Abstract
Purpose
Secretory clusterin (sCLU)/apolipoprotein J is an extracellular chaperone that has been functionally implicated in DNA repair, cell cycle regulation, apoptotic cell death, and tumorigenesis. It exerts a prosurvival function against most therapeutic treatments for cancer and is currently an antisense target in clinical trials for tumor therapy. However, the molecular mechanisms underlying its function remained largely unknown.
Experimental Design
The molecular effects of small interfering RNA-mediated sCLU depletion in nonstressed human cancer cells were examined by focusing entirely on the endogenously expressed sCLU protein molecules and combining molecular, biochemical, and microscopic approaches.
Results
We report here that sCLU depletion in nonstressed human cancer cells signals stress that induces p53-dependent growth retardation and high rates of endogenous apoptosis. We discovered that increased apoptosis in sCLU-depleted cells correlates to altered ratios of proapoptotic to antiapoptotic Bcl-2 protein family members, is amplified by p53, and is executed by mitochondrial dysfunction. sCLU depletion-related stress signals originate from several sites, because sCLU is an integral component of not only the secretory pathway but also the nucleocytosolic continuum and mitochondria. In the cytoplasm, sCLU depletion disrupts the Ku70-Bax complex and triggers Bax activation and relocation to mitochondria. We show that sCLU binds and thereby stabilizes the Ku70-Bax protein complex serving as a cytosol retention factor for Bax.
Conclusions
We suggest that elevated sCLU levels may enhance tumorigenesis by interfering with Bax proapoptotic activities and contribute to one of the major characteristics of cancer cells, that is, resistance to apoptosis.
Secretory clusterin (sCLU) in mammals is a heterodimeric glycoprotein that was initially purified from serum and identified as apolipoprotein J or an extracellular chaperone (1, 2). sCLU is an endoplasmic reticulum (ER)-targeted 449-amino acid polypeptide that represents the predominant translation product of the human CLU gene. Proteolytic removal of the ER-targeting signal peptide and glycosylation results in the ER-associated high mannose form of ~60 kDa (sCLUC). Following further processing in trans-Golgi compartments, sCLUC is transformed to the mature secreted heterodimeric sCLU protein form of ~ 75–80 kDa (sCLUS; ref 3). Another CLU protein form of ~ 55 kDa, designated as nCLU, is translated from an alternatively spliced CLU transcript that bypasses the ER signal peptide (4); nCLU exerts a prodeath function in human cancer cells (4, 5).
sCLU has been functionally implicated in DNA repair, cell cycle regulation, and apoptotic cell death, whereas a prominent feature is its differential expression in many severe physiologic disturbance states, including neurodegeneration, vascular damage, diabetes, and tumor formation (6). Although, in certain cellular contexts, sCLU may suppress cellular growth or promote cell death (7, 8), it mostly exerts a prosurvival role during cell death and confers resistance to cytotoxic agents both in vitro and in vivo (6, 9). Consistent with its prosurvival role is the fact that either antisense oligonucleotides targeting CLU gene or CLU gene expression silencing via small interfering RNA (siRNA) resulted in significant cell sensitization to apoptosis induced by oxidative stress (10), chemotherapeutic drugs (11, 12), ionizing radiation (13), or tumor necrosis factor-related apoptosis-inducing ligands (14). In agreement with these findings, a phase I pharmacokinetic and pharmacodynamic study in prostate cancer patients revealed that CLU antisense oligonucleotides improved the efficacy of therapy by inhibiting expression of CLU and enhancing apoptosis; a phase II trial is currently under way (15). Although sCLU is considered as a secreted protein, emerging evidence indicates that it may also localize in the nucleus (16), mitochondria (17), or cytosol (18) of stressed cells. Interestingly, CLU gene knockdown by siRNAs in nonstressed prostate or osteosarcoma human cancer cells reduced cell proliferation and increased cell death responses (12, 19). In support of these findings, dropout screens for short hairpin RNAs that affect cell proliferation and viability identified (among others) CLU as a gene that is selectively required for proliferation and survival of normal human mammary epithelial cells (20). However, the molecular mechanisms underlying these effects remained largely unknown.
In the current report, we directly addressed the implication of intracellular sCLU in cellular homeostasis. We found that sCLU depletion in nonstressed human cancer cells triggers p53-dependent G1-S growth retardation and increased endogenous mitochondrial apoptosis that is amplified by functional p53 via both transcription-dependent and transcription-independent apoptogenic roles. Our subsequent analyses revealed that stress signals in sCLU-depleted cells originate from several sites because sCLU contributes to the structural integrity of the ER, Golgi apparatus, and mitochondria and also localizes in the nucleocytosolic continuum. We discovered that, in the cytoplasm, sCLU binds and thereby stabilizes the Ku70-Bax complex. This interaction, in turn, suppresses Bax activation and its relocation to mitochondria.
Materials and Methods
Cell lines and culture conditions
Human osteosarcoma U-2 OS cells were purchased from the American Tissue Culture Collection and cultured in DMEM (Invitrogen/Life Technologies) as described previously (12).
Design and synthesis of sCLU siRNA oligonucleotides: siRNA transfection
The Cl-I as well as the Scramble-I (Sc-I) and Scramble-II (Sc-II) siRNA oligonucleotides have been described previously (12). To detect sCLU-specific siRNA oligonucleotides that fulfill the criteria previously set (21), we scanned manually the 99-bp region of exon II that is specific for the sCLU transcript (Supplementary Fig. S1A). A candidate sequence identified four nucleotides downstream of the sCLU gene transcription initiation codon and synthesized by Dharmacon Research (sCLU oligonucleotide; see Supplementary Materials and Methods). siRNA transfection was done as described previously (12). For the transfection mixture, 100 nmol/L siRNA duplex was used for 4 h followed by addition of 1:3 (v/v) medium containing 30% fetal bovine serum for 20 h. At 24 h post-transfection, RNA duplexes were removed and cells were grown in siRNA-free medium. All applied controls (see Supplementary Materials and Methods) verified definitive loss-of-function data. Our previous studies (12), as well as genome-wide microarray analyses,6 clearly showed the complete absence of “off-target” siRNA effects.
Stable overexpression of Bcl-2, wild-type p53, V143A–p53, R273H–p53 plasmids in U-2 OS cells
Bcl-2 and wild-type p53 (WT-p53) inserts were cloned into the pcDNA3.1-Myc-His+ mammalian expression vector (Invitrogen/Life Technologies) and the pCB6+ vector, respectively. Human V143A–p53 and R273H–p53 mutants were excised from the pCMV-NEO vector and subcloned into the pcDNA3.1-Myc-His+ vector. Cells were stably transfected with the plasmids as described previously (22). Selected clones were analyzed for transgene expression levels and homogeneity of expression by immunoblotting and immunofluorescence, respectively.
RNA extraction and real-time PCR analysis
Total RNA was extracted using the RNeasy Mini kit (Qiagen) and converted into cDNA with the ImProm-II Reverse Transcription System (Promega). Real-time PCRs were done as described previously (22). Gene expression was analyzed with Gene Expression Macro version 1.1 (Bio-Rad). The sCLU primers used were forward 5′-ATGATGAAGACTCTGCTGCTG-3′ and reverse 5′-TTCCTGGAGCTCATTGTCTG-3′. GAPDH and actin genes were used as normalizers.
Subcellular fractionation, immunoblotting, and quantitative coimmunoprecipitation
Subcellular fractionation in mitochondrial-cytosolic or nuclear-cytoplasmic preparations was done using Mitochondrial Fractionation or Nuclear Extract kits (Active Motif), respectively, according to the manufacturer’s instructions. To isolate cytosol by hypotonic cell disruption, cells were collected in 10 mmol/L EDTA, 150 mmol/L NaCl, and 40 mmol/L Tris-HCl (pH 7.8) and lysed on ice (1 min) in a buffer containing 1 mmol/L EDTA, 2 mmol/L DDT, 10 mmol/L Tris (pH 7.5), and phosphatase and protease inhibitors (Sigma). Samples were immediately cleared at 120,000 × g for 1 h and both pellet (insoluble membrane fraction) and supernatant (cytosol) were collected. Whole-cell lysates were prepared as described previously (22). Samples were adjusted by Bradford (Bio-Rad) and mixed with reducing Laemmli buffer, and proteins were fractionated by SDS-PAGE followed by immunoblotting (22).
For quantitative coimmunoprecipitation, cells were harvested and resuspended in 1% CHAPS lysis buffer [10 mmol/L HEPES, 150 mmol/L NaCl, 1% CHAPS (pH 7.4)] containing phosphatase and protease inhibitors. Following extraction for 1 h at 4°C, cell lysates or isolated subcellular fractions were precleared with IgG and protein G or protein A Sepharose (Amersham Life Science). Each preparation (250 µg) was then incubated with the primary antibody for 2 h at 4°C followed by addition of the Sepharose beads and overnight incubation at 4°C. The immunoprecipitated complexes were washed and dissolved in reducing or nonreducing Laemmli buffer.
The polyclonal (sc-6419) and monoclonal (sc-5289) antibodies against human sCLU were purchased from Santa Cruz Biotechnology. Both have been exhaustively characterized for their specificity; as shown in Fig. 1A2, within a molecular weight range of ~ 150 to 30 kDa, the sc-6419 antibody cross-reacts with only the sCLUC and sCLUS polypeptides. Antibodies against actin (sc-1616), Bcl-2 (sc-509), cytochrome c (sc-13156), GAPDH (sc-25778), Ku70 (sc-5309 and sc-1487), Ku80 (sc-1485), lamin A/C (sc-7292), p21 (sc-817), p53 (sc-126), poly(ADP-ribose) polymerase (PARP; sc-7150), and proliferating cell nuclear antigen (sc-9857) were purchased from Santa Cruz Biotechnology. Antibodies against Bax (#554104) and OxPhos complex IV (A-21347) were from BD Pharmingen and Invitrogen, respectively. The Bax6A7 monoclonal antibody was a generous gift of Dr. R.J. Youle. Secondary antibodies for immunoprecipitation or immunoblotting were purchased from Jackson ImmunoResearch.
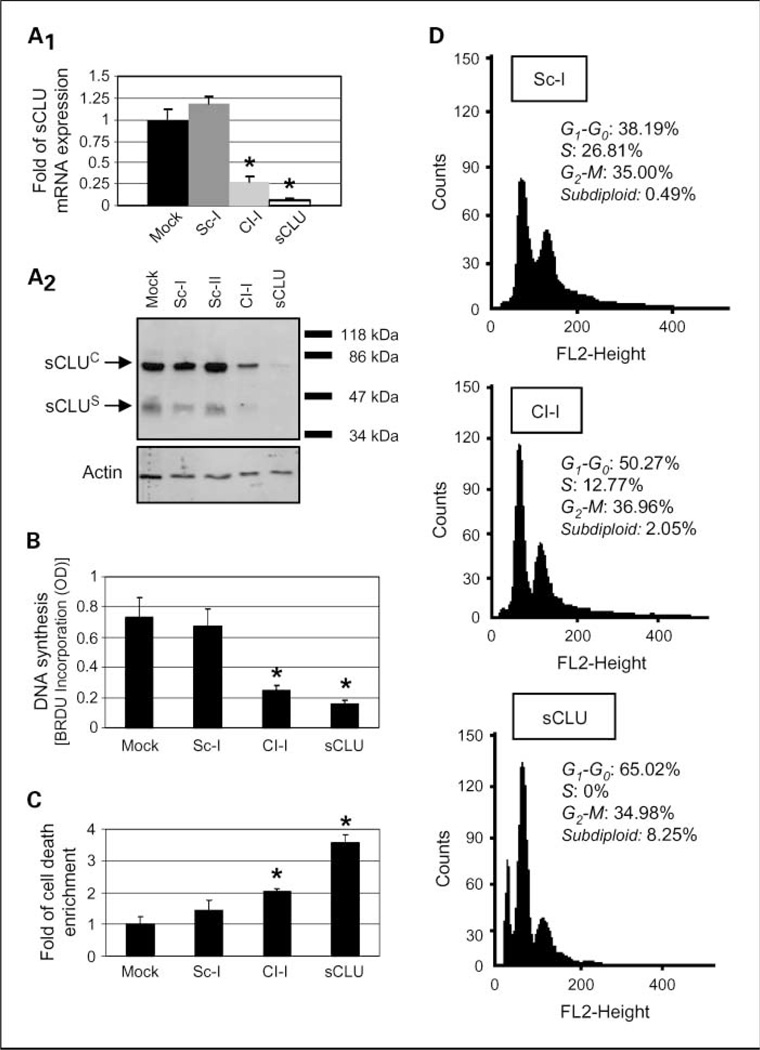
Fig. 1.
sCLU depletion in U-2 OS cells induces growth arrest and increased cell death responses. A1 quantitative PCR analyses (n = 3) of sCLU mRNA levels after cell transfection with indicated siRNA oligonucleotides. A2, immunoblot analyses of sCLU protein levels in whole-cell lysates following transfection with shown siRNAs. Actin probing was used as reference for equal protein loading; molecular weight markers are shown to the right of the blot. B, DNA synthesis measurement (n = 2) after bromodeoxyuridine incorporation. C, cumulative cell death quantitation (n = 3) of DNA fragments following cell transfection with the indicated siRNAs. D, comparative fluorescence-activated cell sorting analysis (representative of two independent experiments) of control (Sc-I), Cl-I, or sCLU siRNA-treated cells. Assays were done 72 h after siRNA cell transfection. Bars, SD. *, P < 0.05.
Cell growth, DNA synthesis, quantification of cell death, and cell cycle analysis
Cell growth was assayed by measuring total cell number with a Coulter counter (Coulter Counter Z). DNA synthesis was measured with a Cell Proliferation ELISA bromodeoxyuridine colorimetric immunoassay method (Roche) according to the manufacturer’s instructions after labeling cells with bromodeoxyuridine for 3 h. For the quantitative analysis of the cytoplasmic (ongoing cell death) or the tissue culture supernatant (cumulative cell death) histone-associated DNA fragments, the Cell Death Detection ELISAPLUS photometric enzyme immunoassay method (Roche) was used as instructed by the manufacturer. Cell death was expressed as “fold of cell death enrichment” and was representative of the biological outcome of the assay. Flow cytometry was performed in a FACS-Calibur flow cytometer as described previously (22).
Caspase inhibitors: measurement of caspase-8 and caspase-9 activity
The pan-caspase (z-VAD-FMK; Biomol) inhibitor was used at a concentration of 50 µmol/L. All other inhibitors [caspase-2 (Z-VDVAD-FMK), caspase-8 (Z-IETD-FMK), caspase-10 (Z-AEVD-FMK), caspase-1 (Z-YVAD-FMK), caspase-9 (Z-LEHD-FMK), caspase-3/7 (Z-DEVD-FMK), and caspase-6 (Z-VEID-FMK)] were from Santa Cruz Biotechnology or Alexis Biochemical and were used at 20 µmol/L concentration. Cells were preincubated with each caspase inhibitor or the solvent (DMSO) for 24 h before siRNA transfection; a fresh amount was added every 24 h to avoid inhibitor inactivation. To measure caspase-8 and caspase-9 activity, 200 µg cell lysates were incubated with the IETD-AFC (50 µmol/L; Santa Cruz Biotechnology) or LEHD-AMC (250 µmol/L; BD Biosciences) substrates, respectively, as suggested by the manufacturer. Emitted fluorescence was measured in a VersaFluor fluorometer (Bio-Rad).
Immunofluorescence staining and confocal laser scanning microscopy
Cell fixation and permeabilization for immunofluorescence was done as described previously (12). For double- or triple-antigen costaining, the primary antibodies (always derived from different species) were mixed and incubated simultaneously on each coverslip for 1 h at room temperature. Affinity-purified, preabsorbed secondary antibodies were applied to coverslips for 1 h at room temperature. Antibodies were obtained from Jackson ImmunoResearch and were conjugated with AMCA (blue), FITCH (green), or Texas red (deep red). Nuclei were stained with 300 nmol/L 4’,6-diamidino-2-phenylindole dihydrochloride (Molecular Probes). Mitochondria were stained by incubating live cells for 30 min at 37°C with 250 nmol/L MitoTracker Deep Red 633 (Molecular Probes). Images of the mounted coverslips were taken by using a Leica TCS SP5 confocal microscope and represent stacks of >10 (~ 1 µm thick) z-sections. Applied controls (12) were negative.
Immunogold localization and transmission electron microscopy
For transmission electron microscopy (TEM) immunogold localization, cells were fixed in 2% formaldehyde (Polysciences), 0.5% glutaraldehyde (Taab Lab) in PBS (pH 7.2) for 1 h at 4°C, dehydrated, and embedded in Unicryl resin (British Biocell International). Thin sections were cut to silver interference colors, floated onto 400 mesh nickel grids (Pelco), blocked with 3% fatty acid-free bovine serum albumin in PBS, and probed with the sCLU antibody. Immunoglobulins or protein A conjugated to 10 or 15 nm gold particles were used to detect primary antibody. Sections were counterstained with 7% uranyl acetate in distilled H2O and examined on a Phillips EM 300 electron microscope operating at 80 kV. Identification of each organelle was based on its morphology, topology, and dimensions. Controls were as for confocal laser scanning microscopy (CLSM) immunofluorescence and were negative.
Software used: statistical analysis
Quantitation of band density in blots was done with the ImageJ 1.38x software7 (NIH). Antigen colocalization in the CLSM micrographs was done by using the Volocity software (Improvision) as described previously (23). Briefly, images were cleared from noise and photobleaching effects and adjusted for brightness and contrast. Threshold levels were set from at least five stained cells. Pixel intensity for each of the two fluorochromes was obtained from >50 cells and plotted in a x–y diagram. Only those pixels that codistribute in the diagonal of the graph and represent the highest values for Pearson’s correlation and overlap coefficient R were selected for calculation of the colocalization coefficients Mx, My. Images are shown as raw data after brightness and contrast adjustment with Adobe Photoshop 6.0. Assays were normalized against their respective controls arbitrarily set to 1 or to 100. Each data point in the graphs corresponds to the mean of the independent experiments; error bars denote SD. Statistical significance was evaluated using the one-way AN OVA. Results at P < 0.05 are shown by an asterisk.
Results
Phenotypic effects induced in U-2 OS cells after sCLU depletion positively correlate with the effectiveness of sCLU gene silencing
We investigated the biological role of intracellular sCLU in the regulation of human cancer cell homeostasis by analyzing the phenotypic effects induced in p53+/+ U-2 OS cells following sCLU depletion by siRNA methodology. To specifically knock down sCLU, we designed siRNA oligonucleotides that specifically targeted exon II of the CLU gene (Supplementary Fig. S1A). sCLU-specific siRNA oligonucleotides (designated as sCLU) were ~ 6- to 8-fold more effective than previously used Cl-I oligonucleotide (12) at knocking down either the sCLU mRNA (Fig. 1A1) or the sCLUC and sCLUS protein isoforms (Fig. 1A2; Supplementary Fig. S1B). The recorded phenotypic effects following sCLU knockdown correlated positively to the effectiveness of sCLU gene silencing. More specifically sCLU depletion induced morphologic alterations, including cell shrinkage, detachment from the substrate, and a significant reduction in cell growth (Supplementary Fig. S1C and S1D). These effects relate to a ~ 5-fold decrease in DNA synthesis (Fig. 1B) and a ~4-fold increase in endogenous cell death (Fig. 1C). In support, fluorescence-activated cell sorting analyses revealed an increase in the subdiploid fraction that was accompanied by a significant decrease of S-phase cells (Fig. 1D).
sCLU depletion activates p53 and alters the ratio of proapoptotic to antiapoptotic Bcl-2 protein family members, triggering mitochondrial dysfunction and apoptosis
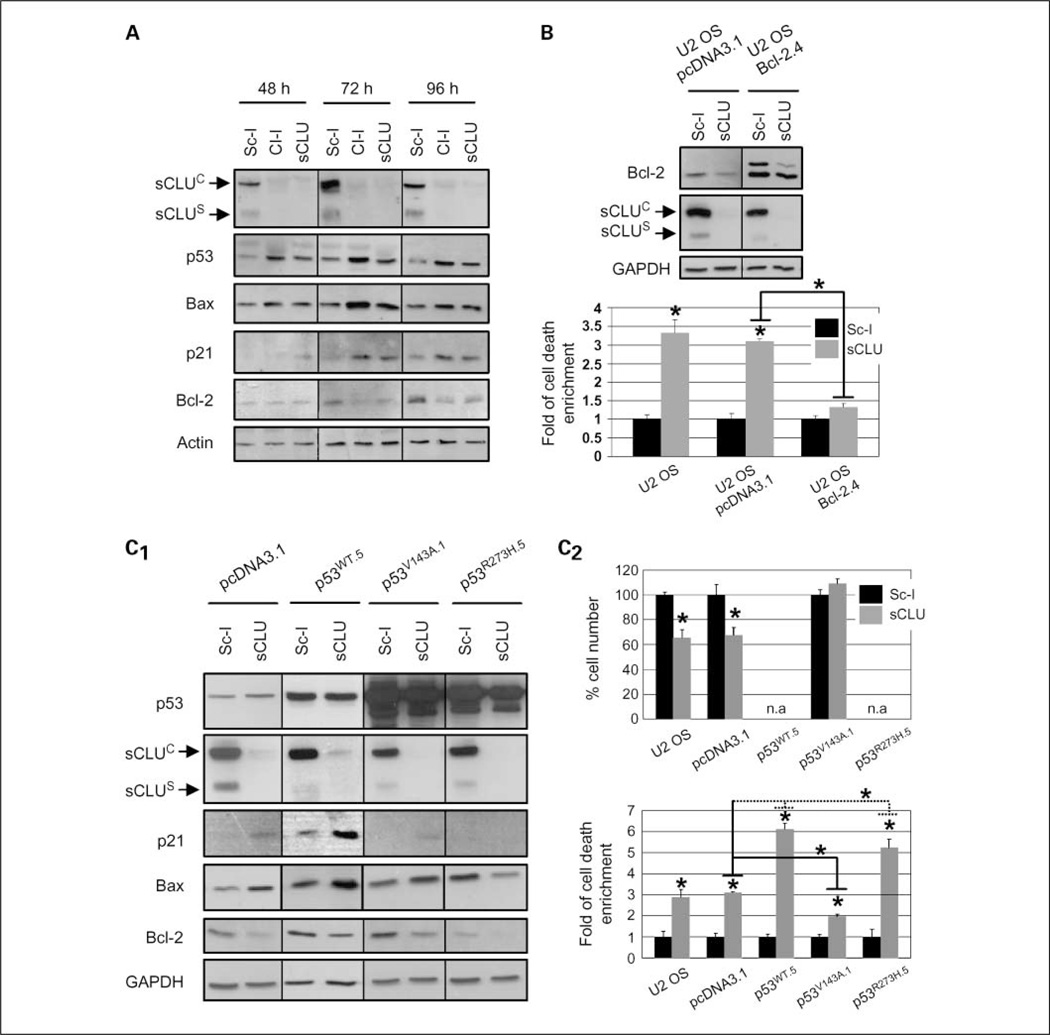
By studying the kinetics and time where sCLU depletion-mediated cell death peaks, in both adherent cells (ongoing cell death) and cell culture supernatant (cumulative death), we found that cells started dying 48 h post-siRNA transfection and cell death levels peaked at 72 h when the siRNA effect was maximized (Supplementary Fig. S2A). To define components of the cellular machinery affected by sCLU depletion, we assayed for expression levels of several proteins that regulate cell growth or death. Immunoblot analyses of cell lysates verified effective sCLU depletion 48 to 96 h post-siRNA transfection and revealed significant accumulation of p53, Bax, and p21Cip1/WAF1 (hereafter p21) proteins along with Bcl-2 down-regulation (Fig. 2A). In addition, sCLU depletion promoted Bcl-XL down-regulation but had no affect on expression levels of the DNA double-strand break repair protein Ku70 or heat shock protein 27, whereas real-time PCR analysis revealed that p53 accumulation after sCLU knockdown did not occur due to mRNA up-regulation and it is thus a consequence of p53 protein stabilization (data not shown). Interestingly, p21 up-regulation was mostly enhanced after moderate (Cl-I mediated; see Fig. 1A) sCLU knockdown (Fig. 2A). The correlation between moderate sCLU gene silencing and p21 up-regulation was supported by titrating the Cl-I and sCLU siRNA concentration. We found that inefficient sCLU depletion resulted in decreased cell death and p21 up-regulation (Supplementary Fig. S2B). To address the functional role of Bcl-2 after sCLU depletion, we stably overexpressed the human Bcl-2 gene in the U-2 OS cells (Fig. 2B, top) and found that Bcl-2 overexpression inhibited cell death (Fig. 2B, bottom). Considering that most proteins found to be differentially regulated after sCLU knockdown are transcriptional targets of p53, we sought to investigate the role of p53 on the molecular effects induced after sCLU depletion in human cancer cells. To this end, we stably overexpressed WT-p53 as well as the highly destabilized structural V143A and DNA contact R273H dominant-negative p53 mutants (24) in U-2 OS cells. Immunoblot analyses revealed that Bcl-2 suppression after sCLU depletion is p53 independent (Fig. 2C1). Interestingly, phenotypic analyses revealed that the V143A–p53 mutant, by suppressing p21 activation, rescued cells from sCLU depletion-mediated growth arrest (Fig. 2C2, top) and, despite residual Bax activation (Fig. 2C1), partially inhibited cell death (Fig. 2C2, bottom). On the other hand, both the WT-p53 and the R273H–p53 mutant augmented sCLU depletion-mediated cell death by ~ 2-fold (Fig. 2C2, bottom). Considering that the highly destabilized V143A mutant unfolds >10 times faster than the WT-p53 core domain, whereas the R273H mutant, which exhibits no DNA-binding activity, has roughly the same half-life as the WT-p53 and retains ~ 98% of the wild-type conformation (24), we concluded that p53 also exerts a direct, transcription-independent, apoptogenic function after sCLU depletion. Similarly to U-2 OS cells, effective sCLU knockdown in other p53+/+ breast (MCF-7; Supplementary Fig. S2C) and lung (A549; Supplementary Fig. S2D) cancer cell lines resulted in a ~ 3-fold increase in spontaneous cell death responses. Knockdown of p53 expression by siRNA in MCF-7 cells could only minimally suppress cell death, which was, however, completely inhibited by a pan-caspase inhibitor (Supplementary Fig. S2C). Next, we asked whether sCLU depletion in a p53−/− cellular context of ovarian and osteosarcoma SKOV-3 and Sa OS cell lines, respectively, could elicit similar responses. We noted that sCLU knockdown in SKOV-3 cells significantly increased spontaneous cell death (Supplementary Fig. S3A). Analyses of the sCLU depletion-related effects in Sa OS cells revealed the down-regulation of the Bcl-2 protein (Supplementary Fig. S3B) as well as moderate enhancement of spontaneous cell death by ~ 1.5- to 2-fold and minimal effects on cell proliferation (Supplementary Fig. S3C). Thus, p53 was not necessary in sCLU knockdown-induced apoptosis but could amplify the cell death responses noted. Consistently, reinforced expression of p53 in tetracycline-inducible p53 wild-type Sa OS cell lines significantly enhanced apoptosis to levels similar to those found in U-2 OS cells (Supplementary Fig. S3D).
Fig. 2.
Down-regulation of Bcl-2 family prosurvival proteins and p53 activation are essential for cell death and growth retardation, respectively, in sCLU-depleted U-2 OS cells. A, immunoblot analyses of whole-cell lysates 48 to 96 h post-transfection with the shown siRNAs. B, whole-cell lysate immunoblot analyses of empty vector (pcDNA3.1) or Bcl-2 stably transfected cells (top) and quantitation of cumulative cell death (bottom; n = 3) in transgenic cells after treatment with either Sc-I or sCLU siRNAs. C, immunoblot analyses of cells stably transfected with vector (pcDNA3.1), WT-p53 (p53WT.5), V143A-p53 (p53v143A.1), and R273H-p53 (p53R273H.5) plasmids (C1) as well as measurement (n = 3) of cellular growth (C2, top) and cumulative cell death (C2, bottom) in the corresponding cell lines after transfection with the indicated siRNAs. All p53WT stable clones showed moderate transgene expression (p53WT.5; C1, top) due to prodeath effect of high p53WT levels. Growth was not assayed (n.a.) in the sCLU-depleted U-2 OS-p53WT.5 and U-2 OS-p53R273H.5 cells due to high death rates. Assays were done 72 h after siRNA transfection unless otherwise indicated. Actin or GAPDH probing was used as protein loading controls. Bars, SD. *, P < 0.05, compared with respective controls.
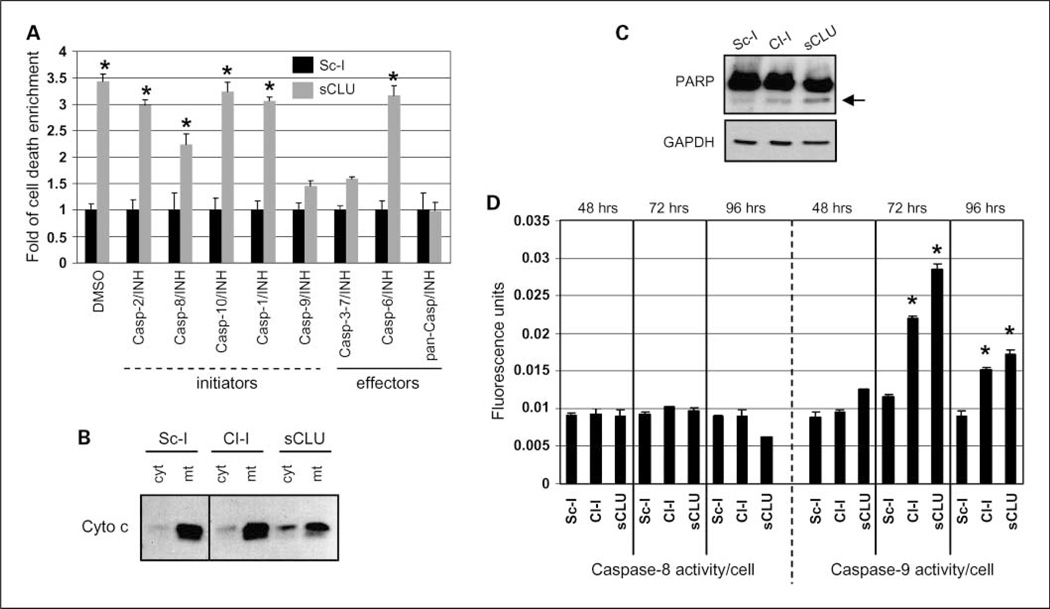
To identify components of the cell death machinery engaged in U-2 OS cells after sCLU depletion, we used a panel of inhibitors targeting most of the known initiator and effector caspases. We showed that, as in MCF-7 cells, a pan-caspase inhibitor completely abolished sCLU depletion-mediated cell death (Fig. 3A). Furthermore, caspase-9 and caspase-3/7 inhibitors also suppressed cell death (Fig. 3A). Thus, we postulated that sCLU depletion induces intrinsic caspase-mediated apoptosis via the mitochondrial apoptotic pathway. This assumption was further verified by showing cytochrome c release from mitochondria (Fig. 3B), PARP cleavage (Fig. 3C), and caspase-9, but not caspase-8, activation (Fig. 3D) in cells depleted of sCLU. Activation of the mitochondrial apoptotic pathway did not solely depend on p53 activity, because sCLU depletion in p53-null Sa OS cells also promoted cytochrome c release in the cytosol as well as caspase-9 activation (Supplementary Fig. S3E).
Fig. 3.
Induction of the mitochondrial pathway of apoptosis in sCLU-depleted U-2 OS cells. A, cumulative cell death measurement (n = 3) after cell transfection with the Sc-I or sCLU siRNAs in the presence of the indicated caspase inhibitors. B, immunoblotting of cytochrome c in cytosolic and mitochondrial cellular fractions following cell transfection with shown siRNAs (for fraction purity, see Fig. 5A). C, immunoblot analysis of control or sCLU-depleted whole-cell lysates probed with antibodies against PARP. Arrow, cleaved 85-kDa apoptotic fragment of PARP. GAPDH probing was used as a protein loading control. D, caspase-8 and caspase-9 activity measurement (n = 2) after incubating whole-cell lysates of sCLU-depleted cells with the caspase-8, IETD-AFC, or the caspase-9, LEHD-AMC, substrates, respectively. Assays were done 72 h after siRNA transfection unless otherwise indicated. Bars, SD. *, P < 0.05.
Intracellular sCLU localizes to the ER-Golgi compartments, mitochondria, and the nucleocytosolic continuum
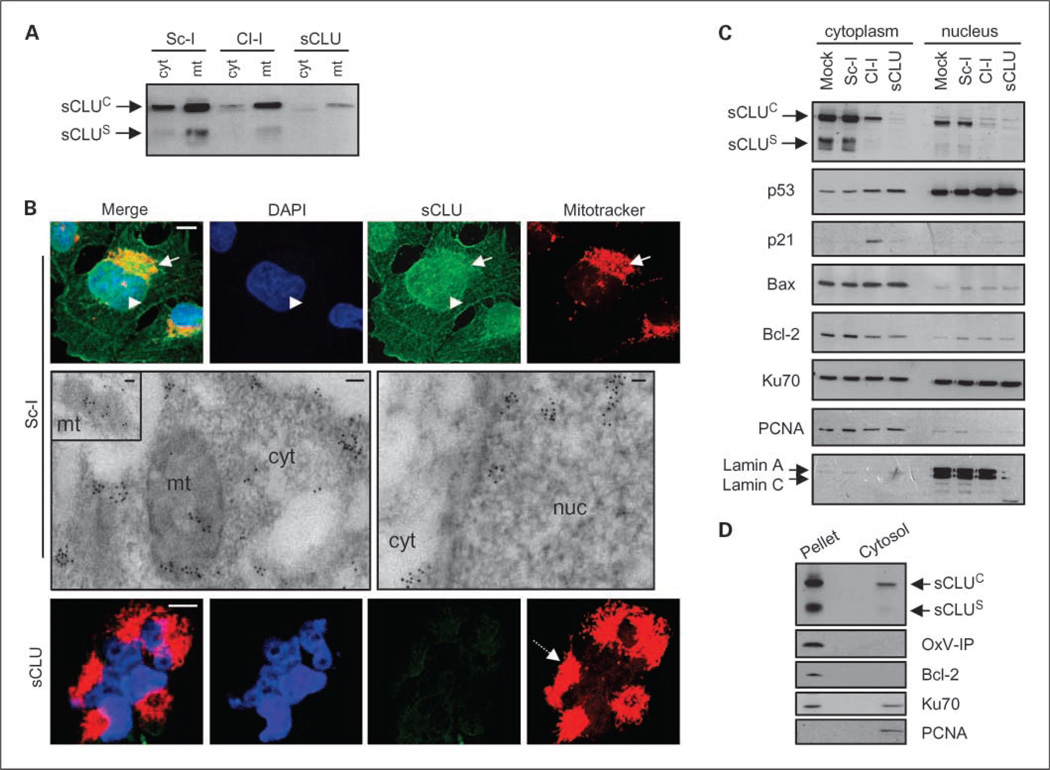
Activation of p53 after sCLU depletion in U-2 OS cells indicated that intracellular sCLU was essential for the integrity of cellular homeostasis and suppressed responses to stress. To identify cellular sites that signal stress after sCLU depletion, we examined endogenous sCLU subcellular localization. As expected, a significant amount of intracellular sCLU was immunolocalized by either CLSM immunofluorescence or TEM immunogold analyses in the membranous secretory cellular compartments, that is, ER Golgi apparatus, and secretory vesicles of U-2 OS (Supplementary Fig. S4A1 and S4B) and Sa OS (data not shown) cells. In both ER and Golgi, sCLU depletion resulted in the formation of foam-like disrupted membranes. The structural abnormalities observed in the ER after sCLU knockdown were sufficient to induce up-regulation of the ER stress marker GRP78 that precedes the unfolded protein response (Supplementary Fig. S4A2). Interestingly, not all intracellular sCLU colocalized with PDI and Golgin-97, ER and Golgi protein markers, respectively. Therefore, we examined in detail the intracellular distribution of endogenous sCLU by cross-verifying subcellular fractionation biochemical techniques and CLSM immunofluorescence or TEM immunogold endogenous sCLU in situ localization. sCLU copurified with the mitochondrial fraction (Fig. 4A) and coimmunolocalized with mitochondria in intact cells after either CLSM immunofluorescence labeling or TEM immunogold localization (Fig. 4B). These findings corroborate a recent report showing that sCLU is a component of mitochondria in fibrosarcoma HT1080 cancer cells (17). Notably, sCLU depletion resulted in mitochondria condensation and aggregation (Fig. 4B). In addition, sCLU copurified with cytosolic (Fig. 4A) and nuclear (Fig. 4C) fractions. Parallel analyses of the subcellular distribution of p53, p21, Bax, Bcl-2, and Ku70 proteins in control and sCLU-depleted U-2 OS cells indicated that, apart from the effects in their differential expression, sCLU depletion did not affect their trafficking between cytoplasm and nucleus. The nuclear and cytosolic distribution of sCLU was further supported by CLSM and TEM in situ immunolocalization (Fig. 4B). To further exclude the possibility that sCLU-containing membranes contaminated our cytosolic biochemical preparations, we isolated cytosol by hypotonic cell lysis. Consistent with our previous findings, a small amount of sCLU was coextracted with the cytosolic fraction (Fig. 4D). Similarly, sCLU was localized in both mitochondria and the nucleocytosolic continuum of Sa OS cells; as in U-2 OS cells, sCLU depletion resulted in mitochondria aggregation (data not shown). To unequivocally verify that sCLU distributes in the cytosol of healthy human cells, we assayed sCLU subcellular localization in an organelle-free cell type, that is, mature RBC; our prior analyses have revealed the complete absence of GRP78 (an ER marker) immunoreactivity in RBCs.6 By combining RBC membrane and cytosol isolation or by TEM immunogold localization, we found that sCLU was a component of mature human RBCs and localized in both the membrane and the cytosol (Supplementary Fig. S4C).
Fig. 4.
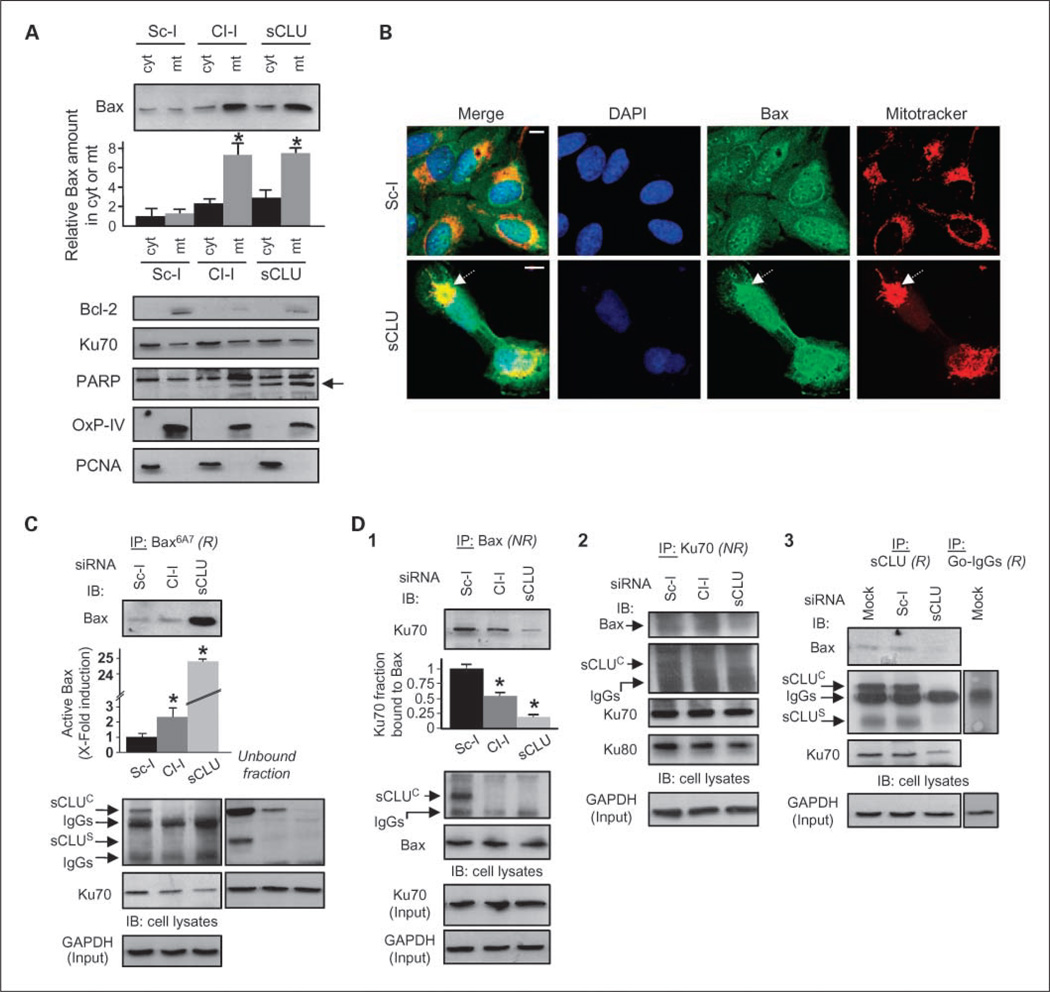
Endogenous sCLU localizes in the mitochondria and the nucleocytosolic continuum of U-2 OS cells. Immunoblot analyses of isolated cytosolic (cyt; A), mitochondrial (mt; A), and cytoplasmic and nuclear fractions (C) probed with an anti-sCLU antibody. Blots in C were also probed with antibodies against p53, p21, Bax, Bcl-2, and Ku70. Fraction purity in A was verified by anti-OxPhos complex IV (OxP-IV) probing (mitochondrial marker; shown in Fig. 5A), and in C by blot probing with anti-proliferating cell nuclear antigen (exclusively cytosolic in osteosarcoma cells) and anti-lamin A/C (nuclear marker). B, CLSM immunofluorescence (top and bottom) or TEM immunogold (middle) localization of sCLU protein in control and sCLU knocked-down cells. For CLSM immunolocalization, cells were costained with an anti-sCLU (green), MitoTracker (mitochondria specific stain; red), and 4’,6-diamidino-2-phenylindole dihydrochloride (blue). Captured images were merged to reveal codistribution sites (yellow for green-red and cyan for green-blue). Top, arrows, mitochondria; arrowheads, nuclei. Bottom, dashed arrow, aggregated mitochondria in sCLU-depleted cells. Middle, sCLU-related gold particles after immunogold TEM localization were found in mitochondria (mt; see also inset), cytosol (cyt), and nucleus (nuc). Bar, 10 µm (CLSM) and 100 nm (TEM). D, immunoblotting of nonextractable pelleted material and the soluble cytosol after hypotonic lysis of control cells. Blots were probed with antibodies against sCLU, OxPhos complex IV, Bcl-2 (membrane bound), Ku70 (nuclear and cytosolic distribution), and proliferating cell nuclear antigen. All assays were done 72 h post-siRNA transfection.
sCLU suppresses Bax activation and relocation to mitochondria by stabilizing the cytoplasmic Ku70-Bax interaction
Our finding that sCLU is distributed in the mitochondria and nucleocytosolic continuum of human cells, and that sCLU depletion can trigger apoptosis in a p53-independent manner, prompted us to ask whether intracellular sCLU modulates the stability of certain protein complexes involved in the regulation of mitochondrial apoptosis. As shown in Fig. 5A, cell fractionation revealed a ~ 7-fold increase in Bax levels that copurified with mitochondria after sCLU knockdown in U-2 OS cells. Also, sCLU depletion induced higher rates of p53 (Supplementary Fig. S5A), but not Bcl-2, Ku70, or PARP, accumulation in mitochondria (Fig. 5A). These observations were verified by CLSM immunofluorescence analyses that revealed massive colocalization of Bax (Fig. 5B; see also Fig. 6A), and to a lesser extent p53 (Supplementary Fig. S5B), with mitochondrial staining in sCLU-depleted cells. The fact that p53 relocates to mitochondria of sCLU-depleted cells provides further evidence for a direct, transcription-independent, p53 apoptogenic role in sCLU-depleted human cells. Supportively, the R273H–p53 mutant, which lacks DNA-binding activity but is structurally stable (24), accumulates in the mitochondria (data not shown) and induces cell death in sCLU-depleted cells as effectively as the WT-p53 (see Fig. 2C2). These observations are consistent with recent studies, which indicated that p53 elicits nontranscriptional Bax-dependent prodeath activities either in the cytosol or directly at the mitochondria (25). The possibility that higher levels of Bax found in U-2 OS mitochondria after sCLU depletion solely originated from p53-dependent transcriptional de novo production of Bax was excluded by directly showing endogenous Bax, but not Ku70, relocation to mitochondria in p53-null Sa OS cells (Supplementary Fig. S6A); interestingly, sCLU depletion had no effect on Bax nuclear localization (Fig. 5B; Supplementary Fig. S6A2). As Bax is kept in its inert cytoplasmic conformation by binding to Ku70 (26), we examined whether sCLU depletion destabilized the endogenous Ku70-Bax protein complex, resulting in Bax activation and translocation to mitochondria. We used the anti-Bax monoclonal antibody clone 6A7 (Bax6A7 antibody), which specifically detected the activated form of Bax (27), to immuno-precipitate endogenous Bax in control and sCLU-depleted U-2 OS cells. Moderate sCLU knockdown (Cl-I siRNA) or effective sCLU depletion (sCLU siRNA) induced ~2- or ~ 25-fold increase, respectively, in the level of Bax immunoprecipitated with Bax6A7 (Fig. 5C). Interestingly, even in the absence of apoptosis (Sc-I-treated cells), the Bax6A7 antibody immunoprecipitated residual Bax levels together with small amounts of endogenous sCLUC and Ku70 proteins (Fig. 5C). Supportively, minimal amounts of sCLUC were previously noted in the cytosol of healthy PC-3M prostate cancer cells; in these cells, the Bax6A7 monoclonal antibody coimmunoprecipitated Bax and sCLUC (17). Concomitant with the high amount of mitochondrial-derived immunoprecipitated Bax in sCLU-depleted cells, Bax-Ku70 complexes were diminished. These data suggested that Ku70 cannot bind active Bax and that the Bax molecules immunoprecipitated in control cells by the Bax6A7 antibody most likely represent inactive Bax.
Fig. 5.
sCLU knockdown in U-2 OS cells promotes Bax activation and translocation to the mitochondria due to destabilization of the cytoplasmic Ku70-Bax complex. A, immunoblot analyses of Bax, Bcl-2, Ku70, and PARP in isolated cytosolic and mitochondrial fractions of control or sCLU knocked-down cells; arrow in PARP immunoblot denotes the apoptotic 85-kDa PARP fragment. Second from top, quantitation (n = 2) of Bax copurification with cytosol or mitochondria. Fraction purity was verified by probing with anti-OxPhos Complex IV and anti-proliferating cell nuclear antigen. B, CLSM immunolocalization of Bax after staining control or sCLU-depleted cells with anti-Bax, MitoTracker, and 4’,6-diamidino-2-phenylindole dihydrochloride. The captured images were merged to reveal antigen colocalization with mitochondria (dashed arrows, bottom). Bar, 10 µm. C and D, quantitative immunoprecipitation analyses of the sCLU, Ku70, and Bax proteins interaction in control and sCLU knocked-down cells. Cells were lysed in CHAPS and lysates were immunoprecipitated (IP; R, reducing conditions; NR, nonreducing conditions) with antibodies against activated Bax (antibody Bax6A7; C), total Bax (D1), Ku70 (D2), and sCLU (D3); the immunoprecipitation with goat IgG (D3) was used to exclude sCLU binding to IgG. Immunoprecipitates were probed (IB) with anti-Bax, anti-sCLU, anti-Ku70, and anti-Ku80. The unbound fraction shown in C depicts the sCLU protein molecules that were not coimmunoprecipitated with the anti-Bax6A7-bead complexes. Quantitative analysis of either Bax activation or Ku70-Bax dissociation (n = 2) after sCLU depletion is shown in C and D1, respectively (second from top). GAPDH or Ku70 probing was used to show equal protein input in the immunoprecipitations. Assays were done 72 h post-transfection with the indicated siRNAs. Bars, SD. *,P <0.05.
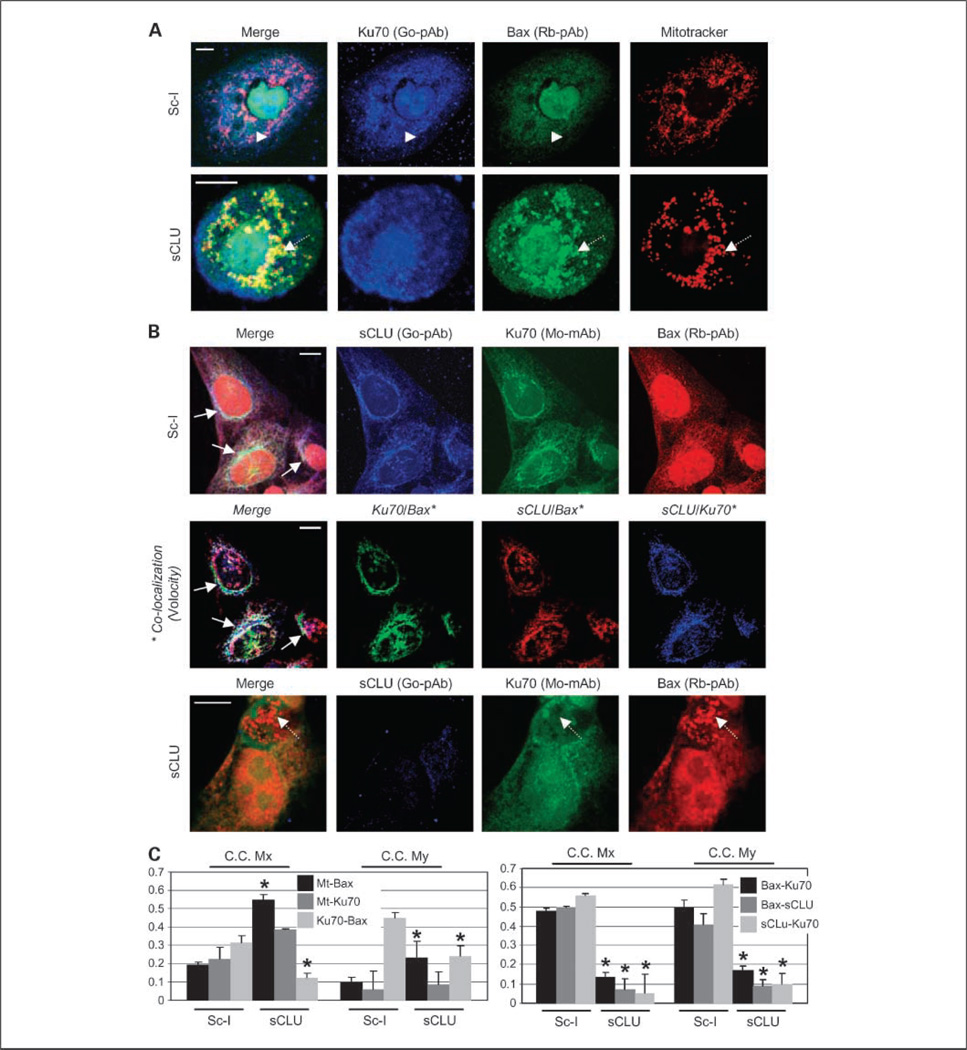
Fig. 6.
sCLU depletion-mediated mitochondrial relocation of Bax in U-2 OS cells relates to the disruption of a cytoplasmic sCLU-Ku70-Bax nexus. A, CLSM staining of control and sCLU-depleted cells with anti-Ku70 (goat polyclonal antibody; blue), anti-Bax (rabbit polyclonal antibody; green), and MitoTracker. Captured images were merged to reveal codistribution sites (yellow for green-red, cyan for green-blue, magenta for red-blue, and white for blue-green-red). Top, arrowheads, cytoplasmic colocalization of Ku70 and Bax in Sc-l-treated cells; bottom, dashed arrows, Bax colocalization with mitochondria after sCLU depletion. B, triple staining of Sc-I or sCLU siRNA-transfected cells with anti-sCLU (goat polyclonal antibody; blue), anti-Ku70 (mouse monoclonal antibody; green), and anti-Bax (rabbit polyclonal antibody; red) antibodies. Middle, digitations of each pair of antigen colocalization in control cells and subsequent superimposition of the three images; the analysis was done by using the Volocity software. Top and middle, arrows, antigen colocalization in the perinuclear cytoplasmic region of control cells; bottom, dashed arrows, elimination of Ku70 and Bax colocalization in sCLU-depleted cells. Assays were done 72 h post-siRNA transfection. Bars, 10 µm (A and B). C, CLSM-derived quantitative analyses of fluorochromes colocalization in preparations of Ku70-Bax-mitochondria (left) and sCLU-Ku70-Bax (right) triple staining. Data were deduced by the Volocity software and values in the graphs indicate the overlapping degree of the two fluorochomes; maximum overlap was set to 1. C.C. Mx and C.C. My are the colocalization coefficients Mx and My (23) and refer to the first and second fluorochromes analyzed, respectively. Thus, for example, the C.C. Mx value for the Ku70-Bax pair in the left denotes the percentage of stained Ku70 that colocalizes with Bax, whereas C.C. My value indicates the percentage of stained Bax that colocalizes with Ku70. Average values of 10 micrographs containing >50 cells are presented in the graphs. Bars, SD. *, P < 0.05.
We hypothesized that sCLU depletion disrupts the Ku70-Bax interaction (rendering Bax in its active state) because sCLU binds to the cytoplasmic Ku70-Bax complex and serves as a stabilizer. Therefore, we immunoprecipitated endogenous Bax, Ku70, and sCLU proteins in U-2 OS cell extracts and assayed for the other two components of the putative trimeric complex. As shown in Fig. 5D1 to D3, antibodies specific for Bax, Ku70, or sCLU coimmunoprecipitated the other two components of the putative sCLU-Ku70-Bax complex. Our immunoprecipitation data confirmed the dissociation of Bax from Ku70 (Fig. 5D1 and D2), but not Ku80 from Ku70 (Fig. 5D2), in sCLU-depleted cells, because <25% Ku70 remained bound to Bax after sCLU depletion (Fig. 5D1). These effects were not cell type specific because Bax was found to interact with both sCLU and Ku70 proteins and was activated by ~ 2.5-fold after sCLU depletion in Sa OS cells (Supplementary Fig. S6B). By investigating the sCLU-Ku70-Bax nexus stability during chemotherapeutic drug-induced mitochondrial apoptosis, we verified the cytoplasmic (and partial nuclear) interaction of sCLU, Ku70, and Bax proteins in nonstressed cells and showed that, during apoptosis, the Ku70-Bax, and to a lesser extent the sCLU-Bax, cytoplasmic interactions were disrupted (Supplementary Fig. S7). Moreover, under these experimental conditions, sCLU binds Bax mostly in the cytoplasm, whereas sCLU-Ku70 and Ku70-Ku80 interactions increased in drug-treated cells; neither Bax nor sCLU bound Ku80.
Ku70-Bax complex disruption and subsequent Bax translocation to mitochondria in sCLU-depleted OS cells was further confirmed by CLSM double immunolocalization of Ku70 and Bax and parallel mitochondria staining. In both U-2 OS (Fig. 6A) and Sa OS cells (Supplementary Fig. S8A), sCLU depletion induced massive relocation of Bax to mitochondria. In agreement with our biochemical data, CLSM-derived quantitative analyses of Bax and Ku70 colocalization with mitochondria revealed a significant increase in the amount of Bax, but not Ku70, which colocalized with mitochondria in sCLU-depleted cells (Fig. 6C; Supplementary Fig. S8C, left). Finally, to directly show sCLU-Ku70-Bax colocalization and identify the subcellular distribution of this trimeric nexus, we applied simultaneous CLSM coimmunolocalization of all three endogenous antigens. Our analyses in control U-2 OS (Fig. 6B, top and second from top) and Sa OS (Supplementary Fig. S8B, top) cells revealed that, despite minimum colocalization in the nucleus, all three antigens colocalize mainly at the cytoplasm. As verified by both direct CLSM immunofluorescence antigen localization (Fig. 6B; Supplementary Fig. S8B, bottom) and CLSM-derived quantitatitive analyses of Bax-Ku70, Bax-sCLU, and sCLU-Ku70 colocalizations (Fig. 6C; Supplementary Fig. S8C, right), depletion of sCLU liberates Bax from Ku70 binding.
Discussion
Despite prior studies, which indicated that sCLU is essential for the maintenance of cellular homeostasis (see Introduction), the role, binding profile, and intracellular site(s) of sCLU function in human cancer cells under physiologic conditions or during cell death execution remained elusive. We report here that siRNA-mediated sCLU depletion in human cancer cells promoted high rates of endogenous mitochondrial apoptosis (Supplementary Fig. S9). We found that, if sCLU depletion occurs in a p53+/+ cellular context, stress signals result in p53 stabilization, which accumulates in mitochondria and nucleus. In the nucleus, p53 activates p21 and Bax genes; p21 arrest cycling cells at the G1-S checkpoint, whereas free Bax localize to mitochondria and trigger apoptosis. Considering that intracellular sCLU contributes to the structural integrity of the ER-Golgi and mitochondria, the stress signals that activate p53 or other, currently unknown, stress sensors following sCLU depletion may originate from several intracellular sites, including these disrupted organelles. The up-regulation of the ER stress marker GRP78 after sCLU depletion provides a link between ER distortion, p53 activation, and the mitochondrial apoptotic pathway (28). Also, ER stress in p53+/+ mouse embryonic fibroblasts activated BH3-only prodeath proteins at the transcription level through p53 (29). On the other hand, ER stress-induced apoptosis in p53−/− cells was only partially suppressed and the p53-independent apoptotic pathway may be mediated by C/EBP homologous protein and caspase-12 (29). More studies are needed to identify the stress sensors that trigger sCLU depletion-mediated cell death in p53-null human cells. Interestingly, although moderate sCLU knockdown (e.g., by the Cl-I siRNA) can sensitize cancer cells to chemotherapeutic drugs (12), it was less effective than under conditions of more complete sCLU depletion at inducing spontaneous apoptosis. It is likely that moderate sCLU knockdown may not allow active Bax to reach levels sufficient to trigger high levels of apoptosis as a substantial amount of Bax remains bound to Ku70 (Fig. 5D1). Moreover, the activated Bax that eventually reaches mitochondria can be effectively inhibited by retained mitochondria-bound sCLUC (ref. 17; Fig. 4A). Consistently, CLU-specific antisense oligonucleotides, which are in general less effective than siRNAs in gene expression silencing,6 sensitized human cancer cells to chemotherapy but did not increase significantly spontaneous cell death responses (11). We propose that sustainable moderate sCLU knockdown brings cells to an “apoptosis-prone” state by lowering the apoptotic machinery threshold. Inducing these conditions in tumor versus normal tissue, along with the expression of functional p53, may have great therapeutic potential in cancer therapies.
One way to understand sCLU function is to map its intracellular distribution. Our novel findings provided direct evidence that both the endogenous sCLUC and sCLUS isoforms are distributed not only in the secretory pathway but also in the soluble nucleocytosolic continuum and in mitochondria of nonstressed human cancer cells as well as in the plasma membrane and cytosol of human RBCs. Our biochemical findings were cross-verified by both CLSM and TEM in situ sCLU localization. Because we entirely focused on endogenous protein levels, the transfer of sCLU from secretory pathway to the nucleocytosolic continuum depends on physiologic conditions and it is not an artificial result caused by sCLU forced overexpression that overrun ER capacity. Moreover, use of TEM overcomes the main drawbacks of the relatively low resolution of CLSM, that is, the blurred outline of the ER and difficulties in detecting cytosolic fluorescence. Our data, along with recent reports showing sCLUC localization in the mitochondria of fibrosarcoma HT1080 cancer cells (17) or sCLUS cytosolic distribution (via a mechanism similar to the ER-associated protein degradation pathway) in stressed human astrocytoma U-251 cells (18), may unravel the mystery of previous reports showing that sCLU binds cytosolic proteins, including the transforming growth factor-β type I and II receptors (30), the cytosolic microtubule-destabilizing stathmin family protein SCLIP (31), and phosphorylated IκBα (32). It appears that localization of endogenous ER-Golgi proteins (such as sCLU) to the cytosol or the mitochondria is not as unusual as once thought. More specifically, analyses of the human chondrocyte mitochondrial proteome revealed that ~ 14% of the identified proteins were functionally distributed in the ER (33). The molecular mechanisms that enable ER-Golgi resident proteins to evade the secretion pathway are largely unknown; however, studies of proteins like sCLU should yield insight into this issue.
The mitochondrial pathway of apoptosis in mammalian cells is mainly regulated by the proapoptotic and antiapoptotic members of the Bcl-2 family (34). Bax is mostly a cytosolic monomer in healthy cells (27), although, as we found in osteosarcoma cells, smaller amounts can localize in the ER, mitochondria, and interphase nuclei (34, 35). During apoptosis, Bax undergoes conformational changes in the cytosol followed by translocation to the outer membrane of mitochondria, where it oligomerizes and promotes mitochondrial outer membrane permeabilization and cell death; on the other hand, Bcl-2 is a membrane-bound protein and a potent inhibitor of Bax oligomerization (34). Bax is kept inactive in the cytosol by binding to Ku70 (26), a protein that has been implicated in the repair of nonhomologous double-stranded DNA via Ku80 binding (36). Our unprecedented findings provide the first paradigm of direct sCLU implication in the regulation of healthy human cells homeostasis by showing that sCLU interferes with molecular events, which regulate Bax localization and function. More specifically, we showed that, via a p53-independent mechanism, sCLU depletion suppresses expression of Bcl-2 and Bcl-XL prosurvival proteins, rendering cells more vulnerable to active Bax; this effect can be reversed by ectopic Bcl-2 overexpression in OS cells. In addition, we provide for the first time evidence that sCLU functions in the cytosol of nonstressed human cancer cells, where it stabilizes the Ku70-Bax complex and inhibits the apoptosis-mediated conformational changes of Bax and its consequent relocation to mitochondria. Although Ku70-deficient cells are sensitive to anticancer drugs, they do not undergo spontaneous apoptosis (37), suggesting that Bax dissociation from Ku70 is not sufficient to trigger its activation or translocation from the cytosol to mitochondria. Bax release from sCLU may, therefore, represent a key step toward its full activation and translocation to mitochondria, suggesting that sCLU binding to the Ku70-Bax complex may be therapeutically regulated in human cancer cells.
Taken together, our results strongly suggest that sCLU, apart from functioning as an extracellular chaperone (2), also acts intracellularly in the membranous secretory pathway, the nucleocytosolic continuum and mitochondria, having a vital role in the maintenance of cellular homeostasis and proteome stability of human cells. Taking into account that sCLU was recently found to be induced by SATB1, a gene that promotes breast tumor growth and metastasis (38), it would be of great interest to address its role in human precancerous lesions where senescence, together with apoptosis, provides a barrier to malignant progression (39), and where sCLU may provide a prosurvival function, acting to enhance cancer progression and metastases.
Supplementary Material
Translational Relevance.
In certain cellular contexts, sCLU can confer resistance to cytotoxic agents both in vitro and in vivo. We found that siRNA-mediated sCLU depletion in nonstressed human cancer cells induces altered ratios of proapoptotic to antiapoptotic Bcl-2 protein family members, triggers p53-dependent G1-S growth retardation, and promotes increased endogenous mitochondrial apoptosis. sCLU depletion-mediated activation of p53 amplifies the apoptotic outcome. We show that, in nonstressed human cells, sCLU binds and thereby stabilizes the Ku70-Bax complex serving as a cytosol retention factor for Bax. Thus, sCLU is a potent regulator of Bax-mediated apoptosis. We suggest that elevated sCLU levels may enhance tumorigenesis by interfering with Bax proapoptotic activities and contribute to one of the major characteristics of cancer cells, that is, resistance to apoptosis. This study is directly linked to sCLU basic, translational, and clinical investigation because sCLU is currently an antisense target in phase II clinical trials for prostate, lung, and breast cancers.
Acknowledgments
We thank N. Chondrogianni and M.N. Alexis (National Hellenic Research Foundation) for critical reading of the article and helpful suggestions during hypotonic cytosol isolation, respectively; S.N. Pagakis (Biomedical Research Foundation- Academy of Athens) for CLSM support; E. Kolettas (University of loannina Medical School) for the Bcl-2 plasmid; B. Vogelstein (Johns Hopkins University) for the V143A–p53 and R273H–p53 plasmids; K. Vousden (Beatson Institute for Cancer Research) for the WT-p53 plasmid; and R.J. Youle (National Institute of Neurological Disorders and Stroke, NIH) for the Bax6A7 antibody.
Grant support: U.S. Department of Energy grant DE-FG02-06ER64186 (CSCN0039;Y. Zou and D.A. Boothman); EU Full-Cost Grant “Genika” (V.G. Gorgoulis); and E.U. Full-Cost Grant “Proteomage”contract LSHM-CT-2005-518230 (I.P. Trougakos, M. Lourda, and E.S. Gonos).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Unpublished data.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.de Silva HV, Harmony JA, Stuart WD, Gil CM, Robbins J. Apolipoprotein J: structure and tissue distribution. Biochemistry. 1990;29:5380–5389. doi: 10.1021/bi00474a025. [DOI] [PubMed] [Google Scholar]
- 2.Wilson MR, Easterbrook-Smith SB. Clusterin is a secreted mammalian chaperone. Trends Biochem Sci. 2000;25:95–98. doi: 10.1016/s0968-0004(99)01534-0. [DOI] [PubMed] [Google Scholar]
- 3.Trougakos IP, Gonos ES. Clusterin/apolipoprotein J in human aging and cancer. Int J Biochem Cell Biol. 2002;34:1430–1448. doi: 10.1016/s1357-2725(02)00041-9. [DOI] [PubMed] [Google Scholar]
- 4.Leskov KS, Klokov DY, Li J, Kinsella TJ, Boothman DA. Synthesis and functional analysis of nuclear clusterin: a cell death protein. J Biol Chem. 2003;278:11590–11600. doi: 10.1074/jbc.M209233200. [DOI] [PubMed] [Google Scholar]
- 5.Caccamo AE, Scaltriti M, Caporali A, et al. Ca2+ depletion induces nuclear clusterin, a novel effector of apoptosis in immortalized human prostate cells. Cell Death Differ. 2005;12:101–104. doi: 10.1038/sj.cdd.4401491. [DOI] [PubMed] [Google Scholar]
- 6.Trougakos IP, Gonos ES. Regulation of clusterin/apolipoprotein J, a functional homologue to the small heat shock proteins, by oxidative stress in ageing and age-related diseases. Free Radic Res. 2006;40:1324–1334. doi: 10.1080/10715760600902310. [DOI] [PubMed] [Google Scholar]
- 7.Scaltriti M, Santamaria A, Paciucci R, Bettuzzi S. Intracellular clusterin induces G2-M phase arrest and cell death in PC-3 prostate cancer cells. Cancer Res. 2004;64:6174–6182. doi: 10.1158/0008-5472.CAN-04-0920. [DOI] [PubMed] [Google Scholar]
- 8.Thomas-Tikhonenko A, Viard-Leveugle I, Dews M, et al. Myc-transformed epithelial cells down-regulate clusterin, which inhibits their growth in vitro and carcinogenesis in vivo. Cancer Res. 2004;64:3126–3136. doi: 10.1158/0008-5472.can-03-1953. [DOI] [PubMed] [Google Scholar]
- 9.Shannan B, Seifert M, Leskov K, et al. Challenge and promise: roles for clusterin in pathogenesis, progression and therapy of cancer. Cell Death Differ. 2006;13:12–19. doi: 10.1038/sj.cdd.4401779. [DOI] [PubMed] [Google Scholar]
- 10.Viard I, Wehrli P, Jornot L, et al. Clusterin gene expression mediates resistance to apoptotic cell death induced by heat shock and oxidative stress. J Invest Dermatol. 1999;112:290–296. doi: 10.1046/j.1523-1747.1999.00531.x. [DOI] [PubMed] [Google Scholar]
- 11.Miyake H, Chi KN, Gleave ME. AntisenseTRPM-2 oligodeoxynucleotides chemosensitize human androgen-independent PC-3 prostate cancer cells both in vitro and in vivo. Clin Cancer Res. 2000;6:1655–1663. [PubMed] [Google Scholar]
- 12.Trougakos IP, So A, Jansen B, Gleave ME, Gonos ES. Silencing expression of the clusterin/apolipoprotein J gene in human cancer cells using small interfering RNA induces spontaneous apoptosis, reduced growth ability, and cell sensitization to genotoxic and oxidative stress. Cancer Res. 2004;64:1834–1842. doi: 10.1158/0008-5472.can-03-2664. [DOI] [PubMed] [Google Scholar]
- 13.Criswell T, Beman M, Araki S, et al. Delayed activation of insulin-like growth factor-1 receptor/Src/ MAPK/Egr-1 signaling regulates clusterin expression, a pro-survival factor. J Biol Chem. 2005;280:14212–14221. doi: 10.1074/jbc.M412569200. [DOI] [PubMed] [Google Scholar]
- 14.Sallman DA, Chen X, Zhong B, et al. Clusterin mediates TRAIL resistance in prostate tumor cells. Mol Cancer Ther. 2007;6:2938–2947. doi: 10.1158/1535-7163.MCT-07-0345. [DOI] [PubMed] [Google Scholar]
- 15.Chi KN, Eisenhauer E, Fazli L, et al. A phase I pharmacokinetic and pharmacodynamic study of OGX-011, a 2′-methoxyethyl antisense oligonucleotide to clusterin, in patients with localized prostate cancer. J Natl Cancer Inst. 2005;97:1287–1296. doi: 10.1093/jnci/dji252. [DOI] [PubMed] [Google Scholar]
- 16.O’Sullivan J, Whyte L, Drake J, Tenniswood M. Alterations in the post-translational modification and intracellular trafficking of clusterin in MCF-7 cells during apoptosis. Cell Death Differ. 2003;10:914–927. doi: 10.1038/sj.cdd.4401254. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Kim JK, Edwards CA, Xu Z, Taichman R, Wang CY. Clusterin inhibits apoptosis by interacting with activated Bax. Nat Cell Biol. 2005;7:909–915. doi: 10.1038/ncb1291. [DOI] [PubMed] [Google Scholar]
- 18.Nizard P, Tetley S, Le Drean Y, et al. Stress-induced retrotranslocation of clusterin/ApoJ into the cytosol. Traffic. 2007;8:554–565. doi: 10.1111/j.1600-0854.2007.00549.x. [DOI] [PubMed] [Google Scholar]
- 19.Lourda M, Trougakos IP, Gonos ES. Development of resistance to chemotherapeutic drugs in human osteosarcoma cell lines largely depends on up-regulation of clusterin/apolipoprotein J. Int J Cancer. 2007;120:611–622. doi: 10.1002/ijc.22327. [DOI] [PubMed] [Google Scholar]
- 20.Schlabach MR, Luo J, Solimini NL, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 22.Trougakos IP, Lourda M, Agiostratidou G, Kletsas D, Gonos ES. Differential effects of clusterin/apolipoprotein J on cellular growth and survival. Free Radic Biol Med. 2005;38:436–449. doi: 10.1016/j.freeradbiomed.2004.10.038. [DOI] [PubMed] [Google Scholar]
- 23.Manders EM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-colour confocal images. J Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 24.Joerger AC, Fersht AR. Structure-function-rescue: the diverse nature of common p53 cancer mutants. Oncogene. 2007;26:2226–2242. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- 25.Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol. 2005;17:631–636. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Cohen HY, Lavu S, Bitterman KJ, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 27.Hsu YT, Youle RJ. Nonionic detergents induce dimerization among members of the Bcl-2 family. J Biol Chem. 1997;272:13829–13834. doi: 10.1074/jbc.272.21.13829. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Lee AS. Stress induction of GRP78/Bi P and its role in cancer. Curr Mol Med. 2006;6:45–54. doi: 10.2174/156652406775574523. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Lee B, Lee AS. Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J Biol Chem. 2006;281:7260–7270. doi: 10.1074/jbc.M509868200. [DOI] [PubMed] [Google Scholar]
- 30.Reddy KB, Karode MC, Harmony AK, Howe PH. Interaction of transforming growth factor β receptors with apolipoprotein J/clusterin. Biochemistry. 1996;35:309–314. doi: 10.1021/bi951880a. [DOI] [PubMed] [Google Scholar]
- 31.Kang SW, Shin YJ, Shim YJ, Jeong SY, Park IS, Min BH. Clusterin interacts with SCLIP (SCG10-like protein) and promotes neurite outgrowth of PC12 cells. Exp Cell Res. 2005;309:305–315. doi: 10.1016/j.yexcr.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Devauchelle V, Essabbani A, De Pinieux G, et al. Characterization and functional consequences of underexpression of clusterin in rheumatoid arthritis. J Immunol. 2006;177:6471–6479. doi: 10.4049/jimmunol.177.9.6471. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz-Romero C, Lopez-Armada MJ, Blanco FJ. Mitochondrial proteomic characterization of human normal articular chondrocytes. Osteoarthritis Cartilage. 2006;14:507–518. doi: 10.1016/j.joca.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 35.Hoetelmans R, van Slooten HJ, Keijzer R, Erkeland S, van deVelde CJ, Dierendonck JH. Bcl-2 and Bax proteins are present in interphase nuclei of mammalian cells. Cell Death Differ. 2000;7:384–392. doi: 10.1038/sj.cdd.4400664. [DOI] [PubMed] [Google Scholar]
- 36.Koike M, Koike A. The Ku70-binding site of Ku80 is required for the stabilization of Ku70 in the cytoplasm, for the nuclear translocation of Ku80, and for Ku80-dependent DNA repair. Exp Cell Res. 2005;305:266–276. doi: 10.1016/j.yexcr.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 37.Kim SH, Kim D, Han JS, et al. Ku autoantigen affects the susceptibility to anticancer drugs. Cancer Res. 1999;59:4012–4017. [PubMed] [Google Scholar]
- 38.Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- 39.Gorgoulis VG, Vassiliou LV, Karakaidos P, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.