Abstract
Objective
Sarcolipin (SLN) regulates muscle energy expenditure through its action on sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) pump. It is unknown whether SLN-dependent respiration has relevance to human obesity, but whole-transcriptome gene expression profiling revealed that SLN was more highly expressed in myocytes from individuals with severe obesity (OB) than in lean controls (LN). The purpose of this study was to examine SLN-dependent cellular respiratory rates in LN and OB human muscles.
Design and Methods
Primary myocytes were isolated from muscle biopsy from seven LN and OB Caucasian females. Cellular respiration was assessed with and without lentivirus-mediated SLN knockdown in LN and OB myocytes.
Results
SLN mRNA and protein abundance was greater in OB compared to LN cells. Despite elevated SLN levels in wildtype OB cells, respiratory rates among SLN-deficient cells were higher in OB compared to LN. Obesity-induced reduction in efficiency of SLN-dependent respiration was associated with altered SR phospholipidome.
Conclusions
SLN-dependent respiration is reduced in muscles from humans with severe obesity compared to lean controls. Identification of molecular mechanism that affects SLN-efficiency might promote an increase in skeletal muscle energy expenditure.
Keywords: Obesity, Energy Expenditure, Respiratory, Phospholipids
Introduction
The recent pandemic in obesity has resulted in exponential growth in the prevalence of metabolic diseases (1, 2). Obesity occurs because of a disruption in energy homeostasis, where excess calories become stored in adipose tissue. While lifestyle interventions such as caloric restriction and physical exercise provide a means to weight loss, their long-term success is uncertain (3). Current medical interventions such as pharmaceutical treatments and bariatric surgery have side effects (4, 5). Although various strategies are employed to limit energy intake, at present physical activity remains the only practical means of increased energy expenditure.
Mitochondrial uncoupling has been well characterized as a site of futile energy expenditure in brown adipocytes (6, 7), muscle (8), and more recently in beige adipocytes (9, 10, 11). Sarcoplasmic reticulum (SR) uncoupling has been proposed as an alternate site of muscle energy dissipation (12, 13, 14). This pathway affects SR calcium cycling dynamics and reduces efficiency of sarco/endoplasmic reticulum Ca2+ ATPases (SERCAs) in a manner analogous to induction of futile proton cycling by mitochondrial uncoupling proteins (UCPs) to reduce energetic efficiency for ATP production by complex V. This SR futile calcium cycling is regulated by the protein sarcolipin (SLN) which appears to uncouple SERCA-dependent ATP hydrolysis from calcium transport into SR (14, 15, 16). SLN-knockout mice have reduced respiratory rates and are more prone to diet-induced obesity than wild type littermates (12).
Skeletal muscle biopsies from human subjects provide valuable insights for cellular processes that are directly relevant to the human population, but they provide only snapshots of the in vivo state and are not well suited for mechanistic studies. Human Skeletal Muscle Cells (HSkMCs) are primary muscle cells isolated from muscle biopsy samples and then induced to differentiate into myocytes, and they retain many metabolic properties of skeletal muscle in vivo, including insulin responsiveness and substrate oxidation (17, 18, 19, 20, 21). Such myocytes can be genetically manipulated and/or treated with pharmacologic agents to study cellular mechanisms (22). Furthermore, potential confounders such as humoral/neural effects and contamination by other cell types can be eliminated in studies using primary cells.
Here we used whole-transcriptome sequencing of HSkMCs from lean humans (LN) and those with severe obesity (OB) in order to compare gene expression profiles and identify candidates that may underlie the obesity metabolic program in these cells. Among 118 genes that were significantly differentially expressed between LN and OB, SLN stood out as a top candidate that was more highly present with obesity. We examined the hypothesis that greater SLN expression in human skeletal muscle with obesity leads to greater SLN-dependent respiration in these myocytes, possibly compensating for substrate overabundance by increasing energy expenditure.
Methods and Procedures
Human subjects, muscle biopsy and HSkMC isolation
The experimental protocol was approved by the Internal Review Board for Human Research at East Carolina University. Informed consent was obtained from all participants. Seven lean subjects without diabetes (LN: BMI < 25 kg/m2) and seven subjects with severe obesity (OB: BMI > 40 kg/m2) were studied (all Caucasian females). A fasting blood sample (glucose and insulin) and muscle biopsy from the vastus lateralis were collected. A portion of the biopsy sample was frozen immediately, and another portion was used to isolate primary muscle cells as previously described.(17)
Cell culture
HSkMCs were maintained in DMEM + 10% fetal bovine serum (FBS) supplemented with 0.5 mg/ml BSA, 0.5 mg/ml fetuin, and 100 µg/ml penicillin/streptomycin. At P2, multiple vials of cells were frozen and stored in liquid nitrogen for subsequent studies. At ~80% confluence, HSkMCs were switched to low-serum (2% horse serum) media to induce differentiation into myocytes. C2C12 cells were maintained in DMEM + 10% FBS. At ~80% confluence, media was changed to DMEM + 2% horse serum to induce differentiation. HEK 293T cells were maintained in DMEM + 10% FBS.
Western blotting
LN and OB HSkMC myocytes were incubated with or without 100 nM of insulin for 15 min. Cells were quickly harvested in a homogenization buffer for the analyses of insulin-stimulated phosphorylation of Akt Ser473 (pAkt: #9271, Cell Signaling Technology, Danvers, MA) by Western blotting as previously described (23). SLN (ABT13, EMD-Millipore), SERCA1 (ab2819, Abcam), SERCA2 (ab3625, Abcam), COXIV (#4844, Cell Signaling), and actin (A2066, Sigma) were also quantified in LN and OB HSkMCs.
Fatty acid oxidation
Rates of fatty acid oxidation were measured (18) by incubating myocytes in media containing 100 µM sodium oleate and 1.0 µCi/ml [14C]-oleate (Perkin Elmer, Waltham, MA) at 37°C. After 3 h, the incubation media was transferred to new dishes and assayed for labeled CO2. Cells were washed in PBS and lysed in homogenization buffer for the measurement of protein concentration by bicinchoninic assay (Thermo, Rockford, IL). All assays were performed in triplicates.
RNA sequencing
Total RNA from three samples per group was isolated by Qiagen RNeasy kit (Venlo, Limburg, Netherlands) and quality was assessed on a bioanalyzer (RIN > 8 for each sample). Poly-A selected cDNA libraries were constructed and sequenced on an Illumina HiSeq2000 by Otogenetics Corp (Norcross, GA) producing at least 20 million 2×101 paired end reads per sample. Reads were aligned to the NCBI GRCh37 USCS hg 19 reference genome with TopHat. RefSeq annotations were used as the gene model and differential expression was assessed with Cufflinks, correcting for multiple comparisons. Gene enrichment and pathway analysis were conducted with the Database for Annotation, Visualization and Integrated Discovery (DAVID). Expressions of select genes were confirmed with quantitative PCR as previously described (24).
Cellular respiratory rates
Cellular respiratory rates were determined with Seahorse Flux Analyzer XF-24 (Seahorse Bioscience, Billerica, MA). HSkMCs (60,000 cells/well) or C2C12 (25,000 cells/well) myocytes were plated and differentiated on a Seahorse culture plate. On the day of the experiment, media was switched to XF Assay Medium Modified DMEM (0 mM glucose) containing 1 mM pyruvate and 0.1% dextrose for 1 h. Oxygen consumption rates (OCR) for different respiratory states were measured using XF-24 by successive injections of oligomycin (1 µM), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) (0.4 µM), and rotenone (0.1 µM)/antimycin A (2 µM). Values for respiratory rates were normalized to cellular protein for each well. For HSkMCs knockdown experiments, values were further normalized for each plate to account for plate-to-plate variability.
Lentivirus-mediated knockdown
Plasmids encoding shRNA for mouse SLN (shSLN: TRCN0000313022) and human SLN (TRCN0000288825) were obtained from Sigma (St. Louis, MO). Packaging vector psPAX2 (ID #12260), envelop vector pMD2.G (ID #12259), and scrambled shRNA plasmid (sc: ID #1864) were obtained from Addgene (Cambridge, MA). 293T cells in 10 cm dishes were transfected using Lipofectamine 2000 with 2.66 µg of psPAX2, 0.75 µg of pMD2.G, and 3 µg of shRNA plasmid. After 48 hours, media were collected, filtered using 0.45 µm syringe filters, and used to treat undifferentiated C2C12 or HSkMCs. After 36 h, target cells were selected with puromycin and were plated on 6-well dishes (for qPCR) or on Seahorse plates (for respiratory rates) and were differentiated. SLN-knockdown did not affect differentiation efficiency.
SR lipidomics
SR fraction was isolated from LN and OB HSkMCs as previously described (24). Lipidomic analyses of LN and OB HSkMCs were conducted in the Department of Medicine Mass Spectrometry Facility of the Washington University School of Medicine as previously described (24). Extracted lipids with internal standards (5 µg [14:0/14:0]-PC ([M+Li]+ m/z 684.58) and 5 µg [14:0/14:0]-PE ([M-H]− m/z 678.72)) were directly infused into the electrospray ion source of a Thermo Vantage triple-quadrupole mass spectrometer. Analyses of [M+Li]+ ions were performed in positive mode with scanning for neutral loss of 183 for PC species, and analyses of [M-H]− ions were performed in negative mode with scanning for the product ions m/z 196 and 153 for PE and PS species, respectively. The amount of each lipid species was normalized to the total protein content of the sample and expressed as nmol/mg protein.
Statistical analyses
Values are expressed as means ± SEM. Statistical comparisons were performed using 2-tailed Student’s t test, or a 2-way ANOVA with Sidak’s multiple comparisons test. P values less than 0.05 were considered significant.
Results
An obesity metabolic program in HSkMCs
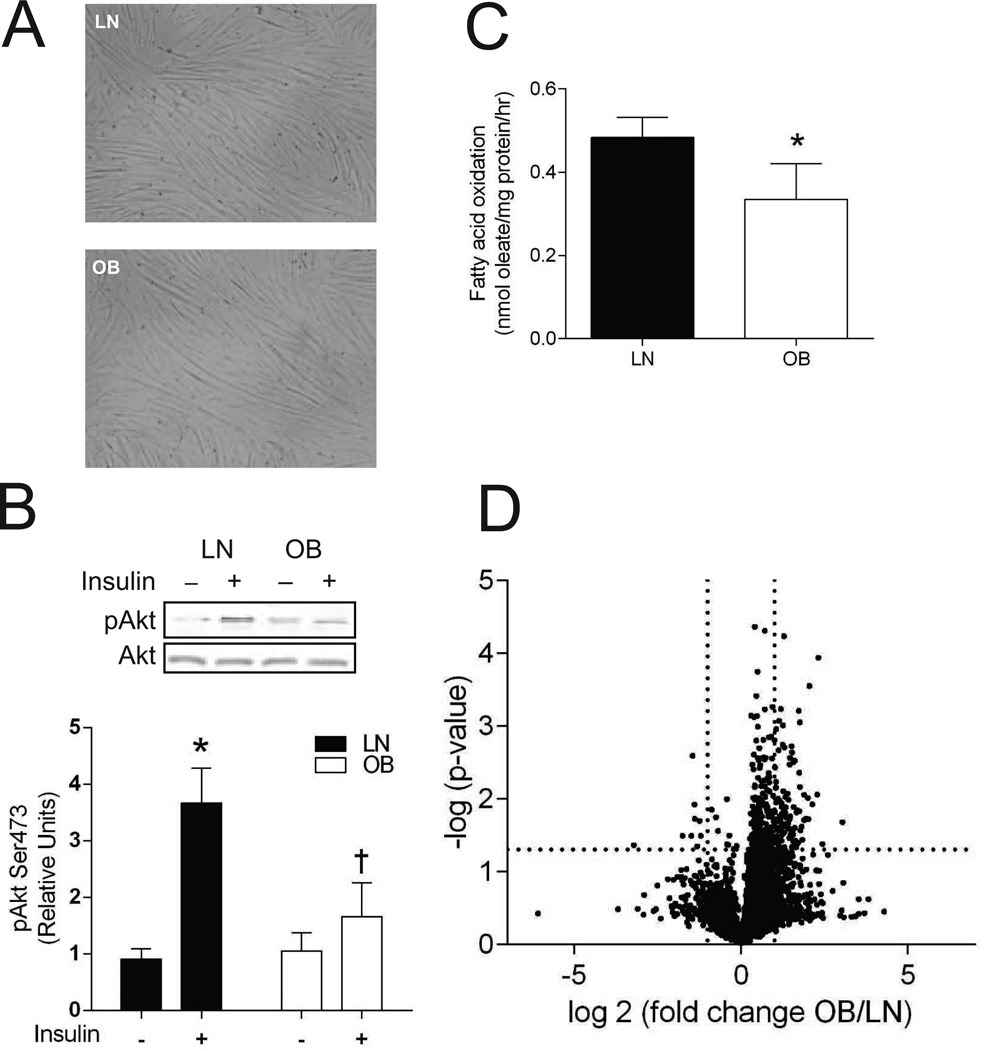
Compared to LN subjects, OB subjects had significantly greater body mass, BMI, fasting insulin and HOMA-IR, but fasting glucose levels were not significantly different between the groups (Table 1). As previously reported (19), HSkMCs myocytes from LN and OB subjects did not have any obvious morphologic differences (Figure 1A). Evidence suggests that ex vivo these cells retain metabolic characteristics of donor skeletal muscle in vivo (18, 19, 20, 21, 22). In the current study, we found that OB HSkMCs are insulin resistant (Figure 1B, reduction in insulin-stimulated pAkt) and have reduced rates of fatty acid oxidation (Figure 1C) compared to LN HSkMCs. These observations support the proposition that HSkMCs derived from human subjects can maintain metabolic traits expressed by skeletal muscle in vivo. HSkMCs thus provide a suitable platform for in vitro mechanistic studies impossible to perform in human subjects.
Table 1.
Subject Demographic
| LN | OB | |
|---|---|---|
| Age (yr) | 30.0 ±3.1 | 37.1 ±2.7 |
| Body Mass (kg) | 63.3 ±3.9 | 144.1 ±8.9* |
| BMI (kg/m2) | 23.2 ±1.1 | 50.1 ±1.7* |
| Fasting Glucose (mM) | 4.8 ±0.2 | 5.1 ±0.1 |
| Fasting Insulin (pM) | 55.6 ±9.7 | 145.8 ±31.3* |
| HOMAIR | 1.75 ±0.3 | 4.7 ±1.0* |
| Total Cholesterol (mM) | 4.06 ±0.28 | 4.54 ±0.36 |
| HDL (mM) | 1.48 ±0.09 | 1.25 ±0.07 |
| LDL (mM) | 2.21 ±0.26 | 2.94 ±0.35 |
| Triglycerides (mM) | 0.82 ±0.14 | 0.79 ±0.10 |
Subject demographic of lean humans (LN) and those with severe obesity (OB) (n=7/group).
BMI: Body mass index. HOMA-IR: homeostatic model assessment of insulin resistance.
: p < 0.05. Data are shown mean ±SEM.
Figure 1. HSkMCs retain metabolic properties ex vivo.
A: Representative images of LN and OB HSkMCs. B: Insulin-stimulated pAkt (n=6/group). *: Effect of insulin, p < 0.05. †: Effect of obesity, p < 0.05. C: Rates of fatty acid oxidation (n=5/group). *: Effect of obesity, p < 0.05. D: Volcano plot of differential expression profile of genes with RPKM > 1 (n=3/group). Dotted lines indicate cutoff lines for fold-difference (2) and p (0.05). All data were collected from day 4 of differentiation. Filled bars: LN, empty bars: OB. Data are shown mean ±SEM.
Because HSkMCs maintain these differential phenotypes in culture in the absence of the hormonal milieu as well as neural inputs, such phenotypes are likely to result from genetic or epigenetic influences, and such differential programming might be expected to be manifest in the gene expression profiles. To explore this possibility, we used whole-genome RNAseq to characterize the transcriptome of LN and OB HSkMCs. Microarray analyses of muscle biopsy samples from groups of subjects with similar demographic profiles have been conducted previously (19). However, muscle biopsies contain contaminating cell types, e.g., adipocytes, endothelial cells and macrophages, and these cell types might influence the transcriptome. Thus, transcriptomic profiling of HSkMCs provides cell-specific information. RNAseq also offers the advantage of wider dynamic range, single-base resolution, comprehensive coverage, and reduced background noise (25).
RNAseq data are displayed in a volcano plot that represents differential gene expression between LN and OB HSkMC transcriptomes (Figure 1D). We identified 118 significantly differentially expressed genes (Benjamini and Hochberg FDR-corrected at 5%, RPKM > 1) whose fold-differences were above 2 (Table 2). As illustrated in the volcano plot, 109 genes were more highly expressed in OB samples whereas only 9 were more highly expressed in LN samples. Pathway analysis revealed enrichment of genes involved in calcium metabolism, including those involved in SR calcium cycling such as SLN, SERCA1 (ATP2A1), SERCA2 (ATP2A2), ryanodine receptor (RYR1), sarcalumenin (SRL) and triadin (TRDN), to be differentially expressed between LN and OB HSkMCs. Because of the newly-identified role of SLN in energy expenditure and obesity (12), we decided to pursue, for the first time, the physiological relevance of this gene in human muscles.
Table 2.
Differential gene expression between LN and OB HSkMCs.
| Gene | OB/LN | Gene | OB/LN | Gene | OB/LN | Gene | OB/LN |
|---|---|---|---|---|---|---|---|
| A2BP1 | 2.01 | DBNDD1 | 3.19 | MRPS26 | 2.10 | STRADB | 2.41 |
| ACSS2 | 2.35 | DEPDC6 | 3.13 | MYL3 | 3.27 | SUGT1P1 | 0.38 |
| ACTN2 | 2.75 | DKK1 | 0.36 | MYL5 | 2.73 | SYNE1 | 2.03 |
| ADRBK2 | 2.76 | DLGAP5 | 3.05 | MYL6B | 2.57 | SYPL2 | 3.05 |
| AFAP1L1 | 2.47 | DNAJA4 | 2.23 | MYLPF | 2.06 | TAL2 | 2.65 |
| ALX1 | 3.71 | DPYSL4 | 2.32 | MYOM1 | 2.28 | TCAP | 3.06 |
| ANGPTL1 | 3.44 | ENO3 | 2.25 | MYOM3 | 2.95 | TCEA3 | 2.16 |
| ANK1 | 3.53 | ETV6 | 2.19 | MYOT | 3.88 | TMEM38A | 2.41 |
| ANXA6 | 2.32 | FGF13 | 2.91 | MYOZ1 | 3.32 | TMEM97 | 2.46 |
| APOBEC2 | 4.41 | FGF9 | 5.00 | NCRNA00162 | 0.30 | TMOD1 | 2.89 |
| ATP2A1† | 5.42 | FITM1 | 3.99 | NDRG2 | 2.52 | TMOD4 | 8.22 |
| ATP2A2† | 2.04 | GABRR1 | 3.02 | NUSAP1 | 2.46 | TNFRSF21 | 2.38 |
| BEST3 | 3.40 | GADD45B | 0.40 | PEG10 | 2.52 | TNNC1 | 2.17 |
| BHMT2 | 2.25 | GATM | 3.36 | POU5F1 | 2.38 | TNNC2 | 3.71 |
| BIN1 | 2.88 | GPM6B | 2.68 | PPAP2A | 2.02 | TNNT3 | 3.14 |
| C17orf103 | 2.19 | GPR137B | 0.42 | PRKAG3 | 2.43 | TPD52L1 | 2.86 |
| C1orf105 | 4.05 | HFE2 | 2.14 | PRKCQ | 2.75 | TPM1 | 2.53 |
| C1orf230 | 2.16 | HRC | 2.00 | PSORS1C1 | 0.11 | TPM2 | 2.03 |
| C6orf134 | 2.45 | ID4 | 0.43 | PVR | 2.29 | TRDN† | 3.40 |
| C6orf142 | 2.44 | IL20RA | 2.07 | RASSF4 | 2.14 | TRIM54 | 2.14 |
| CAPN3 | 2.06 | ISYNA1 | 2.97 | RHOJ | 2.18 | TTC13 | 2.06 |
| CARNS1 | 2.20 | KBTBD5 | 2.31 | RNLS | 2.16 | TTK | 2.31 |
| CASP9 | 2.47 | KCNS3 | 2.40 | RRAGD | 3.59 | TUBA8 | 2.98 |
| CD34 | 2.50 | KIAA0907 | 2.29 | RYR1† | 2.32 | TUSC3 | 2.20 |
| CHRND | 2.19 | KIAA1524 | 2.78 | SCN4A | 2.63 | TXLNB | 2.26 |
| CLEC2B | 2.75 | KRTAP15 | 0.38 | SLN† | 4.13 | VWA5A | 3.17 |
| CORO6 | 2.03 | LAD1 | 2.39 | SMPX | 2.30 | ZFP36L2 | 2.08 |
| CRISPLD2 | 2.25 | LOC100132288 | 0.37 | SOHLH2 | 3.36 | ZNF280A | 2.68 |
| CRMP1 | 2.82 | MAP6D1 | 2.83 | SRL† | 2.23 | ||
| CYSLTR1 | 4.86 | MAPK8 | 2.94 | STAC3 | 2.40 |
A list of differentially expressed genes between LN and OB HSkMCs. OB/LN columns indicate the fold-difference in gene expression between OB and LN groups. Values greater than 1 indicate higher expression in OB myocytes.
Genes involved in SR calcium metabolism.
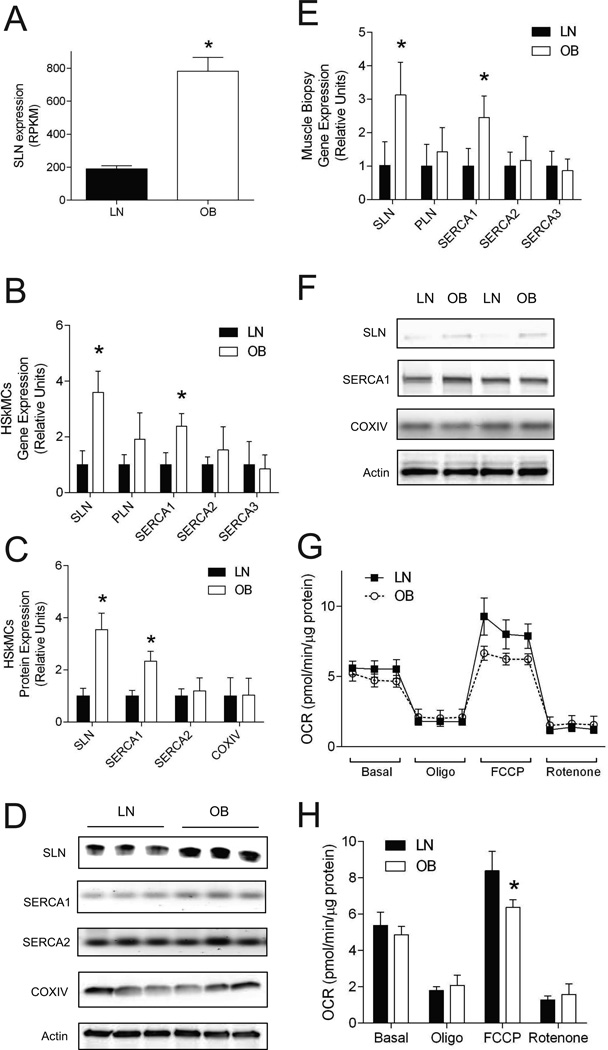
RNAseq data showed that SLN was 4.1-fold greater in OB than in LN (Figure 2A). Quantitative PCR and Western blotting experiments validated these differences in HSkMCs (Figure 2B–D) and the differences were replicated in muscle biopsy samples (Figure 2E and F). SLN exerts its effects on SERCA calcium ion channels in a fashion similar to that of its paralog phospholamban (PLN). Gene expression levels of SERCA1 (but not SERCA2, SERCA3 or PLN) were also elevated in OB HSkMCs and muscle biopsy samples (Figure 2B–E). Because the level of SLN might affect cellular respiration, we measured oxygen consumption in LN and OB HSkMCs. Consistent with previous findings, basal respiratory rates were not different between LN and OB HSkMCs (26, 27), but FCCP-induced respiration was lower in OB HSkMCs (Figure 2G and H). We found abundance of mitochondrial protein COXIV not to be different between LN and OB cells (Figure 2C and D), making it unlikely that lower mitochondrial content to account for reduced FCCP-induced respiration in OB HSkMCs.
Figure 2. Higher SLN expression in OB HSkMCs does not yield elevated respiratory rates.
A: SLN expression by RNAseq. RPKM: reads per kilobase of target region per millions mapped. B: Gene expression of SLN and other SR calcium uptake enzymes in LN and OB HSkMCs by quantitative PCR (n=6/group). C: Protein expression of SLN, SERCA1, SERCA2 and COXIV in LN and OB HSkMCs by western blotting (n=6/group). D: Representative western blots from LN and OB HSkMCs. E: Gene expression of SLN and other SR calcium uptake enzymes in LN and OB muscle biopsies by quantitative PCR (n=6/group). F: Representative western blots of SLN, SERCA1 and COXIV protein from LN and OB muscle biopsies. G: Cellular respiratory rates of LN and OB HSkMCs (n=5/group). Measurements for all 10 samples were performed on a single Seahorse plate. H: Mean values for four phases of respirations in LN and OB HSkMCs (n=5/group). OCR: oxygen consumption rate. Oligo: oligomycin. FCCP: carbonyl cyanide-p-trifluoromethoxyphenylhydrazone. All data were collected from day 4 of differentiation. Filled bars/dots: LN, empty bars/dots: OB. *: p < 0.05. Data are shown mean ±SEM.
Lentivirus-mediated knockdown of SLN reduces cellular respiration
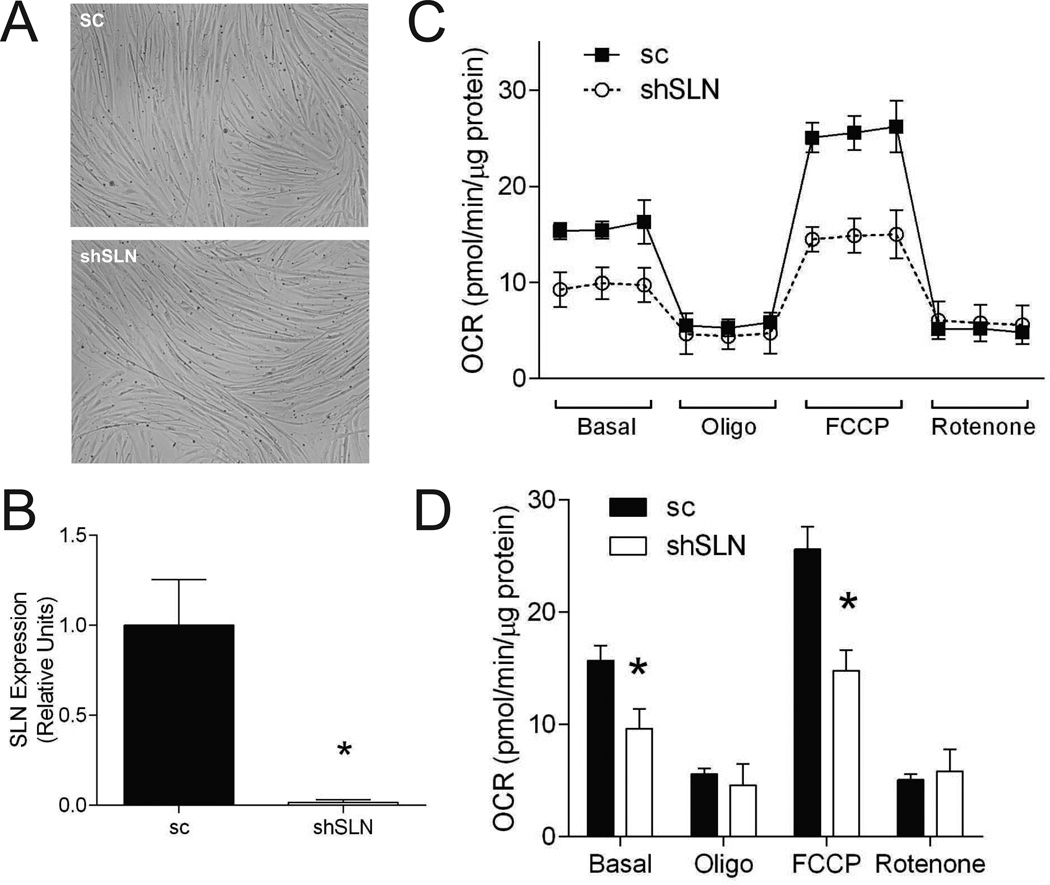
SLN whole-body knockout mice have reduced resting metabolic rate and are prone to diet-induced obesity (12). SLN is highly expressed in skeletal muscle and affects cellular respiration through its action on SERCA (12, 15, 16). To test the possibility that the effect of SLN deletion on muscle respiration is cell-autonomous, lentiviral knockdown of mouse SLN was performed on C2C12 myocytes (Figure 3A and B). Consistent with data from whole-body knockout mice, deletion of SLN in C2C12 cells reduced cellular respiratory rates (basal and FCCP) (Figure 3C and D). Decreased basal respiration suggests sustained decrease in basal metabolic rate, possibly due to reduced ATP demand or mitochondrial proton uncoupling. Lack of a difference in oligomycin-induced respiration suggests that the difference in basal respiration is likely not due to differences in mitochondrial uncoupling. Decreased FCCP respiration reflects reduced maximal respiration, but reduced maximal respiratory capacity alone would not yield lower basal resting respiratory rates as ATP synthesis is regulated by ATP demand, not capacity (28, 29). Thus, loss of SLN function in muscle cells per se appears to reduce basal energy expenditure.
Figure 3. SLN deficiency promotes reduction in cellular respiratory rates.
A: Representative images of sc and shSLN C2C12 cells. SLN knockdown did not affect differentiation efficiency or morphology. B: Quantitative PCR of SLN in sc and shSLN C2C12 myotubes. C: Cellular respiration rates of sc and shSLN C2C12 myotubes. D: Mean values for four phases of respirations. All data were collected from day 4 of differentiation. Filled bars/dots: sc, empty bars/dots: shSLN. sc: scrambled, shSLN: short-hairpin SLN (SLN-knockdown). n=6/group. *: p < 0.05. Data are shown mean ±SEM.
SLN-dependent respiration is blunted with obesity
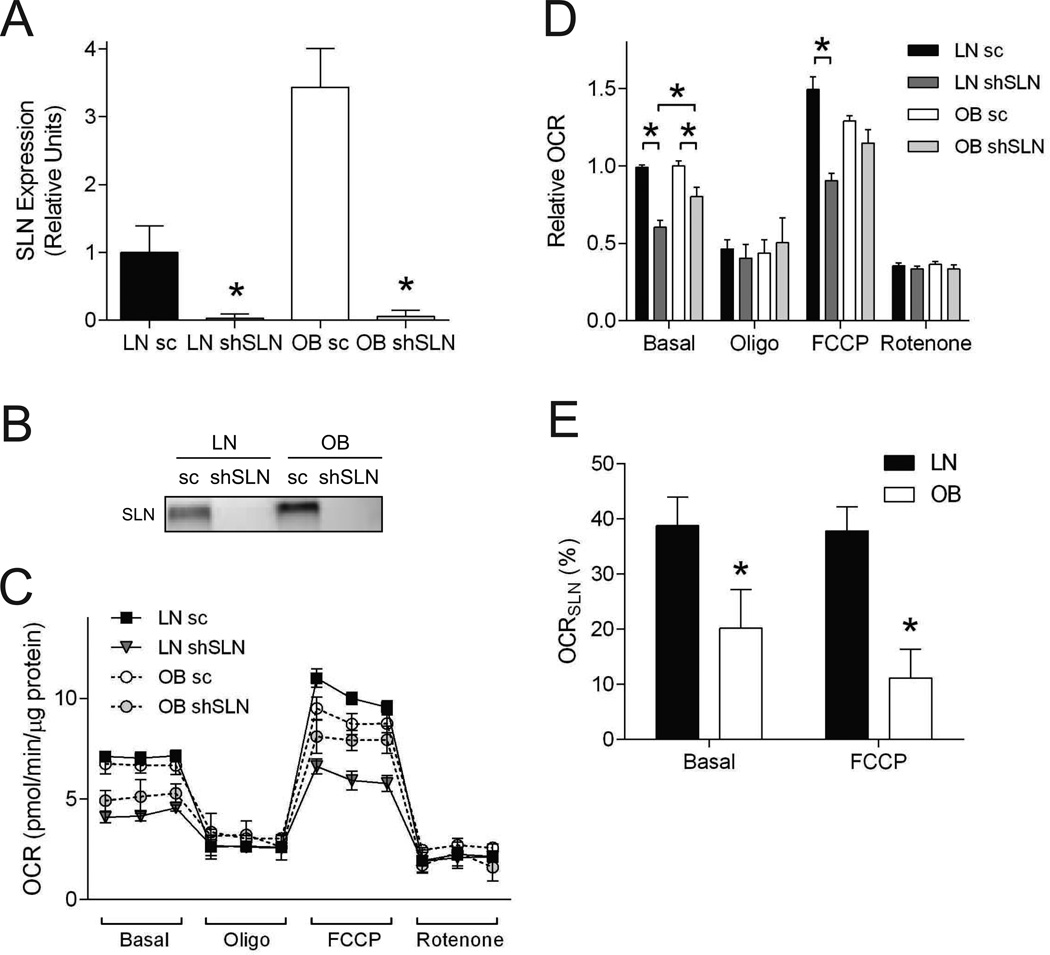
Respiratory rates of OB myocytes were equal to or lower than LN myocytes (Figure 2G and H) even though SLN expression was higher in OB cells (Figure 2A–D). To examine the contribution of SLN on LN and OB HSkMC respiration, lentivirus-mediated knockdown of human SLN was performed (Figure 4A and B). SLN knockdown effectively reduced all SLN mRNA and protein expression to undetectable levels in both LN and OB myocytes. Our initial hypothesis was that SLN knockdown would result in a greater reduction in respiration in OB cells because of their greater basal SLN expression compared to LN cells. In contrast, after SLN knockdown, the basal and FCCP-stimulated respiratory rates were higher, not lower, than those in LN HSkMCs (Figure 4C and D). Myocytes treated with scrambled lentiviral vectors exhibited respiratory rates similar to data in Figure 2G and H.
Figure 4. Reduced SLN efficiency in OB HSkMCs.
A: Quantitative PCR of SLN in LN sc, LN shSLN, OB sc and OB shSLN HSkMCs (n=6/group). B: Representative western blot of SLN protein in LN sc, LN shSLN, OB sc and OB shSLN HSkMCs. C: Cellular respiration rates. Representative data from a single Seahorse plate (n=4 or 5 technical replicates/group). D: Mean values for four phases of respirations (n=6 biological replicates/group). Data from multiple Seahorse plates were each normalized to the first value of LN sc, due to variability in OCR values inherent with HSkMCs and plates. Filled bars/dots: LN sc, dark gray bars/dots: LN shSLN, empty bars/dots: OB sc, light gray bars/dots: OB shSLN. E: OCRSLN: SLN-dependent OCR contribution, calculated with an equation OCRSLN = [OCRsc – OCRshSLN] / [OCRsc]. All data were collected from day 4 of differentiation. Filled bars: LN, empty bars: OB. sc: scrambled, shSLN: short-hairpin SLN (SLN-knockdown). *: p < 0.05. Data are shown mean ±SEM.
To compare the contribution of SLN on total cellular respiration, SLN-dependent respiratory rates (OCRSLN) were calculated by dividing the difference between sc and shSLN OCR values by the sc OCR values according to the equation:
| (eq.1) |
In LN myocytes, OCRSLN was ~40% for both basal and FCCP-stimulated respiration (Figure 4E, filled bars). In contrast, OCRSLN for OB myocytes was significantly lower than that for LN cells under both basal (20%) and FCCP-stimulated (11%) respiration (Figure 4E, empty bars). Notably, the lower OCRSLN for OB cells was observed even though SLN protein abundance was 3.5-fold higher for the OB than for LN cells (Figure 2C). These findings suggest that SLN-dependent oxygen consumption is reduced with obesity.
Differential lipidome in LN and OB HSkMCs
The reduced efficiency of SLN-dependent respiration in OB vs. LN HSkMCs might have been explained by variants (e.g. SNPs) in OB cells that affect SLN enzymatic activity, but sequencing data from our RNAseq studies did not reveal such differences.
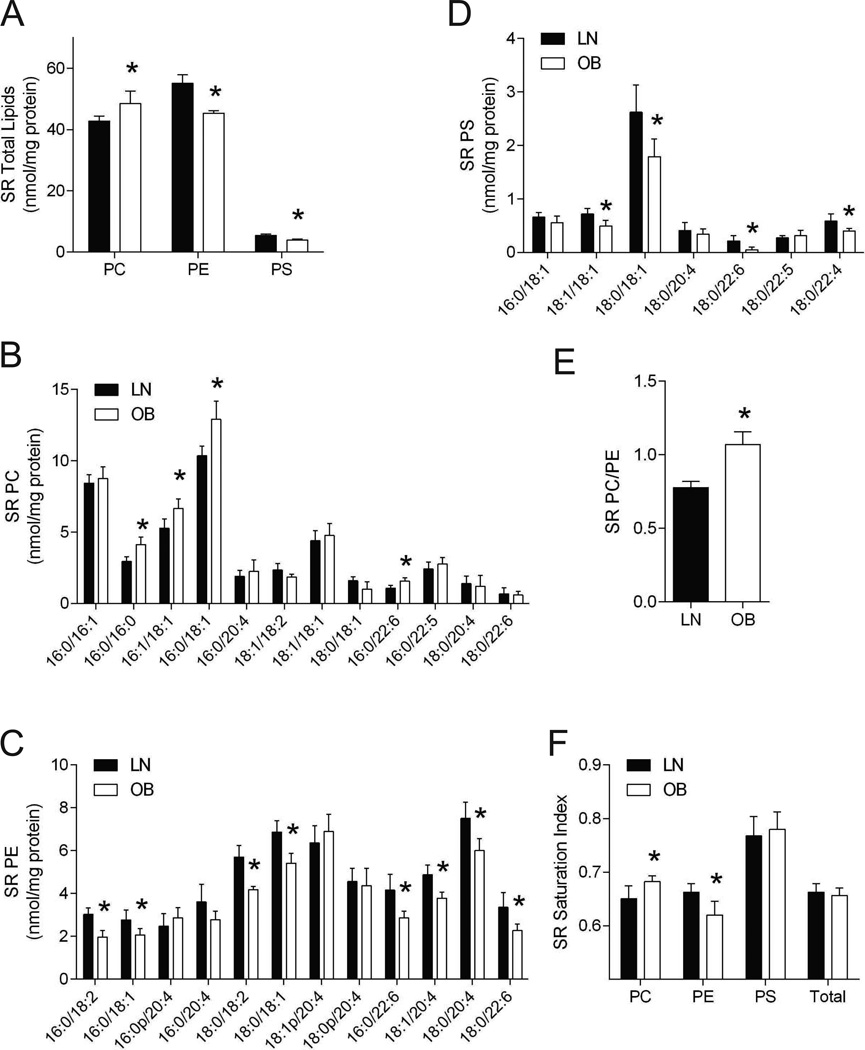
SLN uncouples Ca2+ uptake from ATP consumption by SERCA, and SERCA activity is affected by physiochemical properties of the SR lipid bilayer (24, 30, 31, 32). SR membranes consist mainly of phosphatidylcholine (PC), phosphatidylethanolamine (PE) and phosphatidylserine (PS). We quantified these lipids from isolated SR from LN and OB HSkMCs by mass spectrometry (Figure 5). SR total PC was greater in OB than in LN cells (Figure 5A), specifically for 16:0/16:0-PC, 16:1/18:1-PC, 16:0/18:1-PC, and 16:0/22:6-PC (Figure 5B). SR total PE was lower in OB than in LN cells (Figure 5A), specifically for 16:0/18:2-PE, 16:0/18:1-PE, 18:0/18:2-PE, 18:0/18:1-PE, 16:0/22:6-PE, 18:1/20:4-PE, 18:0/20:4-PE, and 18:0/22:6-PE (Figure 5C). SR total PS was lower in OB than in LN cells (Figure 5A), specifically for 18:1/18:1-PS, 18:0/18:1-PS, 18:0/22:6-PS and 18:0/22:4-PS (Figure 5D). SERCA activity is affected by the properties of phospholipids in the membranes in which it resides, including identities of headgroups and fatty acid substituent chain length and degree of unsaturation (which affect bilayer thickness and membrane fluidity, respectively) (30). Consequences of changes in SR membrane lipid composition on SERCA activity are complex, and several compositional parameters differed between OB and LN HSkMCs. SERCA activity is enhanced by PE compared to PC, and the PC/PE ratio correlates inversely with SERCA activity (24, 30, 32). The PC/PE ratio for OB HSkMCs exceeded that for LN HSkMCs (Figure 5E), and this would be predicted to result in reduced SERCA activity and reduced OCRSLN in OB HSkMCs (Figure 4E). Distribution of the bilayer thickness as estimated from phospholipid fatty acid substituent chain length did not appear to be substantially different between OB and LN HSkMCs (Figure 5B–D). Saturation indexes (nmol saturated acyl-chains/nmol unsaturated acyl-chains) for PC were higher in OB than in LN cells, but this index for PE was lower in OB than in LN cells (Figure 5F). These data suggest that phospholipid composition of SR membranes changes with obesity in a manner that could result in reduced SERCA activity and OCRSLN, thereby reducing the contribution of SLN-dependent oxygen consumption.
Figure 5. Differential SR phospholipidome in LN and OB HSkMCs.
A: Total PC, PE and PS in isolated SR of LN and OB HSkMCs. PC: phosphatidylcholine, PE: phosphatidylethanolamine, PS: phosphatidylserine. B: Individual PC species in isolated SR of LN and OB cells. C: Individual PE species in isolated SR of LN and OB cells. D: Individual PS species in isolated SR of LN and OB cells. E: SR’s PC/PE ratio in LN and OB cells. F: SR lipid saturation index (nmol saturated acyl-chains/nmol unsaturated acyl-chains) in LN and OB cells. N=4/group. All data were collected from day 4 of differentiation. Filled bars: LN, empty bars: OB. *: p < 0.05. Data are shown mean ±SEM.
Discussion
Sarcolipin is a recently-identified enzyme that is involved in the regulation of skeletal muscle energy expenditure (12). The mechanism involved is futile cycling mediated by the uncoupling of Ca2+ transport from ATP consumption by SERCA pump within SR membranes. In this study, characterization of gene expression by primary muscle cells harvested from lean humans and those with severe obesity was profiled using RNA-sequencing technology, revealing many genes involved in SR Ca2+ cycling to be differentially expressed between the two groups. These include SLN, the expression of which was ~4 fold higher in OB than in LN HSkMCs. Nonetheless, knockdown experiments revealed that SLN-dependent respiration (OCRSLN) in skeletal muscle cells from subjects with severe obesity was lower than in such cells from lean controls. SR Lipidomic analyses revealed OB and LN HSkMCs exhibited significant differences in phospholipid composition, including an increased PC/PE ratio and altered unsaturation index for OB HSkMCs that would be predicted to impair SERCA activity. This might represent a mechanism whereby SLN-mediated dissipation of energy is reduced in OB compared to LN HSkMCs and by extrapolation in humans with obesity compared to those that are lean.
Energy dissipation in adipose tissues, both in classical brown fat (6, 7) and beige adipocytes (9, 10), has received much attention because of the potential therapeutic benefits of enhancing energy dissipation and preventing development of obesity. Skeletal muscle is another site of energy expenditure (8). In both adipose tissues and skeletal muscle, uncoupling of proton pumping from ATP generation in mitochondria is one established mechanism for elevating respiration and energy expenditure. Similarly, uncoupling of SR Ca2+ uptake from ATP consumption by SERCA has been proposed as an alternative mechanism for skeletal muscle energy dissipation (12, 13). Our findings suggest that the capacity for SLN-dependent respiration is reduced in skeletal muscles from subjects with obesity compared to lean controls, even though higher expression of SLN is observed in OB than in LN HSkMCs.
The current study does not establish causal relationship between reduced SLN efficiency and obesity. It is conceivable that muscles from individuals with severe obesity are preprogramed in a manner that results in a congenital reduction in muscle SLN efficiency. In that case, reduced SLN efficiency would impair muscle’s ability for futile ATP consumption (which is manifested in augmented basal oxygen consumption) and thereby contribute to obesity. Our data indicating that OB HSkMCs exhibited reduced FCCP-stimulated respiration (Figure 2G and H) might be interpreted to support this possibility, but SLN inefficiency would also be expected to be reflected in effects under basal conditions, which was not observed here or in other studies (26, 27). Reduced FCCP-stimulated respiration phase might reflect reduced oxidative capacity, a phenomenon commonly associated with physical inactivity and/or obesity (33, 34, 35, 36). A more likely scenario is that SLN inefficiency develops as a result of obesity. As we have shown, one possibility for reduced SLN efficiency in these muscles is a change in SR phospholipidome (Figure 5). Lipid flux and organellar lipid composition have been found to change with the development of obesity (24, 28, 37), and this might impair SLN efficiency in promoting energy expenditure by altering the phospholipid milieu in muscle SR membranes that contain SERCAs and SLN.
Human primary muscle cells derived from muscle biopsies were used for the studies described here. These cells are known to mimic under ex vivo conditions many phenotypic properties of muscle metabolism in vivo, and our findings with HSkMCs with respect to SLN-related molecular events might be relevant to human subjects. Genetic knockdown experiments and measurements of respiration described here are only feasible in cell culture, and we have used the best available cell model for human muscles to conduct these experiments. Our finding that SLN knockdown in HSkMCs results in reduced respiration confirms in vivo data previously reported for SLN knockout mice (12). Moreover, SLN expression is increased in human muscle biopsy samples (Figure 2E and F), and muscle basal respiration in vivo is not different between lean humans and those with obesity (38). These findings are compatible with the possibility that SLN efficiency in vivo might be reduced in individuals with obesity.
In conclusion, primary myocytes from humans with severe obesity displayed reduced SLN-dependent respiration compared to lean controls, suggesting inefficiency of SLN-dependent energy expenditure. It is unclear what molecular mechanisms promote SLN inefficiency in obese muscles, but we postulate that a difference in SR membrane phospholipid environment might provide an explanation. Future studies should investigate how alteration in SR phospholipid composition might affect SERCA and SLN enzymatic activities in skeletal muscle.
What is already known about this subject.
Sarcolipin (SLN) is a recently identified regulator of muscle energy expenditure.
Skeletal muscle SLN promotes futile ATP hydrolysis of sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) at sarcoplasmic reticulum (SR).
SLN knockout mice have reduced energy expenditure and are more prone to developing obesity.
What this study adds.
SLN is highly expressed in myocytes from humans with severe obesity
SLN-dependent respiration is reduced in myocytes from humans with severe obesity
Reduction in SLN efficiency might be explained by altered SR phospholipidome
Acknowledgements
CWP and KF conceived the experiments. CWP, ARPV, and TDH conducted cell culture, gene expression and respiratory measurements. CWP and HAL contributed to RNAseq experiments. KZ and SP contributed to fatty acid oxidation measurements. HS and JT conducted lipidomics experiments. TDH, KZ, SP, JAH, and KF conducted the clinical study. ARPV and KF wrote the manuscript. All contributed to manuscript edits.
This work was funded by East Carolina University startup fund and by NIH grants DK095774 to K.F., DK095003 to H.A.L., and DK056112 to J.A.H. The Washington University Medicine Department Mass Spectrometry Facility is supported by United States Public Health Service Grants P41-GM103422, P60-DK20579, and P30-DK56341.
Footnotes
Conflict of Interest
No conflict of interest
References
- 1.CDC. Vital signs: state-specific obesity prevalence among adults --- United States, 2009. Morbidity and Mortality Weekly Report. 2010;59:951–955. [PubMed] [Google Scholar]
- 2.Must A, Spadano J, Coakley EH, Field AE, Colditz G, Dietz WH. The disease burden associated with overweight and obesity. The Journal of the American Medical Association. 1999;282:1523–1529. doi: 10.1001/jama.282.16.1523. [DOI] [PubMed] [Google Scholar]
- 3.Dunkley AJ, Bodicoat DH, Greaves CJ, Russell C, Yates T, Davies MJ, et al. Diabetes prevention in the real world: effectiveness of pragmatic lifestyle interventions for the prevention of type 2 diabetes and of the impact of adherence to guideline recommendations: a systematic review and meta-analysis. Diabetes care. 2014;37:922–933. doi: 10.2337/dc13-2195. [DOI] [PubMed] [Google Scholar]
- 4.Pories WJ. Bariatric surgery: risks and rewards. The Journal of Clinical Endocrinology & Metabolism. 2008;93:S89–S96. doi: 10.1210/jc.2008-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray LJ, Cooper N, Dunkley A, Warren FC, Ara R, Abrams K, et al. A systematic review and mixed treatment comparison of pharmacological interventions for the treatment of obesity. Obesity Reviews. 2012;13:483–498. doi: 10.1111/j.1467-789X.2011.00981.x. [DOI] [PubMed] [Google Scholar]
- 6.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 7.Collins S, Cao W, Robidoux J. Learning new tricks from old dogs: beta-adrenergic receptors teach new lessons on firing up adipose tissue metabolism. Molecular Endocrinology. 2004;18:2123–2131. doi: 10.1210/me.2004-0193. [DOI] [PubMed] [Google Scholar]
- 8.Li B, Nolte LA, Ju JS, Han DH, Coleman T, Holloszy JO, et al. Skeletal muscle respiratory uncoupling prevents diet-induced obesity and insulin resistance in mice. Nature Medicine. 2000;6:1115–1120. doi: 10.1038/80450. [DOI] [PubMed] [Google Scholar]
- 9.Lodhi IJ, Yin L, Jensen-Urstad AP, Funai K, Coleman T, Baird JH, et al. Inhibiting adipose tissue lipogenesis reprograms thermogenesis and PPARgamma activation to decrease diet-induced obesity. Cell metabolism. 2012;16:189–201. doi: 10.1016/j.cmet.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young P, Arch JR, Ashwell M. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS Letters. 1984;167:10–14. doi: 10.1016/0014-5793(84)80822-4. [DOI] [PubMed] [Google Scholar]
- 12.Bal NC, Maurya SK, Sopariwala DH, Sahoo SK, Gupta SC, Shaikh SA, et al. Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals. Nature Medicine. 2012;18:1575–1579. doi: 10.1038/nm.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamu D, Bombardier E, Smith IC, Fajardo VA, Tupling AR. Sarcolipin provides a novel muscle-based mechanism for adaptive thermogenesis. Exercise and Sport Sciences Reviews. 2014;42:136–142. doi: 10.1249/JES.0000000000000016. [DOI] [PubMed] [Google Scholar]
- 14.Maurya SK, Bal NC, Sopariwala DH, Pant M, Rowland LA, Shaikh SA, et al. Sarcolipin is a key determinant of basal metabolic rate and its overexpression enhances energy expenditure and resistance against diet induced obesity. The Journal of biological chemistry. 2015 doi: 10.1074/jbc.M115.636878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahoo SK, Shaikh SA, Sopariwala DH, Bal NC, Periasamy M. Sarcolipin protein interaction with sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA) is distinct from phospholamban protein, and only sarcolipin can promote uncoupling of the SERCA pump. The Journal of Biological Chemistry. 2013;288:6881–6889. doi: 10.1074/jbc.M112.436915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bombardier E, Smith IC, Vigna C, Fajardo VA, Tupling AR. Ablation of sarcolipin decreases the energy requirements for Ca2+ transport by sarco(endo)plasmic reticulum Ca2+-ATPases in resting skeletal muscle. FEBS Letters. 2013;587:1687–1692. doi: 10.1016/j.febslet.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 17.Berggren JR, Tanner CJ, Houmard JA. Primary cell cultures in the study of human muscle metabolism. Exercise and Sport Sciences Reviews. 2007;35:56–61. doi: 10.1249/JES.0b013e31803eae63. [DOI] [PubMed] [Google Scholar]
- 18.Muoio DM, Way JM, Tanner CJ, Winegar DA, Kliewer SA, Houmard JA, et al. Peroxisome proliferator-activated receptor-alpha regulates fatty acid utilization in primary human skeletal muscle cells. Diabetes. 2002;51:901–909. doi: 10.2337/diabetes.51.4.901. [DOI] [PubMed] [Google Scholar]
- 19.Hulver MW, Berggren JR, Carper MJ, Miyazaki M, Ntambi JM, Hoffman EP, et al. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metabolism. 2005;2:251–261. doi: 10.1016/j.cmet.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyle KE, Canham JP, Consitt LA, Zheng D, Koves TR, Gavin TP, et al. A high-fat diet elicits differential responses in genes coordinating oxidative metabolism in skeletal muscle of lean and obese individuals. The Journal of Clinical Endocrinology & Metabolism. 2011;96:775–781. doi: 10.1210/jc.2010-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell JA, Reed MA, Consitt LA, Martin OJ, Haynie KR, Hulver MW, et al. Lipid partitioning, incomplete fatty acid oxidation, and insulin signal transduction in primary human muscle cells: effects of severe obesity, fatty acid incubation, and fatty acid translocase/CD36 overexpression. The Journal of Clinical Endocrinology & Metabolism. 2010;95:3400–3410. doi: 10.1210/jc.2009-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consitt LA, Bell JA, Koves TR, Muoio DM, Hulver MW, Haynie KR, et al. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha overexpression increases lipid oxidation in myocytes from extremely obese individuals. Diabetes. 2010;59:1407–1415. doi: 10.2337/db09-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Funai K, Cartee GD. Inhibition of contraction-stimulated AMP-activated protein kinase inhibits contraction-stimulated increases in PAS-TBC1D1 and glucose transport without altering PAS-AS160 in rat skeletal muscle. Diabetes. 2009;58:1096–1104. doi: 10.2337/db08-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Funai K, Song H, Yin L, Lodhi IJ, Wei X, Yoshino J, et al. Muscle lipogenesis balances insulin sensitivity and strength through calcium signaling. The Journal of Clinical Investigation. 2013;123:1229–1240. doi: 10.1172/JCI65726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle KE, Zheng D, Anderson EJ, Neufer PD, Houmard JA. Mitochondrial lipid oxidation is impaired in cultured myotubes from obese humans. International Journal of Obesity. 2012;36:1025–1031. doi: 10.1038/ijo.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher-Wellman KH, Weber TM, Cathey BL, Brophy PM, Gilliam LA, Kane CL, et al. Mitochondrial respiratory capacity and content are normal in young insulin-resistant obese humans. Diabetes. 2014;63:132–141. doi: 10.2337/db13-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Funai K, Semenkovich CF. Skeletal muscle lipid flux: running water carries no poison. American journal of physiology Endocrinology and metabolism. 2011;301:E245–E251. doi: 10.1152/ajpendo.00152.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holloszy JO. "Deficiency" of mitochondria in muscle does not cause insulin resistance. Diabetes. 2013;62:1036–1040. doi: 10.2337/db12-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gustavsson M, Traaseth NJ, Veglia G. Activating and deactivating roles of lipid bilayers on the Ca(2+)-ATPase/phospholamban complex. Biochemistry. 2011;50:10367–10374. doi: 10.1021/bi200759y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore L, Chen T, Knapp HR, Jr, Landon EJ. Energy-dependent calcium sequestration activity in rat liver microsomes. The Journal of Biological Chemistry. 1975;250:4562–4568. [PubMed] [Google Scholar]
- 32.Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, et al. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida Y, Jain SS, McFarlan JT, Snook LA, Chabowski A, Bonen A. Exercise- and training-induced upregulation of skeletal muscle fatty acid oxidation are not solely dependent on mitochondrial machinery and biogenesis. The Journal of Physiology. 2013;591:4415–4426. doi: 10.1113/jphysiol.2012.238451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holloszy JO, Coyle EF. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. The Journal of Applied Physiology. 1984;56:831–838. doi: 10.1152/jappl.1984.56.4.831. [DOI] [PubMed] [Google Scholar]
- 35.Larsen S, Stride N, Hey-Mogensen M, Hansen CN, Andersen JL, Madsbad S, et al. Increased mitochondrial substrate sensitivity in skeletal muscle of patients with type 2 diabetes. Diabetologia. 2011;54:1427–1436. doi: 10.1007/s00125-011-2098-4. [DOI] [PubMed] [Google Scholar]
- 36.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 37.Watt MJ, Hoy AJ. Lipid metabolism in skeletal muscle: generation of adaptive and maladaptive intracellular signals for cellular function. The American Journal of Physiology - Endocrinology and Metabolism. 2012;302:E1315–E1328. doi: 10.1152/ajpendo.00561.2011. [DOI] [PubMed] [Google Scholar]
- 38.Zurlo F, Larson K, Bogardus C, Ravussin E. Skeletal muscle metabolism is a major determinant of resting energy expenditure. The Journal of Clinical Investigation. 1990;86:1423–1427. doi: 10.1172/JCI114857. [DOI] [PMC free article] [PubMed] [Google Scholar]