Abstract
Fusion transcript formation is one of the fundamental mechanisms that drives the development of prostate cancer. Because of the advance of high-throughput parallel sequencing, many fusion transcripts have been discovered. However, the discovery rate of fusion transcripts specific for prostate cancer is lagging behind the discoveries made on chromosome abnormalities of prostate cancer. Recent analyses suggest that many fusion transcripts are present in both benign and cancerous tissues. Some of these fusion transcripts likely represent important components of normal gene expression in cells. It is necessary to identify the criteria and features of fusion transcripts that are specific for cancer. In this review, we discuss optimization of RNA sequencing depth for fusion transcript discovery and the characteristics of fusion transcripts in normal prostate tissues and prostate cancer. We also propose a new classification of cancer-specific fusion transcripts on the basis of their tail gene fusion protein product and the roles that these fusions may play in cancer development.
Genomic alteration is the hallmark of human malignancies. These alterations are manifested as changes in copy number in a region of chromosome such as amplification, deletion, duplication, or chromosome rearrangement. Single nucleotide mutations that alter the protein structure may also have relevant impact on the development of cancers. The invariable association between human malignancies and these genomic alterations argues strongly that genome alterations, rather than epigenomic changes, are the most critical and common underlying mechanisms for the development of human cancers. The genomic and epigenomic alterations and their corresponding gene expression alteration in prostate cancer were well documented and characterized in the past 15 years.1–11
Studies that used high-throughput genome array and whole-genome sequencing reveal a large number of copy number changes in human prostate cancer genomes, affecting almost all chromosomes.6,12,13 Because of the high level of genomic rearrangement identified in the prostate cancer genome, the term chromoplexy was coined to describe the complexity and abundance of chromosome rearrangements.14 Some of the grave consequences of chromosome rearrangement are the aberrant joining of unrelated genes and the production of a fusion transcript.
The events of joining together two unrelated genes often lead to the creation of a new oncogene that might be analogous to BCR-Abl seen in leukemias. Even though genomic rearrangement in prostate cancer is extensive, only a handful of fusion transcripts related to prostate cancer are properly validated, suggesting that the prevailing approach of fusion transcript discovery is suboptimal. RNA sequencing could be modified to improve the discovery rate of fusion transcripts.
High Coverage of RNA Sequencing Is Necessary to Discover Most Low-Abundance Fusion Transcripts
Abundantly expressed housekeeping genes usually have the dominant presence in whole-transcriptome sequencing. Indeed, we found that the top quartile of expressed genes generally account for 90% of the mapped reads in our transcriptome analyses.15 This leaves few detected reads representing low abundantly expressed genes.
To detect fusion transcripts, accurate detection relies on the finding of split reads that contain fusion-joining sequences of two unrelated genes in a 100-bp sequence when using Illumina HiSeq2500 sequencer. Reads mapped to other regions of a fusion transcript are irrelevant because they overlap a fusion transcript and its wild-type counterpart and thus will be classified as wild-type gene transcripts.
Even though spanning reads that contain pair-end mapped to the head and tail parts of a fusion transcript may provide important support for the presence of a fusion transcript, the rate of validation is low because of uncertainty of the location of the fusion joining point. Thus, the focus of fusion transcript detection is identification of sequences that contain the fusion split juncture. Because of the limit of sequence mapping, the split juncture has to be at least 10 bp away from the end of the read to avoid ambiguous mapping. Sequences not >90 bp from either side of the fusion split juncture will be used when using Illumina 200 cycle paired-end sequencing scheme. As a result, the rate of detecting a split read may be low if the expression of the fusion transcript is not abundant.
In our recent transcriptome sequencing that used 1200× to 1400× coverage per gene,15 of the eight validated fusion transcripts and TMPRSS2-ERG, the numbers of split reads ranged from 2 to 13 (Table 1).15 Most of the fusion transcripts have five or less split reads by using FusionCatcher (https://code.google.com/p/fusioncatcher, last accessed March 12, 2015). This translates to the bottom half of expression levels for all genes. Two of nine fusion transcripts are in the bottom 30% of expressed genes.
Table 1.
Coverage Dilution Simulation to Detect Fusion Transcripts
| Sample | Head gene | Tail gene | Expression percentile | Whole |
1000× |
600× |
300× |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Span | Split | Span | Split | Span | Split | Span | Split | ||||
| 1T | TMPRSS2 | ERG | 30th–40th | 7 | 6 | 6 | 2 | 3 | 1 | 3 | 1 |
| 5T | TMPRSS2 | ERG | 70th–80th | 6 | 13 | 2 | 4 | 0 | 2 | 0 | 0 |
| 3T | TRMT11 | GRIK2 | 50th–60th | 3 | 12 | 5 | 4 | 0 | 0 | 0 | 0 |
| 1T | LRRC59 | FLJ60017 | 30th–40th | 3 | 7 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4T | TMEM135 | CCDC67 | 20th–30th | 3 | 3 | 3 | 1 | 0 | 0 | 0 | 0 |
| 2T | MTOR | TP53BP1 | 50th–60th | 12 | 5 | 10 | 5 | 5 | 5 | 5 | 3 |
| 3T | MAN2A1 | FER | 60th–70th | 3 | 13 | 3 | 8 | 0 | 0 | 0 | 0 |
| 3T | KDM4B | AC011523.2 | 20th–30th | 7 | 2 | 7 | 1 | 0 | 0 | 0 | 0 |
| 3T | CCNH | C5orf30 | 30th–40th | 3 | 5 | 2 | 4 | 0 | 0 | 0 | 0 |
To determine the threshold of coverage to detect these transcripts, we randomized and subsampled the mapped reads to equivalence of 1000×, 600×, 400×, and 200× coverage. Although TMPRSS2-ERG and MTOR-TP53BP1 were consistently detected even when coverage was reduced to 300×, all other fusion transcripts disappeared when sequencing coverage was reduced to 600×. However, when sequencing depth was raised to 1000× coverage, only LRRC59-FLJ60017 was not detectable. Thus, sequencing depth is crucial to detecting novel fusion transcripts.
To detect most fusion transcripts, a minimum of 1000× coverage is required. Because whole-genome sequencing from these prostate cancer samples indicates far more chromosome rearrangements and translocations than the number of fusion transcripts found,14,15 a substantial number of fusion transcripts may have escaped our detection even with 1300× coverage. Quite likely, the undetected fusion transcripts are expressed in low abundance.
Extensive Presence of Fusion Transcripts in Normal OD Prostate Tissues
Similar to nucleotide and copy number variants, most fusion transcripts can be physiologic. Recent transcriptome sequencing of normal prostate tissues from healthy organ donors (OD) revealed numerous fusion transcripts in normal prostate tissues.15 These fusion transcripts can be classified into two categories: The fusion transcripts connect two adjacent transcripts. This type of fusion transcripts appears to result from the splicing of the 5′ end of a head gene and the 3′ end of a tail gene. A chimera mRNA is produced with the arrangement of head–tail gene in the same direction of transcription from the chromosome. Such fusion transcript formation requires no chromosome rearrangement. Some of these transcripts may have a chimera protein translated. The second type of fusion transcript involves the combination of head and tail genes from a far distance, or from different chromosomes, or in different chromosome transcription directions. The formation of such fusion transcripts may require chromosome recombination or may be RNA recombination if the evidence of chromosome rearrangement is absent.
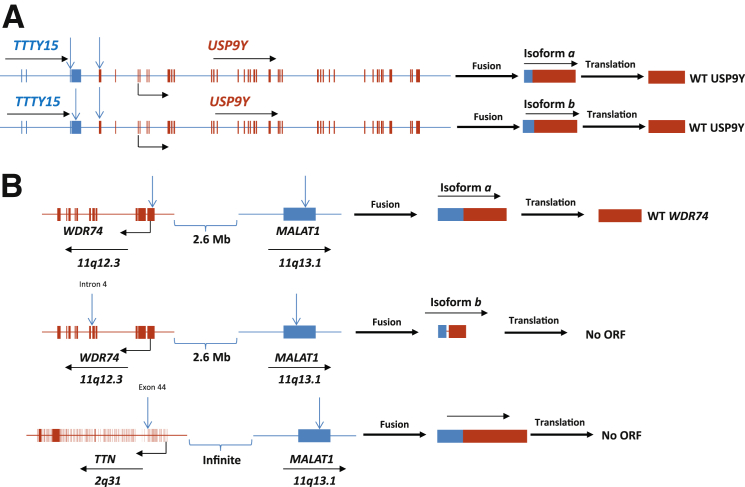
Among the 20 samples of OD prostate samples, >80% of validated fusion transcripts are the results of splicing of a continuous transcript that spans two genes (category 1). The most common iteration of type 1 category 1 fusion is TTTY15-USP9Y from Y chromosome (Table 2). TTTY15 is a testis-specific non-coding RNA with no known function. USP9Y is an ubiquitin-specific peptidase that may be involved in spermatogenesis.16 Two different fusion points in TTTY15 were identified (Figure 1A): one fusion isoform produced chimera RNA with the first 3 exons of TTTY15 connecting with a partial first exon of USP9Y. The first 779 nucleotides of exon 1 in USP9Y were lost. The open reading frame for USP9Y remains intact. It will likely have no impact on the translation of the ubiquitin-specific protease 9, Y chromosome (USP9Y) protein.
Table 2.
Validated Fusion Transcripts Present in Healthy Organ Donors
| Fusion gene | Protein product | Head/tail gene locus | Distance | Head/tail joining sequence | Age/number |
|---|---|---|---|---|---|
| HARS2-ZMAT2 | Head gene-WT | 5q31.3/5q31.3 | 2 kb | 5′-CGTTGTCAAGAG-3′ | 19–71/9 |
| Tail gene-AWTASC | 5′-ACAAAAAACTTG-3′ | ||||
| BAIAP2L2-SLC16A8 | Head gene-WT | 22q13.1/22q13.1 | 1.9 kb | 5′-GGGGTCCGAGAC-3′ | 19–71/9 |
| Tail gene-WT | 5′-CTGCTCCACTTC-3′ | ||||
| KLK4-KLKP1 | Head gene-CTT | 19q13.1/19q13.1 | 20 kb | 5′-TGCTGGCGAACG-3′ | 19–71/10 |
| Tail gene-NC | 5′-ATGCTGTGATTG-3′ | ||||
| AZGP1-GJC3 | Chimera protein | 7q22.1/7q22.1 | 48 kb | 5′-ACGACAGTAACG-3′ | 19–71/10 |
| Chimera protein | 5′-TTCCAGGAAGAA-3′ | ||||
| SLC45A3-ELK4 | Chimera protein | 1q32.1/1q32.1 | 38 kb | 5′-CGGGAGAAGCAG-3′ | 19–71/9 |
| Chimera protein | 5′-CTCATTGCTATGG-3′ | ||||
| PRKAA1-TTC33 | Head gene-CTT | 5p13.1/5p13.1 | 17 kb | 5′-GTAGTATTGATG-3′ | 19–71/10 |
| Tail gene-WT | 5′-AATGGCTTCCTT-3′ | ||||
| C6orf47-BAG6 | Head gene-AWTASC | 6p21.3/6p21.3 | 8.5 kb | 5′-GGTGTGGCCTTG-3′ | 19–71/10 |
| Tail gene-WT | 5′-AGACCTGTCGGC-3′ | ||||
| ADCK4-NUMBL | Chimera protein | 19q13.2/19q13.2 | 6 kb | 5′-TTTGAAACCAAG-3′ | 22–71/7 |
| Chimera protein | 5′-GGCGGACCCCGG-3′ | ||||
| TTTY15-USP9Ya | Head gene-NC | Yq11.1/Yq11.1 | 15 kb | 5′-TTCTGACCTGTG-3′ | 15–75/19 |
| Tail gene-WT | 5′-AGAGCTTGGAG-3′ | ||||
| TTTY15-USP9Yb | Head gene-NC | Yq11.1/Yq11.1 | 15 kb | 5′-GTATTTTGGAAG-3′ | 15–56/16 |
| Tail gene-WT | 5′-AGAGCTTGGAG-3′ | ||||
| MALAT1-TNN | Head gene-NC | 11q13.1/2q31 | Infinite | 5′-ATGGGACAATAG-3′ | 21/1 |
| Tail gene-AWTASC | 5′-TAATGTCGCTGG-3′ | ||||
| MALAT1-WDR74a | Head gene-NC | 11q13.1/11q12.2 | 2.6 mb | 5′-CAGTAAGAAAAA-3′ | 21/1 |
| Tail gene-WT | 5′-ATCAAGTGTAGT-3′ | ||||
| MALAT1-WDR74b | Head gene-NC | 11q13.1/11q12.2 | 2.6 mb | 5′-TGGGTTAGAGAA-3′ | 21/1 |
| Tail gene-AWTASC | 5′-GGAGTCTCACTC-3′ |
Transcripts were validated through RT-PCR and Sanger sequencing.
AWTASC, absence of wild-type ATG start codon; CTT, C-terminal truncation; NC, non-coding; WT, wild-type.
Figure 1.
Fusion transcripts in normal prostate tissues. A: Fusion transcripts result from continuous transcription of two contiguous genes. Schematic diagrams of TTTY15 and USP9Y genomes, fusion transcripts, and predicted translation products. Blue arrow indicates fusion breakpoint in the genome. Transcription direction is indicated by black arrow. Translation start site is indicated by 90° arrow. B: Fusion transcripts result from noncontiguous transcription. Schematic diagrams of MALAT1, WDR74, and TTN genomes, fusion transcripts, and predicted translation products. Blue arrow indicates fusion breakpoint in the genome. Transcription direction is indicated by black arrow. Translation start site is indicated by 90° arrow.
The second isoform has the first three intact exons of TTTY15 but also retains a partial exon 4. The head gene is substantially larger than isoform a but loses 1.7 kb of exon 4. This portion of TTTY15 connects with the same fusion point of USP9Y as isoform a; thus, the isoform has an intact open reading frame for USP9Y. To examine the frequency of TTTY15-USP9Y fusion in healthy persons, RT-PCRs using primers that corresponded to the sequences surrounding the fusion point of TTTY15-USP9Y were performed on 20 samples of OD prostates from persons free of urologic malignancies with ages ranging from 15 to 71 years. Isoform a of TTTY15-USP9Y was readily detected in 19 of 20 OD samples, whereas isoform b was positive in 16 samples. The high frequency rate of TTTY15-USP9Y in donor prostates from persons with wide range of ages suggests that TTTY15-USP9Y fusion is a likely physiologic event, even though it was previously identified in prostate cancer samples.17
A surprising finding of fusion transcripts in normal prostate tissues is the presence of fusions that could not be explained by splicing within the same transcript. This type of fusion makes up <10% of validated fusion transcripts detected in tissues from healthy persons. One of the most frequently detected fusion transcripts of this category is MALAT1 and its associated partners. MALAT1 is a long non-coding RNA with a single exon of 8.7-kb length. MALAT1 appears to involve RNA recombination and is located at the active transcription sites.18 Increase of expression of MALAT1 is associated with poor prognosis of several human malignancies.19–26
Multiple mechanisms were proposed for the oncogenic activity of MALAT1.27–35 A donor prostate from a 21-year-old man was positive for MALAT1 fusion with several gene partners: two isoforms of MALAT1-WDR74 and one MALAT1-TTN were detected (Figure 1B). For MALAT1-WDR74 fusion, the breakpoint of fusion isoform a occurred in cDNA position 7106 bp of MALAT1 and the first 25 nucleotide of WDR74. The resulting fusion transcript contains an intact open reading frame for WDR74 and thus likely retains function to translate a wild-type WD repeat-contained protein 74 (WDR74) protein. The breakpoint of MALAT1-WDR74 isoform b, however, occurred in the exon of MALAT1 and intron 4 of WDR74. Such fusion eliminates the ATG translation start codon for WDR74; thus, no protein will be produced from this template.
These fusions cannot be explained by continuous transcription through these two genes because the directions of these two transcripts are opposite and are away from each other. MALAT1-TTN fusion occurred between MALAT1 exon and TTN partial exon 44. The resulting transcript will have ATG translation start site deleted for TTN; thus, no titin (TTN) protein will likely be translated. Multiple fusion partners of MALAT1 fusion in the same sample (Table 2) suggest that these fusion transcripts occurred at the RNA level. Even though transgene splicing is a distinct possibility, the lack of splicing donor or acceptor sequences at some of the fusion breakpoints makes it an unlikely mechanism. Because MALAT1 plays a critical role in RNA splicing through RNA–RNA interaction,36 MALAT1 RNA may be involved in RNA cleavage and rejoining process. Probably a yet-to-be-discovered RNA recombination mechanism drives such epigenetic (post-transcriptional) process.
The discovery of a substantial number of fusion transcripts in normal tissues from healthy persons of various ages suggests that many gene fusions are likely to be physiologic processes. Most of these fusion transcripts do not produce chimeric proteins. Instead, some of these fusion transcripts produce a truncated head gene-driven transcription of a tail gene. Protein truncations or complete elimination of open reading frame are frequent. The repeated presence of specific fusion transcripts in normal prostate tissues suggests that these fusions are not random events. Rather, fusion transcripts add a new sphere of diversity and complexity of protein and gene expression regulation. Fusion transcripts because of RNA recombination may allow cells to destroy unwanted transcripts and prevent them from translating into proteins that might substantially alter the differentiation paths of these cells. Truncated proteins may interfere or weaken the function of the wild-type proteins by competing for the same substrates or binding molecules. A head gene-driven tail gene transcript may have the effect of weakening the expression of the head gene protein because of truncation, but enhancing the expression of a tail gene protein when open reading frame is intact, because of transcripts from both the head and tail gene promoters. The investigation of the mechanisms that drive these physiologic fusion formations and of the biological role of these fusion transcripts will give us important new insight into the epigenetic regulation of gene/protein expressions.
Fusion Transcripts in Prostate Cancer
Classification
In general, fusion transcripts in prostate cancer can be classified into three categories on the basis of the protein structure of the head gene and the tail gene in the fusion transcripts (Table 3): chimera protein-forming fusion transcripts, independent wild-type tail gene fusion transcripts, and non–fusion-forming fusion transcripts. For chimera protein-forming fusion transcript, translation of a fusion protein occurs at the ATG start codon of the head gene and connect it in frame with the C-terminus of the tail gene. For independent wild-type tail gene fusion transcript, the fusion point occurs at the 5′ untranslated region of the tail gene mRNA such that both the ribosomal binding site and the ATG start codon are preserved. The consequence of such fusion is independent translation of the tail gene in the transcript, whereas the expression of the tail gene is driven by the head gene promoter. The head gene may or may not express a truncated protein. For non–fusion-forming fusion transcript, the ATG translation start codon and the ribosome binding site of the tail gene are deleted, whereas the head gene has C-terminus truncation or has lost the open reading frame all together. In rare occasion, a wild-type head gene is preserved in non–fusion-forming fusion transcript.
Table 3.
Fusion Transcripts that Result from Chromosome Rearrangement
| Fusion gene | Head gene locus | Tail gene locus | Fusion protein product |
|---|---|---|---|
| Chimera protein fusion | |||
| ALG5-PIGU∗ | 13q13.3 | 20q11.22 | ALG5(aa1-157)-PIGU(aa209-434) |
| PIGU-ALG5∗ | 20q11.22 | 12q13.3 | PIGU(aa1-208)-ALG5(aa158-294) |
| MAN2A1-FER | 5q21-22 | 5q21 | MAN2A1(aa1-703)-FER(aa572-822) |
| SLC45A2-AMACR | 5p13.2 | 5p13 | SLC45A2(aa1-187)-AMACR(aa84-394) |
| DDX5-ETV4∗ | 17q21 | 17q21 | DDX5(aa1-102)-ETV4(aa67-484) |
| HNRNPA2B1-ETV1 | 7p15 | 7p21.3 | HNRNPA2B1(aa1-2)-ETV1(aa16-477) |
| TMPRSS2-ETV5∗ | 21q22.2 | 3q28 | TMPRSS2(aa1-116)-ETV5(b) (aa1-510) |
| TMPRSS2 (aa1-18)-ETV5(a) (aa1-510) | |||
| Independent wild-type tail gene protein fusion | |||
| TMPRSS2-ERG(b) | 21q22.2 | 21q22.3 | TMPRSS2, CTT; ERG, WT |
| CCNH-C5orf30 | 5q13.3-14 | 5q21.1 | CCNH, CCT; C5orf30, WT |
| TNPO1-IKBKB∗ | 5q13.2 | 8p11.2 | TNPO1, CCT; IKBKB, WT |
| Non-chimera protein-forming fusion | |||
| TMPRSS2-ERG(a) | 21q22.2 | 21q22.3 | TMPRSS2, CTT; ERG, AWTASC |
| TMPRSS2-ETV1 | 21q22.2 | 7p21.3 | TMPRSS2, CTT; ETV1, AWTASC |
| TMPRSS2-ETV4 | 21q22.2 | 17q21 | TMPRSS2, CTT; ETV4, AWTASC |
| SLC45A3-ERG∗ | 1q32.1 | 21q22.3 | NK |
| SLC45A3-ETV5∗ | 1q32.1 | 3q28 | SLC45A3, AWTASC; ETV5, AWTASC |
| ACSL3-ETV1∗ | 2q34-35 | 7p21.3 | ACSL3, CTT; ETV1, AWTASC |
| HERV-K-ETV1∗ | 22q11.23 | 7p21.3 | HERV-K, CTT; ETV1, AWTASC |
| FLJ35294-ETV1 | 17p13.1 | 7p21.3 | FLJ35294, CTT; ETV1, AWTASC |
| FOXP1-ETV1∗ | 3p13 | 7p21.3 | FOXP1, CTT; ETV1, AWTASC |
| C15orf21-ETV1 | 15q21.1 | 7p21.3 | C15orf21, AWTASC; ETV1, AWTASC |
| KLK2-ETV4∗ | 19q13.33 | 17q21 | KLK2, CTT; ETV4, AWTASC |
| KLK2-ETV1 | 19q13.33 | 7p21.3 | KLK2, CTT; ETV1, AWTASC |
| CANT1-ETV4 | 17q25.3 | 17q21 | CANT1, CTT; ETV4, AWTASC |
| TMEM135-CCDC67 | 11q14.2 | 11q21 | TMEM135, CTT; CCDC67, AWTASC |
| LRRC59-FLJ60017 | 17q21.33 | 11q12.3 | LRRC59, CTT; FLJ60017, AWTASC |
| TRMT11-GRIK2 | 6q22.33 | 6q16.3 | TRMT11, CTT; GRIK2, AWTASC |
| MTOR-TP53BP1 | 1p36.2 | 15q15.21 | MTOR, CTT; TP53BP1, AWTASC |
| KDM4B-AC011523.1 | 19p13.3 | 19p12 | MTOR, CTT; tail gene unknown |
AWTASC, absence of wild-type ATG start codon; CCT, C-terminus truncation; NK, not known; WT, wild-type.
Identified in only one case.
ETS Gene Fusions
E26 transformation-specific (ETS) gene fusions are some of the most studied fusion transcripts in prostate cancer.37,38 ETS genes belong to one type of transcription factor that contains a domain binding to DNA sequences with a central GGA(A/T) motif. There are 12 subfamilies.39 ETS was coined because transcription of some of its members can be activated by leukemia virus, E26.
Fusion transcripts that involve ETS gene family members represent the largest collection of fusion gene in prostate cancer.37 With the use of expression outlier and RNA–exon quantification analyses, Tomlins et al40 identified the presence of TMPSS2-ERG and TMPRSS2-ETV1 in prostate cancer cell lines and primary prostate cancer samples. Since then, TMPRSS2-ETV4 and TMPRSS2-ETV5 were also discovered.41–44
ETS gene fusions tend to have multiple head gene partners. Many of these head genes are driven by promoters that respond to androgen stimulation. Several TMPRSS2-ERG fusion variants were discovered.45–47 Although most variants produce no functional chimera protein, some produce a wild-type ERG. Up to 65% of prostate cancers were detected to contain TMPRSS2-ERG fusion.40
ETV1, ETV4, and ETV5 have substantially more head gene partners, whereas only 1% to 5% prostate cancer are positive for ETV fusion. Most ETV fusion genes produce no chimera protein, whereas HNRNPA2B1-ETV1 produces a chimera protein that resembles a full-length ETV1, losing only 15 amino acids at its N-terminus. Thus, both the regulatory domain and the DNA binding domain of ETV1 are likely preserved. TMPRSS2-ETV5 preserves the entire ETS translocation variant 5 (ETV5) protein, whereas it has a giant truncation on TMPRSS2 gene. These fusions likely retain the full function of the tail genes.
For non–fusion-forming type of ETS fusions, an alternative ATG start codon for the tail genes has to be used if truncated proteins were to be expressed in cancer samples, because the wild-type ATG start codon of the tail gene is deleted in these fusion transcripts. But it is not clear which alternative translation start codon is used and what the efficiency of such translation initiation is, if such translation does exist in primary cancer samples. Correlation of protein expression of ERG and TMPRSS-ERG rearrangement yielded conflicting results,48 suggesting the efficiency of the alternative start codon may not be high. In general, the consequences of most of these chromosome rearrangements that involve ERG, ETV1, ETV4, and ETV5 are probably functional loss of these genes and their head gene partners.
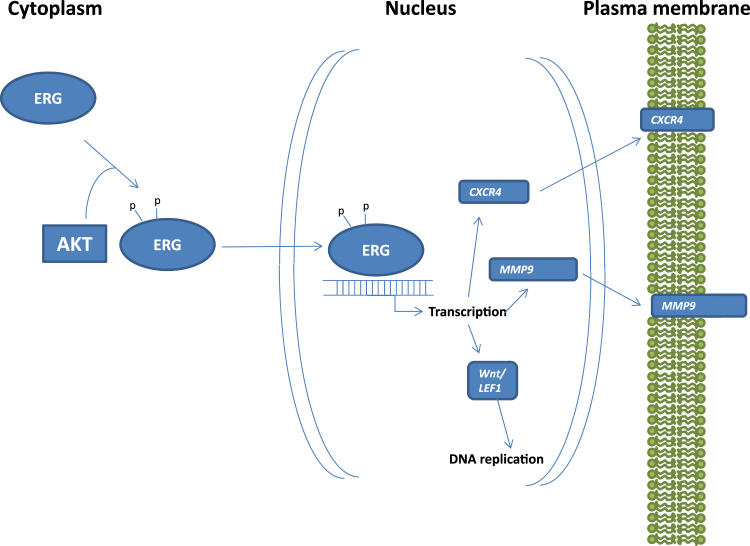
Some studies indicate that expression of ERG or ETV1 in transgenic mice under an androgen-responding promoter leads to prostatic intraepithelial neoplasia but not frank invasive prostate cancer.49–53 However, conflicting results were observed in other groups.52,54 ERG expression accelerated the carcinogenesis development in Pten knockout mice. When combining with androgen receptor overexpression, high levels of ERG produced invasive prostate cancer.53 These analyses suggest that ERG may have an oncogenic role in assisting the development of prostate cancer. Because ERG genome rearrangement was identified in up to 20% high-grade prostatic intraepithelial neoplasia, TMPRSS2-ERG would be considered as an early event in prostate carcinogenesis.55–57 However, conflicting results were reported in its association with the aggressive behavior of prostate cancer.58–69 These conflicting results suggest that other genetic factors such as Pten deletion, may play a major role in determining the clinical outcomes of prostate cancer. In addition, samples with isoform a of TMPRSS2-ERG would have a different implication in ERG protein expression in comparison with samples with isoform b. Such complexity may contribute to the variation of the analyses. ERG is a transcription factor that is activated by phosphorylation at its serine residues 81 and 215 by AKT kinase70 (Figure 2). Presumably, the activated ERG induces the expression of several critical proteins that mediates cell growth71 and invasion.70,72 CXCR4 and MMP9 are of particular interest because they were well documented as direct mediators of adhesion and invasion, respectively. Increased levels of CXCR4 and MMP9 may co-operate to enhance the invasion of prostate cancer.
Figure 2.
Schematic diagram of ERG activation. ERG protein is activated with phosphorylation of serine residues 81 and 215 by AKT kinase. The activated ERG protein facilitates the transcription of Wnt/LEF1, MMP9, and CXCR4.
Fusion Genes Containing Functional Chimera Proteins
One of the examples of chimera protein is ALG5-PIGU, a chimeric protein between dolichyl-phosphate beta-glucosyltransferase (ALG5) and phosphatidylinositol glycan anchor biosynthesis, class U (PIGU). ALG5-PIGU or PIGU-ALG5 fusion is the result of balanced translocation between chromosomes 20 and 13.73 ALG5-PIGU largely retains the glycosyl-transferase domain in ALG5 but has its GPI transamidase domain severely truncated in PIGU. It is not clear whether ALG5-PIGU chromosome rearrangement is recurrent. Recently, genomic and transcript sequencing allow for comprehensive analysis of genome and RNA sequencing. Our group recently uncovered several fusion transcripts by using these methods.15
Among these transcripts, fusion transcripts mannosidase, α, class 2A, member 1-fer (fps/fes related) tyrosine kinase (MAN2A1-FER), and solute carrier family 45, member 2-α-methylacyl-CoA racemase (SLC45A2-AMACR) fusion transcripts express chimera proteins. MAN2A1-FER fusion results from chromosome rearrangement that leads to the loss of the genome sequence corresponding to the 5′ end of FER. Whole-genome sequencing revealed that the breakpoint of MAN2A1-FER was located in intron 13 of MAN2A1 and intron 14 of FER.
MAN2A1 is a Golgi enzyme required for conversion of high mannose to complex type structure of N-glycan for mature glycosylation of a membrane protein.74 Little is known about its relation with human malignancies. However, FER, a tyrosine kinase, is a well-documented oncogene.75 Several studies showed that FER activates androgen receptor by phosphorylating Tyr223 in androgen recetpor76 and is essential for NF-κB activation of epidermal growth factor receptor.77 Some studies indicate that FER is an essential component of stem cell tyrosine kinase 177 and mast cell growth factor receptor (kit)78 signaling. Overexpression of FER is associated with poor clinical outcomes of several human malignancies.79–82
The fusion transcript contains the first 13 exons from MAN2A1 and the last 6 exons from FER. The fusion transcript is predicted to produce a chimera mannosidase α class 2A member (MAN2A1)-FER protein of 954 amino acids. The chimera protein retains the N-terminal glycoside hydrolase domain and α-mannosidase domain but loses the C-terminal glycoside hydrolase domain from MAN2A1. The fusion between MAN2A1 and FER also results in a large truncation of the FER regulatory domains, including SH2 and FCH domains, but retains an intact tyrosine protein kinase domain in the chimera protein. The absence of SH2 domain may convert a normally inactive FER kinase to a constitutively activated protein kinase. Furthermore, the signal peptide in MAN2A1 N-terminus may bring MAN2A1-FER to the Golgi apparatus. A misplaced and activated FER kinase may ectopically phosphorylate glycoproteins in the Golgi. This may lead to alterations of multiple signaling pathways initiated by transmembrane proteins. Only 5% of prostate cancers are positive for MAN2A1-FER fusion transcripts. However, 80% of prostate cancers positive for MAN2A1-FER experienced prostate cancer recurrence.15
Similar to MAN2A1-FER, SLC45A2-AMACR fusion also results from chromosome rearrangement of the short arm of chromosome 5. Fluorescence in situ hybridization indicates a loss of chromosome sequence corresponding to the 5′ end of AMACR in SLC45A2-AMACR fusion transcript-positive prostate cancer samples. SLC45A2 is a member of solute carrier that is highly expressed in melanoma. It is thought that SLC45A2 is involved in melanin metabolism. Mutation of SLC45A2 is associated with albinism.74 Little is known about its relation with other human malignancies. However, AMACR, a racemase involved in branch fatty acid metabolism, is a well-documented progrowth cancer marker.75,83 High levels of expression of AMACR were identified in numerous human malignancies, including prostate cancer.84–93 Knockdown of AMACR induced cell growth arrest and apoptosis.94
In SLC45A2-AMACR transcript, the first two exons of SLC45A2 join exons 2 to 5 of AMACR to produce a fusion transcript. The fusion transcript translates into a chimera protein that contains the first 187 amino acids from SLC45A2 at its N-terminus and 311 amino acids from AMACR at its C-terminus. The chimera protein has 7 of 12 transmembrane domains from SLC45A2 truncated but retains a largely intact racemase domain. In contrast to AMACR which is a protein residing in mitochondria, truncation of its signal peptide in the N-terminus of AMACR and the presence of plasma membrane localization signal from SLC45A2 in SLC45A2-AMACR imply a likely membranous and cytoplasmic localization of the chimera protein. The presence of an ectopic racemase may affect the fatty acid-related signaling in the cells. Because AMACR is one of the most frequent discovered biomarkers for human malignancies, it raises the possibility that some of the AMACR elevations are the results of fusion gene formation of AMACR attaching to a highly expressed housekeeping gene. Approximately 6.5% of prostate cancers are positive for SLC45A2-AMACR fusion, and 87% of these patients experienced chemical recurrence. Such low prevalence rate of SLC45A2-AMACR fusion suggests that AMACR may have other head gene partners to drive its expression.
Fusion Transcripts that Produce Wild-Type Tail Gene Protein but Have Truncation of Head Gene Protein
Several rare isoforms of TMPRSS2-ERG and TMPRSS2-ETV4 fusions and of TNPO1-IKBKB belong to this category. Such arrangement allows the tail gene expression under the control of head gene promoter. Because TMPRSS2 is driven by the promoter responding to androgen, the expression of ERG and ETV4 in these prostate cancers may play a bigger role in prostate cancer development. It would be of interest to compare treatment responses of these prostate cancers with cancers containing TMPRSS2-ERG and TMPRSS2-ETV4 fusions that result in functional loss of the proteins. CCNH-C5orf30 is another example of a fusion gene expressing the intact tail gene. Cyclin H (CCNH) belongs to the cyclin family. This cyclin binds CDK7 and MAT1 to form a complex to activate CDK2,95 which, in turn, promotes cell cycle entry into the S phase. Other studies suggest that CCNH also partners with RNA polymerase II and TFIIH and participates in transcription process.96,97 CCNH-C5orf30 fusion results in loss of 17 amino acids from the C-terminus of CCNH. This may have an impact on its activity in promoting cell cycle progression and transcription-related activity. C5orf30 encodes a novel protein of 206 amino acids that have high homology with UNC119 binding protein. Its function is largely unknown. Its sequence is conserved in most mammals. Thus, important physiologic function of this gene is expected.
Fusion Transcripts that Result in the Loss of Tail Gene Protein and the Truncation of Head Gene Protein
Most fusion transcripts do not result in in-frame translation of both head gene and tail gene partners into a chimera protein. The consequence of such fusion is the loss of tail gene protein because of the deletion of the ATG translation start codon in the tail gene and the C-terminal truncation of the head gene protein because of the loss of the 3′ end of the mRNA. This category of fusion transcripts includes most of the variants of ETS fusion transcripts and several newly discovered fusion genes. One important example is TRMT11-GRIK2, that is, a fusion between tRNA methyltransferase and glutamate receptor. The fusion creates a giant C-terminal truncation on TRMT11. Only 24 amino acids are translated from TRMT11. As a result, most of the methyl-transferase motifs in TRMT11 are lost. Because the ATG start codon of GRIK2 is deleted in the fusion transcript, there is likely no protein translation occurring for GRIK2 from the fusion gene RNA template. Interestingly, in samples in which TRMT11-GRIK2 was detected, there was no wild-type mRNA for either TRMT11 or GRIK2, whereas normal prostate tissues have moderate amounts of RNA for both,15 suggesting that the transcription of GRIK2 or TRMT11 comes from a single allele. TRMT11-GRIK2 fusion essentially results in functional loss of both genes. The presence of TRMT11-GRIK2 in prostate cancer signals high aggression of prostate cancer. Among the 20 patients of prostate cancers that were positive for TRMT11-GRIK2 fusion, no patient survived 5 years without prostate cancer recurrence. Most of these patients experienced rapid doubling of prostate-specific antigen during their recurrence. It is unclear whether this aggressive behavior is because of the loss of both TRMT11 and GRIK2 expression or because of the truncation of tRNA [guanine(10)-N2]-methyltransferase homolog (TRMT11) protein.
Another example of the loss of function of a tail gene is MTOR-TP53BP1. This fusion results from translocation of the short arm of chromosome 1 to the long arm of chromosome 15.15 The resulting MTOR-TP53BP1 transcript is predicted to produce a C-terminus–truncated mammalian target of rapamycin (MTOR) protein. The remaining 926 amino acids from the N-terminus of MTOR do not contain a known functional domain in the protein, whereas phosphatidylinositol 3-kinase and rapamycin-binding domains are deleted from the protein because of truncation. The fusion transcript has the ATG start codon deleted from the TP53BP1 gene; thus, it unlikely has viable translation occurring for the tumor suppressor p53-binding protein 1 (TP53BP1) protein, unless it proves that an alternative ATG start site exists for TP53BP1 translation. Approximately 4% prostate cancer samples are positive for MTOR-TP53BP1 fusion transcript. All proved highly aggressive and recurrent. Because MTOR is thought to be a central signaling molecule for cell growth and proliferation, the lack of MTOR signaling in these cancers suggests that cancer cells have an alternative mechanism to bypass the MTOR signaling pathway. MTOR-TP53BP1 fusion also represents a novel mechanism for inactivation of tumor suppressor TP53BP1.
Features that Distinguish Transcripts Resulting from Malignant Transformation from Those from Benign Tissues
Abundant evidence mentioned in Fusion Transcripts in Prostate Cancer suggests that fusion transcripts are present in normal prostate or other tissues.98 The presence of these fusion transcripts in healthy young adults indicates that these fusion transcripts may be physiologic and may perform important functions in prostate gland development. Many more fusion transcripts in normal OD prostate tissues were detected by deep transcriptome sequencing, but they have yet to be validated. If some of these fusion transcripts turn out to be authentic, this raises the question on how to classify fusion transcripts. Indeed, some of the previously thought cancer fusion transcripts such as TTTY15-USP9Y and SLC45A3-ELK4 are present in healthy young adults. It is important to identify the characteristics of fusion transcripts that are pathologic and are important for cancer development.
So far, the main features of cancer-specific fusion transcripts are the results of preformed chromosome rearrangement. This could be because of chromosome translocation, deletion, or isochromosome formation. Because most of the genome rearrangements occur in the intron regions, proper splicing is found in most cancer-specific fusion transcripts. Most cancer-specific fusion transcripts do not contain known intron sequences. In contrast to the features seen in cancer-related gene fusions, no known fusion transcripts detected in healthy persons contain chromosome rearrangement, even though cryptic chromosome rearrangement was identified in some of the healthy adult population.99 Many of the fusion transcripts represent continuous transcription of two adjacent genes. Even when trans fusion occurs in healthy tissues, there is no evidence of accompanying chromosome rearrangement.98 In some instances, intron or partial exon sequence is present in the transcript. This clearly indicates that fusion transcripts in normal tissues are the results of post-transcription events.
To classify cancer-specific fusion transcripts versus physiologic ones, one might consider the following criteria. i) For cancer-specific transcripts, the evidence of chromosome rearrangement involving the gene partners of the fusion transcript should be present, as provided by hybridization or sequencing results. ii) The breakpoint of a fusion transcript should not contain known intron or partial exon sequence, because the presence of intron sequence suggests post-transcription and RNA processing events rather than DNA-based alterations and is likely transient. iii) The fusion transcript is not known to be present in healthy persons. iv) If the fusion direction is cis, the distance between the breakpoints of the head gene and the tail gene should be not <100 kb, because transcripts overriding intergenic brakes are common in physiologic condition. For trans direction, no distance limitation is required.
Perspective of the Importance of Discovery of Fusion Transcripts in Prostate Cancer
Most fusion transcripts in prostate tissues may be physiologic, unless they are the products of chromosome rearrangement. The presence of these fusion transcripts increases the diversity of human gene structure and gene function. We speculate that mechanisms exist in the prostate tissues to regulate the formation of these fusion transcripts, and they may play important roles in organ development of the prostate gland. The most common fusion transcript TMPRSS2-ERG was found in up to 60% of prostate cancer and high-grade prostatic intraepithelial carcinoma. Recently, TMPRSS2-ERG fusion was also found in benign prostatic hyperplasia.100 This suggests that TMPRSS2-ERG chromosome rearrangement occurs in the early stage of prostate cancer development. Interestingly, with a few exceptions, most fusion transcripts discovered so far were found to lose the original ATG start site for the tail gene and had no chimera protein formation because of frame shift from the head gene translation or concomitant loss of the ATG start codon of the head gene. As a result, it is unlikely that an efficient translation would occur for the tail gene protein. Such arrangement suggests that only a limited amount of truncated protein or no protein for the tail gene would be produced. Because most head genes experience a large loss of amino acid sequences, it implies that most fusion transcripts in cancer cells represent shell RNA such that no effective translation can occur. These findings argue that the consequence of most fusion transcripts discovered in prostate cancer is the functional loss of both head gene and tail gene. Indeed, even with fusion protein formation, most functional domains of head gene or tail gene are eliminated after the fusion. The functions of these proteins are severely compromised or altered. The loss of two genes in one hit may produce a moderate or even serious imbalance in homeostasis of a cell, depending on the function of the genes. Additional hits are likely to be required for the development of prostate cancer. These hits could be additional chromosome rearrangement, mutations, copy number alterations, or epigenetic changes. Overall, regardless of whether fusion genes are loss or gain of functions, abnormal joining of two genes represents the most important and fundamental changes in the genome of cancer cells. They are probably the foundation and hallmark of prostate cancer development.
The challenge of interpreting the importance of gene fusions for the homeostasis of any cell type is immense. Analysis of the regulation of the regularly structured genome has exceeded the capabilities of most computer-based models tried to date. The added analysis of the importance of new abnormally present genes that result from fusion of normal genes adds a huge but exciting challenge. The understanding of this new complexity is likely to have strong implications for normal cell biology, for neoplastic development, and hopefully for development of new cancer treatment modalities. We should not forget that true cancer-driver gene fusions occur only in the cancer cells. This uniqueness is potentially the basis for new therapeutic applications, based on genome editing approaches or on new drugs that may finally succeed in affecting only the cancer cells, leaving normal cells intact.
Acknowledgments
We thank George Michalopoulos and Joel Nelson for their constructive comments and suggestions on the manuscript, Amantha Martin for editing, and Songyang Zheng for technical support.
J.-H.L. takes responsibility for the integrity of the article.
Footnotes
Supported by NIH grant RO1 CA098249 (J.-H.L.), American Cancer Society grant RSG-08-137-01-CNE (Y.P.Y.), and a grant from the University of Pittsburgh Cancer Institute.
Disclosures: None declared.
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Luo J.H., Yu Y.P., Cieply K., Lin F., Deflavia P., Dhir R., Finkelstein S., Michalopoulos G., Becich M. Gene expression analysis of prostate cancers. Mol Carcinog. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- 3.Luo J.H., Yu Y.P. Genetic factors underlying prostate cancer. Expert Rev Mol Med. 2003;5:1–26. doi: 10.1017/S1462399403006057. [DOI] [PubMed] [Google Scholar]
- 4.Yu Y.P., Landsittel D., Jing L., Nelson J., Ren B., Liu L., McDonald C., Thomas R., Dhir R., Finkelstein S., Michalopoulos G., Becich M., Luo J.H. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 5.Yu Y.P., Paranjpe S., Nelson J., Finkelstein S., Ren B., Kokkinakis D., Michalopoulos G., Luo J.H. High throughput screening of methylation status of genes in prostate cancer using an oligonucleotide methylation array. Carcinogenesis. 2005;26:471–479. doi: 10.1093/carcin/bgh310. [DOI] [PubMed] [Google Scholar]
- 6.Yu Y.P., Song C., Tseng G., Ren B.G., Laframboise W., Michalopoulos G., Nelson J., Luo J.H. Genome abnormalities precede prostate cancer and predict clinical relapse. Am J Pathol. 2012;180:2240–2248. doi: 10.1016/j.ajpath.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo J.H., Ding Y., Chen R., Michalopoulos G., Nelson J., Tseng G., Yu Y.P. Genome-wide methylation analysis of prostate tissues reveals global methylation patterns of prostate cancer. Am J Pathol. 2013;182:2028–2036. doi: 10.1016/j.ajpath.2013.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y.P., Ding Y., Chen R., Liao S.G., Ren B.G., Michalopoulos A., Michalopoulos G., Nelson J., Tseng G.C., Luo J.H. Whole-genome methylation sequencing reveals distinct impact of differential methylations on gene transcription in prostate cancer. Am J Pathol. 2013;183:1960–1970. doi: 10.1016/j.ajpath.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo J.H. Gene expression alterations in human prostate cancer. Drugs Today (Barc) 2002;38:713–719. doi: 10.1358/dot.2002.38.10.704653. [DOI] [PubMed] [Google Scholar]
- 10.Ren B., Yu Y.P., Tseng G.C., Wu C., Chen K., Rao U.N., Nelson J., Michalopoulos G.K., Luo J.H. Analysis of integrin alpha7 mutations in prostate cancer, liver cancer, glioblastoma multiforme, and leiomyosarcoma. J Natl Cancer Inst. 2007;99:868–880. doi: 10.1093/jnci/djk199. [DOI] [PubMed] [Google Scholar]
- 11.Lin F., Yu Y.P., Woods J., Cieply K., Gooding B., Finkelstein P., Dhir R., Krill D., Becich M.J., Michalopoulos G., Finkelstein S., Luo J.H. Myopodin, a synaptopodin homologue, is frequently deleted in invasive prostate cancers. Am J Pathol. 2001;159:1603–1612. doi: 10.1016/S0002-9440(10)63006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S., Arora V.K., Kaushik P., Cerami E., Reva B., Antipin Y., Mitsiades N., Landers T., Dolgalev I., Major J.E., Wilson M., Socci N.D., Lash A.E., Heguy A., Eastham J.A., Scher H.I., Reuter V.E., Scardino P.T., Sander C., Sawyers C.L., Gerald W.L. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes S., Yoshimoto M., Beheshti B., Houlston R.S., Squire J.A., Evans A. The use of whole genome amplification to study chromosomal changes in prostate cancer: insights into genome-wide signature of preneoplasia associated with cancer progression. BMC Genomics. 2006;7:65. doi: 10.1186/1471-2164-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baca S.C., Prandi D., Lawrence M.S., Mosquera J.M., Romanel A., Drier Y. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Y.P., Ding Y., Chen Z., Liu S., Michalopoulos A., Chen R., Gulzar Z.G., Yang B., Cieply K.M., Luvison A., Ren B.G., Brooks J.D., Jarrard D., Nelson J.B., Michalopoulos G.K., Tseng G.C., Luo J.H. Novel fusion transcripts associate with progressive prostate cancer. Am J Pathol. 2014;184:2840–2849. doi: 10.1016/j.ajpath.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K.H., Song G.J., Kang I.S., Kim S.W., Paick J.S., Chung C.H., Rhee K. Ubiquitin-specific protease activity of USP9Y, a male infertility gene on the Y chromosome. Reprod Fertil Dev. 2003;15:129–133. doi: 10.1071/rd03002. [DOI] [PubMed] [Google Scholar]
- 17.Ren S., Peng Z., Mao J.H., Yu Y., Yin C., Gao X., Cui Z., Zhang J., Yi K., Xu W., Chen C., Wang F., Guo X., Lu J., Yang J., Wei M., Tian Z., Guan Y., Tang L., Xu C., Wang L., Gao X., Tian W., Wang J., Yang H., Wang J., Sun Y. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012;22:806–821. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji P., Diederichs S., Wang W., Boing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E., Thomas M., Berdel W.E., Serve H., Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 20.Han Y., Liu Y., Nie L., Gui Y., Cai Z. Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology. 2013;81:209.e1–209.e7. doi: 10.1016/j.urology.2012.08.044. [DOI] [PubMed] [Google Scholar]
- 21.Jiang Y., Li Y., Fang S., Jiang B., Qin C., Xie P., Zhou G., Li G. The role of MALAT1 correlates with HPV in cervical cancer. Oncol Lett. 2014;7:2135–2141. doi: 10.3892/ol.2014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G., Zhang H., Wan X., Yang X., Zhu C., Wang A., He L., Miao R., Chen S., Zhao H. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014;2014:780521. doi: 10.1155/2014/780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutschner T., Hammerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M., Zornig M., MacLeod A.R., Spector D.L., Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H.M., Yang F.Q., Chen S.J., Che J., Zheng J.H. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947–2955. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 25.Luo J.H., Ren B., Keryanov S., Tseng G.C., Rao U.N., Monga S.P., Strom S., Demetris A.J., Nalesnik M., Yu Y.P., Ranganathan S., Michalopoulos G.K. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu C., Yang M., Tian J., Wang X., Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–175. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 27.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., Blencowe B.J., Prasanth S.G., Prasanth K.V. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripathi V., Shen Z., Chakraborty A., Giri S., Freier S.M., Wu X., Zhang Y., Gorospe M., Prasanth S.G., Lal A., Prasanth K.V. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown J.A., Valenstein M.L., Yario T.A., Tycowski K.T., Steitz J.A. Formation of triple-helical structures by the 3'-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang F., Yi F., Han X., Du Q., Liang Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013;587:3175–3181. doi: 10.1016/j.febslet.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 31.Wu X.S., Wang X.A., Wu W.G., Hu Y.P., Li M.L., Ding Q., Weng H., Shu Y.J., Liu T.Y., Jiang L., Cao Y., Bao R.F., Mu J.S., Tan Z.J., Tao F., Liu Y.B. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15:806–814. doi: 10.4161/cbt.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalik K.M., You X., Manavski Y., Doddaballapur A., Zornig M., Braun T., John D., Ponomareva Y., Chen W., Uchida S., Boon R.A., Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 33.Ji Q., Liu X., Fu X., Zhang L., Sui H., Zhou L., Sun J., Cai J., Qin J., Ren J., Li Q. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/beta-catenin signal pathway. PLoS One. 2013;8:e78700. doi: 10.1371/journal.pone.0078700. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Wang J., Su L., Chen X., Li P., Cai Q., Yu B., Liu B., Wu W., Zhu Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68:557–564. doi: 10.1016/j.biopha.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Dong Y., Liang G., Yuan B., Yang C., Gao R., Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 36.Wilusz J.E., JnBaptiste C.K., Lu L.Y., Kuhn C.D., Joshua-Tor L., Sharp P.A. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark J.P., Cooper C.S. ETS gene fusions in prostate cancer. Nat Rev Urol. 2009;6:429–439. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- 38.Feng F.Y., Brenner J.C., Hussain M., Chinnaiyan A.M. Molecular pathways: targeting ETS gene fusions in cancer. Clin Cancer Res. 2014;20:4442–4448. doi: 10.1158/1078-0432.CCR-13-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunn M.F., Seeburg P.H., Moscovici C., Duesberg P.H. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature. 1983;306:391–395. doi: 10.1038/306391a0. [DOI] [PubMed] [Google Scholar]
- 40.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J.E., Shah R.B., Pienta K.J., Rubin M.A., Chinnaiyan A.M. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 41.Iljin K., Wolf M., Edgren H., Gupta S., Kilpinen S., Skotheim R.I., Peltola M., Smit F., Verhaegh G., Schalken J., Nees M., Kallioniemi O. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res. 2006;66:10242–10246. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]
- 42.Attard G., Clark J., Ambroisine L., Mills I.G., Fisher G., Flohr P., Reid A., Edwards S., Kovacs G., Berney D., Foster C., Massie C.E., Fletcher A., De Bono J.S., Scardino P., Cuzick J., Cooper C.S., Transatlantic Prostate Group Heterogeneity and clinical significance of ETV1 translocations in human prostate cancer. Br J Cancer. 2008;99:314–320. doi: 10.1038/sj.bjc.6604472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han B., Mehra R., Dhanasekaran S.M., Yu J., Menon A., Lonigro R.J., Wang X., Gong Y., Wang L., Shankar S., Laxman B., Shah R.B., Varambally S., Palanisamy N., Tomlins S.A., Kumar-Sinha C., Chinnaiyan A.M. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68:7629–7637. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomlins S.A., Mehra R., Rhodes D.R., Smith L.R., Roulston D., Helgeson B.E., Cao X., Wei J.T., Rubin M.A., Shah R.B., Chinnaiyan A.M. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 45.Hermans K.G., Bressers A.A., van der Korput H.A., Dits N.F., Jenster G., Trapman J. Two unique novel prostate-specific and androgen-regulated fusion partners of ETV4 in prostate cancer. Cancer Res. 2008;68:3094–3098. doi: 10.1158/0008-5472.CAN-08-0198. [DOI] [PubMed] [Google Scholar]
- 46.Hermans K.G., van der Korput H.A., van Marion R., van de Wijngaart D.J., Ziel-van der Made A., Dits N.F., Boormans J.L., van der Kwast T.H., van Dekken H., Bangma C.H., Korsten H., Kraaij R., Jenster G., Trapman J. Truncated ETV1, fused to novel tissue-specific genes, and full-length ETV1 in prostate cancer. Cancer Res. 2008;68:7541–7549. doi: 10.1158/0008-5472.CAN-07-5930. [DOI] [PubMed] [Google Scholar]
- 47.Jhavar S., Reid A., Clark J., Kote-Jarai Z., Christmas T., Thompson A., Woodhouse C., Ogden C., Fisher C., Corbishley C., De-Bono J., Eeles R., Brewer D., Cooper C. Detection of TMPRSS2-ERG translocations in human prostate cancer by expression profiling using GeneChip Human Exon 1.0 ST arrays. J Mol Diagn. 2008;10:50–57. doi: 10.2353/jmoldx.2008.070085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Udager A.M., Shi Y., Tomlins S.A., Alva A., Siddiqui J., Cao X., Pienta K.J., Jiang H., Chinnaiyan A.M., Mehra R. Frequent discordance between ERG gene rearrangement and ERG protein expression in a rapid autopsy cohort of patients with lethal, metastatic, castration-resistant prostate cancer. Prostate. 2014;74:1199–1208. doi: 10.1002/pros.22836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomlins S.A., Laxman B., Dhanasekaran S.M., Helgeson B.E., Cao X., Morris D.S., Menon A., Jing X., Cao Q., Han B., Yu J., Wang L., Montie J.E., Rubin M.A., Pienta K.J., Roulston D., Shah R.B., Varambally S., Mehra R., Chinnaiyan A.M. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 50.Carver B.S., Tran J., Gopalan A., Chen Z., Shaikh S., Carracedo A., Alimonti A., Nardella C., Varmeh S., Scardino P.T., Cordon-Cardo C., Gerald W., Pandolfi P.P. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klezovitch O., Risk M., Coleman I., Lucas J.M., Null M., True L.D., Nelson P.S., Vasioukhin V. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.King J.C., Xu J., Wongvipat J., Hieronymus H., Carver B.S., Leung D.H., Taylor B.S., Sander C., Cardiff R.D., Couto S.S., Gerald W.L., Sawyers C.L. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zong Y., Xin L., Goldstein A.S., Lawson D.A., Teitell M.A., Witte O.N. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci U S A. 2009;106:12465–12470. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carver B.S., Tran J., Chen Z., Carracedo-Perez A., Alimonti A., Nardella C., Gopalan A., Scardino P.T., Cordon-Cardo C., Gerald W., Pandolfi P.P. ETS rearrangements and prostate cancer initiation. Nature. 2009;457:E1. doi: 10.1038/nature07738. discussion E2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cerveira N., Ribeiro F.R., Peixoto A., Costa V., Henrique R., Jeronimo C., Teixeira M.R. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8:826–832. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perner S., Mosquera J.M., Demichelis F., Hofer M.D., Paris P.L., Simko J., Collins C., Bismar T.A., Chinnaiyan A.M., De Marzo A.M., Rubin M.A. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 57.Mosquera J.M., Perner S., Genega E.M., Sanda M., Hofer M.D., Mertz K.D., Paris P.L., Simko J., Bismar T.A., Ayala G., Shah R.B., Loda M., Rubin M.A. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin Cancer Res. 2008;14:3380–3385. doi: 10.1158/1078-0432.CCR-07-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrovics G., Liu A., Shaheduzzaman S., Furusato B., Sun C., Chen Y., Nau M., Ravindranath L., Chen Y., Dobi A., Srikantan V., Sesterhenn I.A., McLeod D.G., Vahey M., Moul J.W., Srivastava S. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 59.Winnes M., Lissbrant E., Damber J.E., Stenman G. Molecular genetic analyses of the TMPRSS2-ERG and TMPRSS2-ETV1 gene fusions in 50 cases of prostate cancer. Oncol Rep. 2007;17:1033–1036. [PubMed] [Google Scholar]
- 60.Saramaki O.R., Harjula A.E., Martikainen P.M., Vessella R.L., Tammela T.L., Visakorpi T. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14:3395–3400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- 61.Lapointe J., Li C., Giacomini C.P., Salari K., Huang S., Wang P., Ferrari M., Hernandez-Boussard T., Brooks J.D., Pollack J.R. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- 62.Tu J.J., Rohan S., Kao J., Kitabayashi N., Mathew S., Chen Y.T. Gene fusions between TMPRSS2 and ETS family genes in prostate cancer: frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissues. Mod Pathol. 2007;20:921–928. doi: 10.1038/modpathol.3800903. [DOI] [PubMed] [Google Scholar]
- 63.Perner S., Demichelis F., Beroukhim R., Schmidt F.H., Mosquera J.M., Setlur S., Tchinda J., Tomlins S.A., Hofer M.D., Pienta K.G., Kuefer R., Vessella R., Sun X.W., Meyerson M., Lee C., Sellers W.R., Chinnaiyan A.M., Rubin M.A. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 64.Gopalan A., Leversha M.A., Satagopan J.M., Zhou Q., Al-Ahmadie H.A., Fine S.W., Eastham J.A., Scardino P.T., Scher H.I., Tickoo S.K., Reuter V.E., Gerald W.L. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–1406. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J., Cai Y., Ren C., Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 66.Nam R.K., Sugar L., Yang W., Srivastava S., Klotz L.H., Yang L.Y., Stanimirovic A., Encioiu E., Neill M., Loblaw D.A., Trachtenberg J., Narod S.A., Seth A. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97:1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajput A.B., Miller M.A., De Luca A., Boyd N., Leung S., Hurtado-Coll A., Fazli L., Jones E.C., Palmer J.B., Gleave M.E., Cox M.E., Huntsman D.G. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60:1238–1243. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Attard G., Clark J., Ambroisine L., Fisher G., Kovacs G., Flohr P., Berney D., Foster C.S., Fletcher A., Gerald W.L., Moller H., Reuter V., De Bono J.S., Scardino P., Cuzick J., Cooper C.S., Transatlantic Prostate Group Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshimoto M., Joshua A.M., Cunha I.W., Coudry R.A., Fonseca F.P., Ludkovski O., Zielenska M., Soares F.A., Squire J.A. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451–1460. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 70.Singareddy R., Semaan L., Conley-Lacomb M.K., St John J., Powell K., Iyer M., Smith D., Heilbrun L.K., Shi D., Sakr W., Cher M.L., Chinni S.R. Transcriptional regulation of CXCR4 in prostate cancer: significance of TMPRSS2-ERG fusions. Mol Cancer Res. 2013;11:1349–1361. doi: 10.1158/1541-7786.MCR-12-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu L., Zhao J.C., Kim J., Jin H.J., Wang C.Y., Yu J. ERG is a critical regulator of Wnt/LEF1 signaling in prostate cancer. Cancer Res. 2013;73:6068–6079. doi: 10.1158/0008-5472.CAN-13-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tian T.V., Tomavo N., Huot L., Flourens A., Bonnelye E., Flajollet S., Hot D., Leroy X., de Launoit Y., Duterque-Coquillaud M. Identification of novel TMPRSS2:ERG mechanisms in prostate cancer metastasis: involvement of MMP9 and PLXNA2. Oncogene. 2014;33:2204–2214. doi: 10.1038/onc.2013.176. [DOI] [PubMed] [Google Scholar]
- 73.Pflueger D., Terry S., Sboner A., Habegger L., Esgueva R., Lin P.C., Svensson M.A., Kitabayashi N., Moss B.J., MacDonald T.Y., Cao X., Barrette T., Tewari A.K., Chee M.S., Chinnaiyan A.M., Rickman D.S., Demichelis F., Gerstein M.B., Rubin M.A. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moremen K.W., Robbins P.W. Isolation, characterization, and expression of cDNAs encoding murine alpha-mannosidase II, a Golgi enzyme that controls conversion of high mannose to complex N-glycans. J Cell Biol. 1991;115:1521–1534. doi: 10.1083/jcb.115.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hao Q.L., Heisterkamp N., Groffen J. Isolation and sequence analysis of a novel human tyrosine kinase gene. Mol Cell Biol. 1989;9:1587–1593. doi: 10.1128/mcb.9.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rocha J., Zouanat F.Z., Zoubeidi A., Hamel L., Benidir T., Scarlata E., Brimo F., Aprikian A., Chevalier S. The Fer tyrosine kinase acts as a downstream interleukin-6 effector of androgen receptor activation in prostate cancer. Mol Cell Endocrinol. 2013;381:140–149. doi: 10.1016/j.mce.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 77.Guo C., Stark G.R. FER tyrosine kinase (FER) overexpression mediates resistance to quinacrine through EGF-dependent activation of NF-kappaB. Proc Natl Acad Sci U S A. 2011;108:7968–7973. doi: 10.1073/pnas.1105369108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Voisset E., Lopez S., Dubreuil P., De Sepulveda P. The tyrosine kinase FES is an essential effector of KITD816V proliferation signal. Blood. 2007;110:2593–2599. doi: 10.1182/blood-2007-02-076471. [DOI] [PubMed] [Google Scholar]
- 79.Ivanova I.A., Vermeulen J.F., Ercan C., Houthuijzen J.M., Saig F.A., Vlug E.J., van der Wall E., van Diest P.J., Vooijs M., Derksen P.W. FER kinase promotes breast cancer metastasis by regulating alpha6- and beta1-integrin-dependent cell adhesion and anoikis resistance. Oncogene. 2013;32:5582–5592. doi: 10.1038/onc.2013.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miyata Y., Kanda S., Sakai H., Greer P.A. Feline sarcoma-related protein expression correlates with malignant aggressiveness and poor prognosis in renal cell carcinoma. Cancer Sci. 2013;104:681–686. doi: 10.1111/cas.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ahn J., Truesdell P., Meens J., Kadish C., Yang X., Boag A.H., Craig A.W. Fer protein-tyrosine kinase promotes lung adenocarcinoma cell invasion and tumor metastasis. Mol Cancer Res. 2013;11:952–963. doi: 10.1158/1541-7786.MCR-13-0003-T. [DOI] [PubMed] [Google Scholar]
- 82.Li H., Ren Z., Kang X., Zhang L., Li X., Wang Y., Xue T., Shen Y., Liu Y. Identification of tyrosine-phosphorylated proteins associated with metastasis and functional analysis of FER in human hepatocellular carcinoma cells. BMC Cancer. 2009;9:366. doi: 10.1186/1471-2407-9-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krolewski J.J., Lee R., Eddy R., Shows T.B., Dalla-Favera R. Identification and chromosomal mapping of new human tyrosine kinase genes. Oncogene. 1990;5:277–282. [PubMed] [Google Scholar]
- 84.Luo J., Zha S., Gage W.R., Dunn T.A., Hicks J.L., Bennett C.J., Ewing C.M., Platz E.A., Ferdinandusse S., Wanders R.J., Trent J.M., Isaacs W.B., De Marzo A.M. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- 85.Lee Y.E., He H.L., Lee S.W., Chen T.J., Chang K.Y., Hsing C.H., Li C.F. AMACR overexpression as a poor prognostic factor in patients with nasopharyngeal carcinoma. Tumour Biol. 2014;35:7983–7991. doi: 10.1007/s13277-014-2065-z. [DOI] [PubMed] [Google Scholar]
- 86.Tretiakova M.S., Sahoo S., Takahashi M., Turkyilmaz M., Vogelzang N.J., Lin F., Krausz T., Teh B.T., Yang X.J. Expression of alpha-methylacyl-CoA racemase in papillary renal cell carcinoma. Am J Surg Pathol. 2004;28:69–76. doi: 10.1097/00000478-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 87.Fellegara G., Gabba S., Dorji T., De Luca G., Colecchia M. Observations on Aron et al's “Utility of a triple antibody cocktail intraurothelial neoplasm-3 (IUN-3 CK20/CD44s/p53) and alpha-methylacyl-CoA racemase (AMACR) in the distinction of urothelial carcinoma in situ (CIS) and reactive urothelial atypia”. Am J Surg Pathol. 2014;38:1013–1015. doi: 10.1097/PAS.0000000000000191. [DOI] [PubMed] [Google Scholar]
- 88.Shilo K., Dracheva T., Mani H., Fukuoka J., Sesterhenn I.A., Chu W.S., Shih J.H., Jen J., Travis W.D., Franks T.J. Alpha-methylacyl CoA racemase in pulmonary adenocarcinoma, squamous cell carcinoma, and neuroendocrine tumors: expression and survival analysis. Arch Pathol Lab Med. 2007;131:1555–1560. doi: 10.5858/2007-131-1555-MCRIPA. [DOI] [PubMed] [Google Scholar]
- 89.Scheil-Bertram S., Lorenz D., Ell C., Sheremet E., Fisseler-Eckhoff A. Expression of alpha-methylacyl coenzyme A racemase in the dysplasia carcinoma sequence associated with Barrett's esophagus. Mod Pathol. 2008;21:961–967. doi: 10.1038/modpathol.2008.73. [DOI] [PubMed] [Google Scholar]
- 90.Huang W., Zhao J., Li L., Huang Y., Yang X., Wang J., Zhang T. a-Methylacyl coenzyme A racemase is highly expressed in the intestinal-type adenocarcinoma and high-grade dysplasia lesions of the stomach. Histol Histopathol. 2008;23:1315–1320. doi: 10.14670/HH-23.1315. [DOI] [PubMed] [Google Scholar]
- 91.Li W., Cagle P.T., Botero R.C., Liang J.J., Zhang Z., Tan D. Significance of overexpression of alpha methylacyl-coenzyme A racemase in hepatocellular carcinoma. J Exp Clin Cancer Res. 2008;27:2. doi: 10.1186/1756-9966-27-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ha Y.S., Kim Y.W., Min B.D., Lee O.J., Kim Y.J., Yun S.J., Lee S.C., Kim W.J. Alpha-methylacyl-coenzyme a racemase-expressing urachal adenocarcinoma of the abdominal wall. Korean J Urol. 2010;51:498–500. doi: 10.4111/kju.2010.51.7.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang Z., Fanger G.R., Banner B.F., Woda B.A., Algate P., Dresser K., Xu J., Reed S.G., Rock K.L., Chu P.G. A dietary enzyme: alpha-methylacyl-CoA racemase/P504S is overexpressed in colon carcinoma. Cancer Detect Prev. 2003;27:422–426. doi: 10.1016/j.cdp.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 94.Zha S., Ferdinandusse S., Denis S., Wanders R.J., Ewing C.M., Luo J., De Marzo A.M., Isaacs W.B. Alpha-methylacyl-CoA racemase as an androgen-independent growth modifier in prostate cancer. Cancer Res. 2003;63:7365–7376. [PubMed] [Google Scholar]
- 95.Fisher R.P., Morgan D.O. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 96.Shiekhattar R., Mermelstein F., Fisher R.P., Drapkin R., Dynlacht B., Wessling H.C., Morgan D.O., Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 97.Makela T.P., Parvin J.D., Kim J., Huber L.J., Sharp P.A., Weinberg R.A. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc Natl Acad Sci U S A. 1995;92:5174–5178. doi: 10.1073/pnas.92.11.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jividen K., Li H. Chimeric RNAs generated by intergenic splicing in normal and cancer cells. Genes Chromosomes Cancer. 2014;53:963–971. doi: 10.1002/gcc.22207. [DOI] [PubMed] [Google Scholar]
- 99.Gajecka M., Glotzbach C.D., Jarmuz M., Ballif B.C., Shaffer L.G. Identification of cryptic imbalance in phenotypically normal and abnormal translocation carriers. Eur J Hum Genet. 2006;14:1255–1262. doi: 10.1038/sj.ejhg.5201710. [DOI] [PubMed] [Google Scholar]
- 100.Velaeti S., Dimitriadis E., Kontogianni-Katsarou K., Savvani A., Sdrolia E., Pantazi G., Stefanakis S., Trangas T., Pandis N., Petraki K. Detection of TMPRSS2-ERG fusion gene in benign prostatic hyperplasia. Tumour Biol. 2014;35:9597–9602. doi: 10.1007/s13277-014-2250-0. [DOI] [PubMed] [Google Scholar]