of HIV status in a clinical trial setting: antiretroviral drug screening can help distinguish between newly-diagnosed and previously-diagnosed HIV infection. Clin Infect Dis. 2014;58(1):117–120. doi: 10.1093/cid/cit672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piwowar-Manning E, Fogel JM, Laeyendecker O, et al. Failure to identify HIV-infected individuals in a clinical trial using a single HIV rapid test for screening. HIV Clin Trials. 2014;15(2):62–68. doi: 10.1310/hct1502-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen MS, Chen YQ, McCauley M, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365(6):493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitoria M, Vella S, Ford N. Scaling up antiretroviral therapy in resource-limited settings: adapting guidance to meet the challenges. Curr Opin HIV AIDS. 2013;8(1):12–18. doi: 10.1097/COH.0b013e32835b8123. [DOI] [PubMed] [Google Scholar]
- 36.World Health Organization (WHO) Use of antiretroviral drugs for treating pregnant women and preventing HIV infection in infants. Programmatic update. [Accessed: July 24, 2014];2012 Available at: http://whqlibdoc.who.int/hq/2012/WHO_HIV_20126_eng.pdf?ua=1.
- 37.Mullis CE, Munshaw S, Grabowski MK, et al. Differential specificity of HIV incidence assays in HIV subtypes A and D-infected individuals from Rakai, Uganda. AIDS Res Hum Retroviruses. 2013;29(8):1146–1150. doi: 10.1089/aid.2012.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longosz AF, Morrison CS, Chen PL, et al. Immune responses in Ugandan women infected with subtypes A and D HIV using the BED capture immunoassay and an antibody avidity assay. J Acquir Immune Defic Syndr. 2014;65(4):390–396. doi: 10.1097/QAI.0000000000000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemelaar J, Gouws E, Ghys PD, et al. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011;25(5):679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klarkowski D, O'Brien DP, Shanks L, et al. Causes of false-positive HIV rapid diagnostic test results. Expert Rev Anti Infect Ther. 2014;12(1):49–62. doi: 10.1586/14787210.2014.866516. [DOI] [PubMed] [Google Scholar]
- 41.Everett DB, Baisely KJ, McNerney R, et al. Association of schistosomiasis with false-positive HIV test results in an African adolescent population. J Clin Microbiol. 2010 May;48(5):1570–1577. doi: 10.1128/JCM.02264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swaminathan S, Hanna LE, Sundaramurthi JC, et al. Prevalence and pattern of cross-reacting antibodies to HIV in patients with tuberculosis. AIDS Res Hum Retroviruses. 2008 Jul;24(7):941–946. doi: 10.1089/aid.2007.0211. [DOI] [PubMed] [Google Scholar]
- 43.Trama AM, Moody MA, Alam SM, et al. HIV-1 envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe. 2014 Aug 13;16(2):215–226. doi: 10.1016/j.chom.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


