Abstract
Cell fusion likely drives tumor evolution by undermining chromosomal and DNA stability and/or by generating phenotypic diversity; however, whether a cell fusion event can initiate malignancy and direct tumor evolution is unknown. We report that a fusion event involving normal, nontransformed, cytogenetically stable epithelial cells can initiate chromosomal instability, DNA damage, cell transformation, and malignancy. Clonal analysis of fused cells reveals that the karyotypic and phenotypic potential of tumors formed by cell fusion is established immediately or within a few cell divisions after the fusion event, without further ongoing genetic and phenotypic plasticity, and that subsequent evolution of such tumors reflects selection from the initial diverse population rather than ongoing plasticity of the progeny. Thus, one cell fusion event can both initiate malignancy and fuel evolution of the tumor that ensues.
The multiple genetic changes that convert a normal cell to a malignant cell likely occur in one of the following two pathways: the pathway involving the accretion of point mutations with or without ensuing chromosomal damage over time1–4 or the pathway involving a catastrophic event causing manifold genetic changes, including those underlying malignant transformation.5–7 Inherited defects in DNA repair, exposure to ionizing radiation, and infection with oncogenic viruses accelerate the accumulation of multiple discrete mutations or DNA damage and hence the development of cancer.4 However, inheritance, infection, or instantaneous exposure to an environmental carcinogen cannot explain the inception of most cancers. Hence, identification of discrete events that cause normal cells to undergo oncogenesis remains a compelling challenge.
For many years, cell fusion has been considered in theory an appealing explanation for oncogenesis. Cell fusion can be detected in existing cancers.8–10 Cell fusion can generate aneuploidy, chromosomal instability, and DNA damage, all of which might cause multiple genetic changes and cancer.11–19 Cell fusion might explain how terminally differentiated, nonproliferating cells initiate tumors.11,13,20
However, cell fusion by itself has never been proven to initiate malignancy. Lack of such proof reflects the exigencies of experimental systems used for analysis of karyotype and malignant transformation (ie, proliferation of parent and fused cells over multiple generations). Formation of tumors has never been found to occur as a consequence of spontaneous fusion of cells in whole animal systems.14,15,21–25 Therefore, the question of whether cell fusion can initiate cancer remains a matter of speculation.
We tested whether cell fusion can initiate tumors using IE-6 cells. Originally isolated as outgrowths from fragments of rat intestine,26 IEC-6 cells are considered the archetype of normal intestinal crypt epithelial cells.26–28 As in normal crypt epithelium, the proliferation and differentiation of IEC-6 cells are likely governed by the caudal type homeobox genes Cdx1 and Cdx2,29–31 and the cells have a stable karyotype, nontransformed phenotype and are unable to form tumors at repeated passage in cultures.26–28 To generate sufficient numbers of fused and unfused cells, we used polyethylene glycol (PEG), which does not promote and in fact might inhibit oncogenesis,32 to facilitate fusion and isolated and cloned the ensuing hybrids. Analysis of these clones reveals that cell fusion engenders aneuploidy, DNA damage, phenotypic heterogeneity, transformation, and the capacity to form tumors and that these properties are established immediately or within a few cell divisions after the fusion event.
Materials and Methods
Animal Study
The use of animals was in accord with all guidelines of and was approved by the University of Michigan Committee on Use and Care of Animals and the National Institutes of Health.
Cell Cultures
IEC-6 cells (ATCC, Manassas, VA) cultured from epithelial cell outgrowths from fragments of intestine of germ-free rats as epithelial cell cultured in the presence of collagenase were used.26 The cells were cultivated in Dulbecco’s modified Eagle’s medium with 0.1 U/mL of bovine insulin and 10% heat inactivated fetal bovine serum in a humidified, 37°C incubator containing 10% carbon dioxide. HeLa cells (ATCC) from adenocarcinoma of the cervix33 were cultivated in Dulbecco’s modified Eagle’s medium with 10% heat inactivated fetal bovine serum.
Cell Fusion and Sorting
Cells were grown to approximately 80% confluence and then labeled with 10 μM carboxyfluorescein diacetate succinimidyl ester (CFSE) or with carboxylic acid acetate succinimidyl ester (SNARF-1) (Invitrogen, Grand Island, NY), which have distinct spectral properties. After entering cells, CFSE and SNARF-1 undergo chemical reactions that cause the dyes to be retained in cytoplasm. Labeled cells were released with trypsin, admixed, and cultured in petri dishes for 4 hours to allow the cells to adhere and spread. Mixtures of differentially labeled cells examined at this time revealed no evidence of admixing of the dyes. To facilitate fusion, 50% PEG (USB Corp, Cleveland, OH) was added for 1 minute, and the cells were washed three times with Dulbecco’s modified Eagle’s medium. Cells were then released with trypsin and analyzed by fluorescence activated cell sorting (FACS) (FACSDiVa Cell Sorter, BD Biosciences San Jose, CA).34 Cells with a relatively large postfusion size were gated using forward scatter (FSC) A versus side scatter (SSC) A plot. After eliminating doublets on FSC-A vs FSC-W plots, fused cells emitting both carboxyfluorescein diacetate succinimidyl ester and SNARF-1 were single cell sorted by FACS and placed into individual wells. Nonfused cells, defined as PEG-treated cells emitting fluorescence of either CFSE or SNARF-1 with the size of individual IEC-6 cells, were also sorted and placed in individual wells. The presence of both dyes within fused cells and one dye in nonfused cells was confirmed by fluorescence microscopy (DMI 6000B; Leica, Buffalo Grove, IL). In some experiments, IEC-6 cells cultured as described above but not treated with PEG were used as controls.
Chromosomal Labeling and Fusion of HeLa Cells
To trace the fate of chromosomal DNA after fusion of cells, biosynthetically labeled HeLa cells were used in lieu of IEC-6 cells, which are irreversibly damaged by labeling. HeLa cells were synchronized at S phase by treatment with 5 mmol/L thymidine (Sigma-Aldrich, St Louis, MO) for 15 hours and then biosynthetically labeled with 0.05 mmol/L Cy3- or Cy5-dUTP (GE/Amersham, Piscataway, NJ) by exposure to approximately 200-μm glass beads (Sigma-Aldrich) and scraping.35 Labeled cells were co-cultured and fused with PEG and fusion ascertained by fluorescence microscopy (ApoTome; Zeiss, Oberkochen, Germany).
Live Cell Imaging
After fusion with PEG, labeled HeLa cells were transferred to the WeatherStation environmental chamber (37°C, 5% carbon dioxide) of the DeltaVision RT Image Restoration Microscopy System (Applied Precision, Issaquah, WA). Time-lapse images were acquired every 10 minutes using SoftWoRx software version 3.5.1 (Applied Precision).
Karyotype Analysis
Cells were pretreated with 0.1 μg/mL of demecolcine (Sigma-Aldrich) for 2 hours and then trypsinized and suspended in 0.56% (w/v) potassium chloride for 15 minutes. The cells were fixed in methanol and acetic acid (3:1), stained with DAPI, to approximate G-band pattern.36 Images were acquired with a fluorescence microscope (DMI 6000B; Leica).
Analysis of Cell Transformation
Cell transformation was evaluated by assays for loss of contact inhibition (focus formation) and acquisition of ability to proliferate unattached to a surface (colony formation in soft agar). The analysis of focus formation assay was performed using cells cultured for 2 weeks after confluence. Soft agar assay was performed by suspending 1 × 104 cells in 0.5% agar per well in 6-well plates and culturing for 3 weeks. Foci and colonies were photographed through an inverted microscope (DM IL; Leica) and a digital camera (QICAM; QImaging, Surrey, BC).
Immunofluorescence Analysis
To evaluate DNA damage and the cellular response it can provoke, we assayed and localized phosphohistone H2A.X (γ-H2AX), which marks double-strand DNA breaks, tumor protein p53, and caspase 3 by indirect immunofluorescence. Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline for 15 minutes, permeabilized with 0.1% Triton X-100 for 5 minutes, blocked in 2% bovine serum albumin, and incubated with mouse monoclonal anti–γ-H2AX (JBW301; Millipore, Billerica, MA) or anti-p53 (PAb 240; BD Biosciences) or with rabbit anti-cleaved caspase 3 (ab52181; Abcam, Cambridge, MA). Binding was detected using Alexa-488 or -594 conjugated goat anti-mouse IgG (Invitrogen). Images were acquired using a fluorescence microscope (DMI 6000B; Leica). Controls were prepared as described except primary antibodies were omitted.
Single-Cell Gel Electrophoresis (Comet Assay)
DNA damage was assayed in part by neutral comet assay that detects migration of DNA fragments in an electrical field. When DNA damage has occurred, the fragments resemble the tail of a comet. The neutral comet assay was performed according to the manufacturer's instructions (Trevigen, Gaithersburg, MD). Cells suspended in LMAgarose at 37°C were loaded onto CometSlides. After solidification at 4°C in the dark, cells were treated with cold lysis buffer for 1 hour and then with cold neutral electrophoresis buffer for 30 minutes. Electrophoresis was performed at 21 V for 45 minutes at 4°C. Nuclei and broken DNA fragments were visualized using SYBR Green by fluorescence microscopy (DMI 6000B; Leica). Images were analyzed with the ImageJ software version 1.44p (NIH, Bethesda, MD).
Analysis of Tumor Formation
Female immunodeficient mice (NOD.Cg-PrkdcscidIl2rgtm1Sug/JicTac, NOG; Taconic, Hudson, NY)37 were injected subcutaneously with 2 × 106 cells suspended in 67% serum-depleted Matrigel (BD Biosciences). Tumor dimensions were measured weekly with calipers and volumes estimated by the modified ellipsoidal formula38: .
Mice were sacrificed at 12 weeks or when tumor volume approached 1 cm.3
Histologic Analysis
Pieces of tumor were fixed in buffered formalin and embedded in paraffin. Sections prepared and stained with hematoxylin and eosin.
Results
Generation and Cloning of Fused and Nonfused Cells
To explore cell fusion and oncogenesis, we used a line of rat intestinal epithelial cells (IEC-6) that exhibits the phenotype and functions of normal intestinal epithelium.26 The cells retain the appearance and behavior of primary explants of crypt epithelial cells, including a stable diploid karyotype (42 chromosomes), anchorage-dependent growth, and arrest of growth at confluence during repeated passages for 6 months, and do not initiate tumors in syngeneic animals.
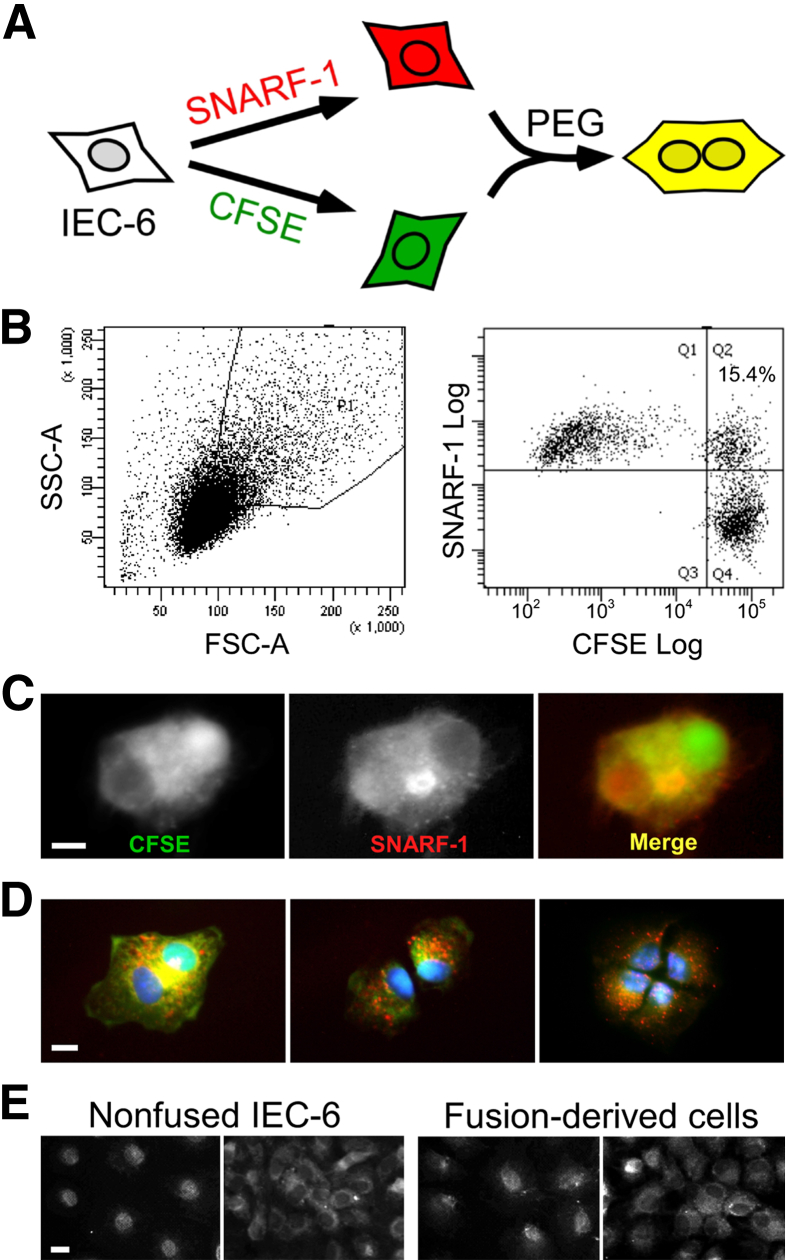
IEC-6 cells were labeled with CFSE or SNARF-1 and fused by brief exposure to 50% PEG (Figure 1A). After treatment with PEG and washing, the cells were analyzed by FACS. Cells emitting fluorescence both of CFSE (green) and SNARF-1 (red) were single cell sorted by FACS and placed into individual wells. Dual labeling was verified by fluorescence microscopy (Figure 1, B and C). In control preparations, differentially labeled cells in confluent cultures remained distinct, and repeated harvesting and replating in the absence of PEG generated no appreciable admixture of dyes.
Figure 1.
Identification, isolation, and cloning of fused cells. Rat intestinal epithelial cells (IEC-6 cells) were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) (green) or carboxylic acid acetate succinimidyl ester (SNARF-1) (red) and fused using 50% polyethylene glycol (PEG). Fused and nonfused cells identified based on size, nuclear contents, and fluorescence were isolated and single-cell sorted by fluorescence activated cell sorting (FACS). A: Schematic illustrating isolation of fused cells. B: Identification and sorting by FACS of fused and nonfused cells. Cells with a relatively large postfusion size were gated using forward scatter (FSC)-A versus side scatter (SSC)-A plots (left panel). After eliminating doublets on FSC-A versus FSC-W plots (not shown), fused cells emitting both CFSE and SNARF-1 were single-cell sorted by FACS and placed into individual wells (right panel). C: Typical image of fused cell sorted by FACS and visualized by fluorescence microscopy. The fused cell emits both CFSE and SNARF-1 fluorescence; the image represents at least 50 cells examined per experiment. D: Mitosis of fused cells. Images are of a fused cell undergoing initial two cell divisions are representative of 20 cells per experiment. Green, CFSE; red, SNARF-1; blue, DAPI. E: Localization of p53 protein in clones of nonfused and fusion-derived IEC-6 cells. Early passage cells from nonfused clones and fusion-derived clones were stained with monoclonal antibodies specific for p53. Representative images of p53 staining at low cell density (left panel) or high cell density (right panel). Scale bars: 5 μm (C and D); 10 μm (E).
We explored the proliferative potential of cells that had undergone fusion. Repeated examination of fused cells by fluorescence microscopy revealed that 62% of 92 fused IEC-6 cells completed one cell division within 24 hours and 9% completed two cell divisions within 48 hours (Figure 1D). These results are consistent with observations that although many tetraploid cells often do not proliferate, owing to failed chromosomal pairing in mitosis,39,40 a fraction of tetraploid mammalian cells can nonetheless complete mitosis.41,42
Ability of Fused Cells to Establish Clones
Nineteen percent (248 of 1305) of fused IEC-6 cells sorted as single cells by FACS into conditioned medium established clones, hereafter termed fusion-derived clones.
PEG-treated IEC-6 cells emitting fluorescence of one fluorophore with normal size (ie, smaller than fused cells) were single cell sorted and cloned. These clones are hereafter termed nonfused clones. The nonfused clones may have been originated by fusion of cells carrying the same fluorophore and were excluded based on the fluorescence intensity and size (fused cells emit more fluorescence and are larger).
Expression and Localization of Tumor Suppressor p53 after Cell Fusion
Because p53 controls the fate of tetraploid and aneuploid cells40,43,44 and regulates cell cycling in IEC-6 cells,27,45 we examined expression and localization of this protein in fusion-derived and nonfused clones at low and high density. Fusion-derived and nonfused clones had detectable p53 protein localized to the nucleus at low cell density and to the cytoplasm at high cell density, typical of normal cells (Figure 1E).
Cell Fusion and Aneuploidy
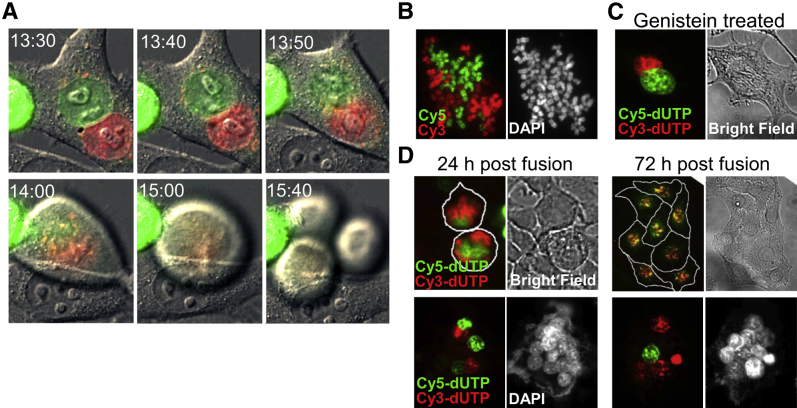
To determine how fusion-derived cells inherit chromosomes from parental cells, we biosynthetically labeled parental chromosomes with either Cy3- or Cy5-dUTP, fused the labeled cells, and monitored nuclear fluorescence for up to 72 hours. These experiments were performed using HeLa cells in lieu of IEC-6 cells because few IEC-6 cells survive labeling in this way. Fusion of differentially labeled HeLa cells generated cells that contained two distinct nuclei, the DNA of which remained entirely separate until mitosis had begun and the nuclear envelope was disassembled (Figure 2, A and B). Mitosis of the fused cells generated synkaryons, that is, mononuclear cells with chromosomal DNA from both fusion partners. Mitosis of fused cells was sometimes asymmetric, the daughter cells receiving unbalanced division of parental chromosomes, possibly owing to multipolar cell division (Figure 2A).
Figure 2.
Admixture of chromosomes and formation of synkaryons require mitosis. HeLa cells were biosynthetically labeled with either Cy3-dUTP or Cy5-dUTP delivered by transient disruption of cell membranes during exposure to glass beads. Samples of differentially labeled cells were mixed and fused with polyethylene glycol (PEG). A: Time-lapse images show changes in nuclear morphology typically observed after fusion of cells. The two nuclei did not fuse during interphase, but during prometaphase the nuclear envelopes fragmented simultaneously (13:50) and only then did the nuclear contents intermix (14:00). The mixed chromosomes were then segregated into three daughter cells. The images are representative of observations of 20 fusion events in two independent experiments. B: Representative images of metaphase chromosome spread of fusion-derived cells: the merged image of Cy3 and Cy5 florescence (right panel) and the DAPI chromosome counterstain (left panel). Images are representative of 50 cells studied in two independent experiments. C: Impact of cell cycle arrest on admixture of chromosomes after cell fusion. HeLa cells were labeled and fused as described and then cultured for 2 days with 60 μmol/L genistein to arrest cell cycle progression at G2/M. Nuclear fusion and/or admixing of chromosomal DNA is not observed in cell cycle–arrested fusion cells in 50 fusion events studied in two independent experiments: the merge of Cy3 and Cy5 florescence imaging (right panel) and bright field of the same fused cell (left panel). D: Impact of the number of cell fusion events on the fate of fused cells. HeLa cells were labeled with Cy3-dUTP or Cy5-dUTP, fused with PEG as described, and studied by fluorescence and bright field microscopy at various times thereafter. The top panel shows two synkaryons (or daughter cells of one synkaryon) that contain nuclear dyes at 24 hours; at 72 hours the cells have proliferated, and the eight daughter cells contain both dyes. The bottom panel shows a multinucleated giant cell formed by fusion of differentially labeled parental cells. At 24 hours, the giant cell has multiple nuclei; some are distinctly labeled, some are unlabeled, but none of the nuclei contain both labels, indicating the nuclear envelopes are intact. At 72 hours this (or a similar) cell contains multiple nuclei; some are labeled, and some are unlabeled and exhibit no admixture of the labels. Red, Cy3-dUTP; green, Cy5-dUTP. The images, and particularly lack of admixture of labels, are typical of observations of 60 fusion events studied in three independent experiments.
To determine whether admixture of chromosomal DNA and formation of synkaryons requires mitosis or might instead reflect fusion of nuclei, we tested whether genistein, which arrests cell cycle progression at G2/M,46 would impede synkaryon formation. Fused cells treated with genistein maintained intact nuclei and did not form synkaryons (Figure 2C). The experiments also indicated that the proliferative potential of fusion-derived cells depended on the number of nuclei. Synkaryons could proliferate and initiate clones; however, fusion-derived cells with more than two nuclei rarely underwent mitosis, and the nuclear contents remained separate (Figure 2D).
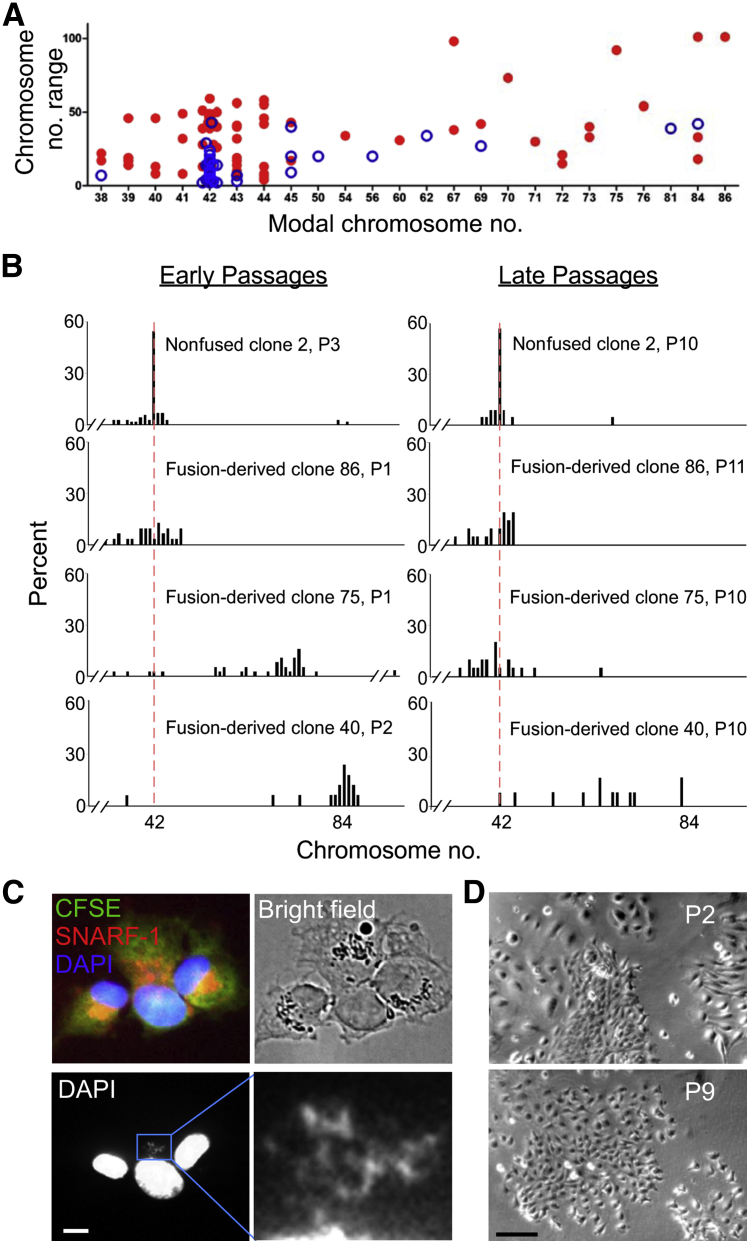
We next asked whether cell fusion engenders aneuploidy by analyzing karyotypes of newly established clones. Of 79 fusion-derived (IEC-6) clones studied, 32 (41%) were aneuploid, 44 (56%) near diploid (modal chromosome numbers 40 to 44), and 3 (4%) tetraploid (modal chromosome number 84) (Figure 3A). In contrast, nonfused clones were predominantly diploid [62 (86%) of 72 clones], with only eight (11%) being aneuploid and one tetraploid and one hypodiploid (Figure 3A). Fusion-derived clones also exhibited a greater difference between the maximum and the minimum numbers of chromosomes than nonfused clones (P < 0.0001, t-test) (Figure 3A). Fusion-derived clones with a modal chromosome number of 42 (2N) had, on average, a range of 30 chromosomes, indicating the presence of frequent aneuploid cells in each clone (Figure 3A). Nonfused clones with modal 2N = 42 had on average a range of 9.9 chromosomes (P < 10−10, t-test). Gross chromosomal translocations, however, were not observed in the newly established clones derived from fused or nonfused cells (data not shown).
Figure 3.
Chromosome number and cell morphology during serial passage of fusion-derived clones and nonfused clones. A: Range versus modal numbers of chromosomes in newly established nonfused and fusion-derived clones. IEC-6 cells have a stable mode of 2N = 42 (not shown). At least 15 metaphase spreads were evaluated for each clone. The range of chromosome numbers was taken as the difference between the maximum and minimum chromosome numbers of 90% of metaphase chromosome spreads in a clone. Blue circles represent nonfused clones (n = 72). Red dots represent fusion-derived clones (n = 79). B: Changes in chromosome number during repeated passage of fusion-derived clones and nonfused clones. Chromosome numbers at passages 1 to 3 (P1, P2, or P3) and passages 10 or 11 (P10 or P11) are shown. The red dashed line denotes 2N (42). Twenty metaphase spreads were analyzed for each clone. Chromosome numbers >95 are not shown in the plots. C: Unequal segregation during the second mitotic division of a fused-cell clone. Box denotes region shown at higher magnification. Green, carboxyfluorescein diacetate succinimidyl ester (CFSE); red, carboxylic acid acetate succinimidyl ester (SNARF-1); blue DAPI. D: Variation in size and morphology of cells in fusion-derived clones. At passage 2 (P2), cells from fusion-derived clone 75 exhibit three distinct sizes and morphologies; large cells on the left appear separated, smaller cells in the middle are closely clustered, and cells of intermediate size and spindle shape are on the right. At passage 9 (P9), the cells exhibit uniform size and appearance. Scale bars: 5 μm (C); 100 μm (D). Original magnification: ×866 (C, boxed area); ×72 (D).
We next determined whether chromosome number remained stable over time. Nonfused clones generally exhibited a normal karyotype at early (passages 1 to 3) and later (passages 10 to 11) passages (Figure 3B), like unmanipulated IEC-6 cells. Fusion-derived clones that were near diploid at early passage generally remained so during 10 to 11 passages (Figure 3B). In contrast, the number of chromosomes in fusion-derived clones near triploid or tetraploid at early passage usually decreased with repeated passage, with 6 of 15 becoming near diploid (Figure 3B), 9 of 15 having >2N chromosomes but fewer than the number at early passage (Figure 3B). These results indicate that cell fusion generates karyotypic instability, with the changes sometimes resolving toward near diploidy but often persisting. Whether reversion of the karyotype toward diploidy reflects selection or ongoing plasticity is not clear.
Chromosome missegregation could underlie the transition from near tetraploidy to aneuploidy after cell fusion.40,42,47,48 Consistent with this possibility, chromosomes in fusion-derived cells did not always segregate evenly into daughter cells. Figure 3C shows an example of unequal segregation during the second division of a fused cell: one daughter cell inherited most of the chromosomes, forming a much larger nucleus, whereas the other inherited only a few loosely packed chromosomes.
The volume of mammalian cells is thought to be proportional to ploidy.49 To test whether that relationship might apply with dynamic change in chromosome numbers, we examined the size of cells in clone 75, which had 36 to 126 chromosomes (mode 75) at passage 1 and a near-diploid number at passage 10 (Figure 3B). Consistent with the concept, clone 75 exhibited at least three distinct cell sizes and shapes at early passage but a predominantly small size at late passage (Figure 3D).
Cell Fusion and DNA Double-Strand Breaks
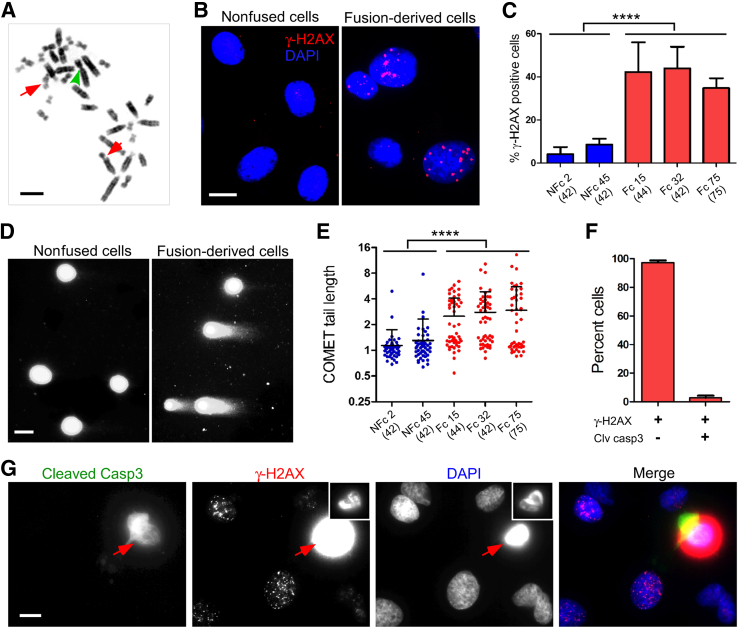
Aneuploidy and chromosomal rearrangements are commonly seen in cancer.50 The newly established fusion-derived clones often exhibited aneuploidy, as indicated above, but not gross chromosomal rearrangements. However, at passage 11 of clone 15, Robertsonian translocations, fusion of the long arms of two acrocentric chromosomes, were observed in 9 (11%) of 83 of cells (Figure 4A) and chromosomal breaks (red arrows) and translocation were observed in 2 cells (2%).
Figure 4.
Association of chromosome rearrangement and DNA damage in fusion-derived cells. A: Representative image of chromosomal aberrations. The metaphase chromosome spread was prepared from fusion-derived clone (Fc) 15 at passage 11. The arrowhead points to fusion between the long arms of two acrocentric chromosomes (ie, Robertsonian translocation); arrows denote chromosome breaks. Robertsonian translocations were observed in 9 (11%) of 83 cells and chromosomal breaks and translocation in 2 cells (2%). B: DNA damage detected by localization of phosphohistone H2A.X (γ-H2AX) in nonfusion clones (NFcs) and Fcs. Cells at early passage were fixed and stained with antibodies specific for γ-H2AX and studied by immunofluorescence microscopy. Nuclei were counterstained with DAPI. C: Frequency of DNA damage. DNA damage was detected by nuclear staining for γ-H2AX in NFcs andFcs. The bars indicate means ± SD of the percentage of γ-H2AX–positive cells in nine images from three independent experiments, analyzing 156 to 360 cells in total for each clone. The number in parentheses is the modal chromosome number. D: DNA double-strand breaks detected by neutral comet assay. Cells from NFcs and Fcs were embedded in agarose, lyzed, subjected to single-cell electrophoresis, and stained with CYBR gold. Representative images are shown. E: DNA damage detected by comet assay. NFcs and Fcs were compared using comet assay. Comet tail length is expressed as units relative to tail length/mean radius of intact nuclei in nonfused cells. Fifty randomly selected cells of NFcs or Fcs were analyzed in two independent experiments. F: Relationship between binding of γ-H2AX and presence of cleaved caspase 3 (casp3) in fusion-derived cells. Cells of Fc32 were fixed and stained with antibodies specific for γ-H2AX and with antibodies specific for cleaved casp3. Three hundred γ-H2AX–positive cells were examined for cleaved casp3 staining in three independent experiments. The results indicate that few cells with bound γ-H2AX are undergoing apoptosis. G: Appearance of cells positive for γ-H2AX and cleaved casp3. The photomicrograph shows a cell typical of those positive for cleaved casp3 and diffuse γ-H2AX (arrows). Condensed chromatin probably reflects apoptosis (insets: reduced exposure of γ-H2AX and DAPI staining). Cells exhibiting speckled staining for γ-H2A, no detectable cleaved caspase 3, and fine chromatin are far more common. ∗∗∗∗P < 0.0001, one-way analysis of variance, indicating a significant difference between NFcs and Fcs.
Aneuploidy in tumors is often associated with DNA damage,48,51 which could presage rearrangement. To test whether fusion-derived clones sustained DNA damage, we determined the frequency and extent of double-strand DNA breaks in fusion-derived clones with modal chromosome numbers of 42, 44, and 75 at early passage. Phosphorylated (Ser 139) γ-H2AX, which clusters at the sites of double-strand DNA breaks, was localized using monoclonal antibodies.52 Multiple clusters of γ-H2AX were detected in the nuclei of 35% to 42% of cells from fusion-derived clones, whereas only 4% to 9% of cells of nonfused clones had such clusters (P < 0.0001, one-way analysis of variance) (Figure 4, B and C). This finding was confirmed by neutral comet assays, which detect migration of DNA fragments in an electrical field (Figure 4D).53 On average, 48% of fusion-derived cells exhibited a prominent COMET tail (>2-tail unit), representing migration beyond the nucleus, as against only 5% of nonfused cells. The comet tails were more frequent in fusion-derived clones, and the comet tail lengths were on average 2.3-fold longer than the lengths measured in nonfused clones (P < 0.0001, one-way analysis of variance) (Figure 4, D and E).
DNA damage exceeding the capacity for repair causes apoptosis. To determine whether the DNA damage revealed by γ-H2AX and comet assays reflected apoptosis, we assayed caspase 3 activation. Only 3% of γ-H2AX–positive cells in fused cell clones contained activated caspase 3, and these had condensed chromatin and more intense and diffuse staining for γ-H2AX (Figure 4, F and G). Thus, the DNA damage detected in fusion-derived cells did not reflect apoptosis.
Cell Fusion and Transformation
Chromosomal instability is clearly associated with cell transformation and oncogenesis5,42; however, whether cell fusion incites, marks, or even suppresses transformation is unclear.54,55 Because unmanipulated IEC-6 cells are nontransformed and have a stable karyotype, whereas fused IEC-6 cells exhibit chromosomal instability, we asked whether fusion also induces transformation.
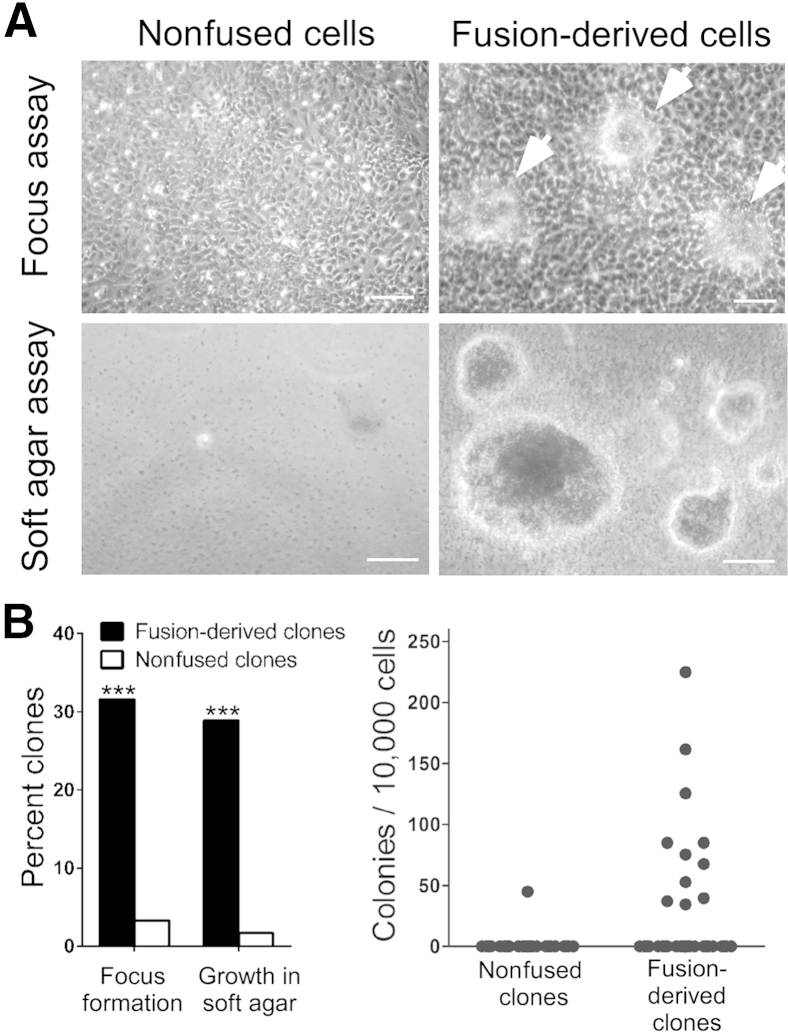
Twelve (32%) of 38 fusion-derived clones, after 12 passages at low cell density, lost cell contact inhibition, assayed by focus formation, forming discrete foci of stacked cells (Figure 5). Similarly, 11 (29%) of 38 exhibited anchorage-independent growth, assayed by colony formation in soft agar. In contrast, only 2 (3%) of 60 nonfused clones formed foci (P < 0.001, χ2 test), and only 1 exhibited anchorage-independent growth (Figure 5). However, transformation was not apparently caused by aneuploidy because clones with high modal chromosome numbers at early passage were no more likely to be transformed than clones with near diploidy (P = 0.142, χ2 test). Thus, after cell fusion, aneuploidy and transformation are independent events, at least in this system.
Figure 5.
Cell fusion and transformation. A: Comparison of nonfused cells and fusion-derived cells in focus formation and soft agar colony formation assays for cell transformation. In focus formation assays performed at passage day 12, 14 days after confluence, cells from most fusion-derived clones usually form foci (arrowheads); cells from nonfused clones do not. In colony formation assays performed 3 weeks after seeding in soft agar, cells from fused cell clones form colonies (representing subclones), cells from nonfused clones do not. B: Frequency of focus formation and soft agar colony formation of nonfused clones and fusion-derived clones. Percentage of 60 nonfused clones and 38 fusion-derived clones exhibiting focus formation or formation of colonies (>100 μm) in soft agar (left panel). Numbers of colonies (>100 μm) in soft agar after inoculating 104 cells per clone (right panel). ∗∗∗P < 0.001, χ2 test, indicating the difference between nonfused clones and fusion-derived clones. Scale bar = 100 μm (A).
Although p53 protein was detected early after cell fusion (Figure 1E), censoring of DNA damage and/or chromosomal instability might have been compromised by mutation of TP53. To address that possibility, as much as possible, we compared the region of rat Tp53 homologous to human exons 5 to 8 in which mutations are usually found in tumors.56 The sequences from the nine colonies were identical with wild type, making it highly improbable that variants caused transformation in these cells. Consistent with this conclusion and with the images in Figure 1E, the levels of p53 protein in transformed fusion-derived cells were equivalent to those in nonfused IEC-6 cells (data not shown).
Cell Fusion and Tumor Formation
We next asked whether cell fusion promotes tumor formation. Two million cells from a pool of fused, but not cloned, IEC-6 cells were injected in the flanks of immunodeficient (NOD.Cg-PrkdcscidIl2rgtm1Sug/JicTac) mice,37 and the mice were monitored for 12 weeks for formation of tumors. Of 18 such injections, 11 (61%) generated tumors (Figure 6A). In contrast, neither 2 × 106 unmodified IEC-6 cells nor 2 × 106 cells from each of three nonfused clones formed tumors [P < 0.001, χ2 (1, N = 36) = 15.84] (Figure 6A). Thus, cell fusion is associated with oncogenesis.
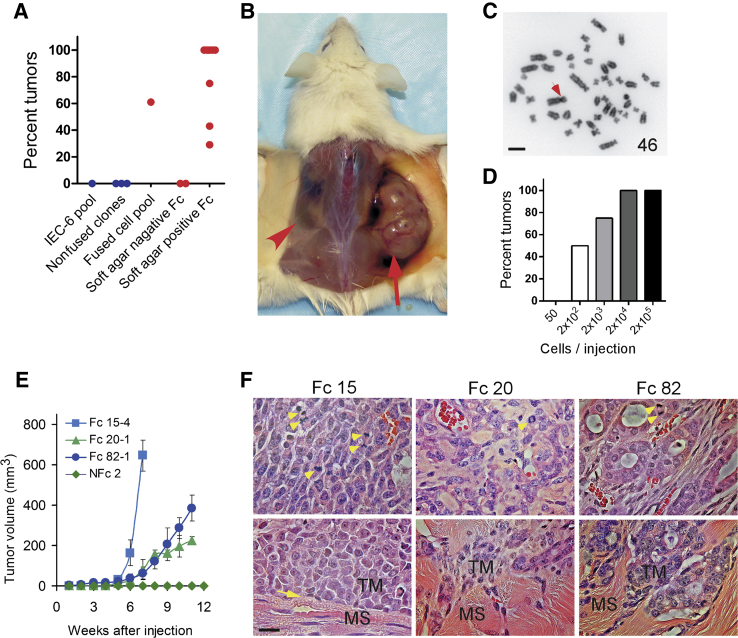
Figure 6.
Cell fusion and tumor formation. A: Frequency of tumor formation after injection of 2 × 106 cells from fused or nonfused clones in immunodeficient mice. Unmanipulated IEC-6 cells, nonfused clones, a pool of fused cells, and fusion-derived clones that did or did not form colonies in soft agar were inoculated subcutaneously into flanks and axillae of immunodeficient (NOD.Cg-PrkdcscidIl2rgtm1Sug/JicTac) mice, and the frequency of tumors, identified by palpation and confirmed by histology, within 12 weeks was noted. Each dot represents at least four injection sites and indicates the percentage of injection sites at which tumors developed. Results for nonfused cells and clones are shown in blue. Results for fused cells and clones are shown in red. Results represent two independent experiments. B: Tumor (arrow) 7 weeks after injection of 2 × 106 cells from fusion clone 15. No tumor formed in the opposite flank injected with of 2 × 106 nonfused clone 28 cells (arrowhead). C: Cytogenetic analysis of cells isolated from the tumor shown in B. The number indicates chromosome counts. The arrow denotes a Robertsonian translocation. Chromosomes from 20 cells at metaphase were analyzed. D: Frequency of tumor formation as a function of number of cells injected. Bars represent the percentage of injection sites at which tumors formed within 12 weeks after injection of various numbers of cells from fusion clone 15. At least four sites were injected for each number of cells tested. E: Rate of growth of tumors formed from various clones of fused and nonfused cells. Labeled IEC-6 cells were treated with polyethylene glycol, and fused and nonfused cells were isolated and cloned by fluorescence activated cell sorting. Samples of each clone (2 × 106 cells) were injected into each of at least four sites, and the size of the ensuing tumors was measured weekly. Tumor volumes were estimated as 1/2 (length × width2) and depicted as means ± SD. F: Histology and invasiveness of tumors originated by cell fusion. Tumors formed after implantation of fusion-derived clones 15 (Fc 15), 20 (Fc 20), and 82 (Fc 82) were sectioned and stained with hematoxylin and eosin. Upper panels show red blood cells in each tumor and moderate focal glandular differentiation in the tumor formed by Fc 82. Arrowheads indicate mitotic cells. Lower panels show invasiveness of tumors. Note a clear border (arrow) between muscle fibers in tumor derived from Fc 15: invasiveness of Fc 20 and Fc 82 tumor cells into muscle layers. MS, muscle fibers; TM, tumor cells.
We next asked whether the capacity of fused cell clones to form tumors preceded or followed introduction of the cells into immunodeficient hosts. Nine fusion-derived clones that had undergone transformation generated tumors within 12 weeks, six at every injection site (Figure 6, A and B). In contrast, two fusion-derived clones that were demonstrably not transformed failed to generate tumors at any site [P < 0.001, χ2 (1, N = 60) = 25.91] (Figure 6A). The tumors did not appear to result from cytogenetic changes arising during culture or after injection (including fusion with murine cells) because the karyotypes of tumor cells were the same as those of the fusion-derived clones used. Figure 6C provides one example in which tumor cells exhibited the same aberrations in chromosome number and structure as the fusion-derived clone from which the tumor originated. These results indicate that cell fusion leading to transformation generates a high probability of tumor formation, whereas cell fusion not leading to transformation does not. The results also suggest that ongoing chromosomal instability is neither common nor essential for tumor formation once transformation has occurred.
We next evaluated the relative frequency of oncogenesis in fusion-derived cells. Aliquots of 200 cells generated tumors in approximately 50% and aliquots of 2 × 104 cells in 100% of injection sites within 12 weeks (Figure 6D). This frequency of tumor formation suggests that cell fusion generates populations of cells in which approximately 1 of 200 can have the capacity to form tumors.
The aggressiveness of tumors and capacity to metastasize appear to evolve over time.4,57–60 Such evolution might reflect i) ongoing chromosomal genomic instability and phenotypic plasticity; ii) subsequent catastrophic events, such as homotypic or heterotypic fusion; or iii) ongoing selection from a diverse population of cells established during oncogenesis. To determine whether one or several of these mechanisms operate in the system used, we asked whether the biological properties of tumors become more diverse, reflecting the first two mechanisms, or less diverse, reflecting the third, over time.
As one test, we examined the growth rates and histology of tumors formed by cells harvested from soft agar colonies (subclones) of three fusion-derived clones. Tumors arising from the same subclone exhibited similar rates of growth (n = 6 for each subclone) and histology (n = 6 for each subclone), whereas tumors arising from different subclones exhibited distinct rates of growth and histology (Figure 6, E and F). For example, 2 × 106 cells expanded from a soft agar colony of fusion-derived clone 15 generated rapidly growing tumors at all injected sites, whereas 2 × 106 cells from clones 20 and 82 formed slowly growing tumors. Samples from fusion clone 15 generated high-grade undifferentiated tumors, consisting of cells with large nuclei and little stroma and exhibiting little or no local invasiveness (a clear border between tumor cells and muscle fibers) at each of eight injection sites (Figure 6F). Fusion clone 20 formed tumors that grew more slowly than tumors that originated from clone 15 and exhibited a high degree of invasiveness (Figure 6F). Clone 20 generated poorly differentiated tumors that contained cells with large nuclei surrounded by a large amount of stroma. Fusion clone 82 generated tumors that also grew more slowly than tumors from clone 15 and exhibited moderate focal glandular differentiation, consistent with low-grade adenocarcinomas but a high degree of invasiveness (Figure 6F). Mitotic figures and angiogenesis, small capillaries invading the tumor, were observed in all tumors (Figure 6F). The distinct and stable properties of tumors generated by different clones (established at the time of fusion) suggested these properties were established by events taking place at fusion or during early passages, rather than by ongoing diversification and evolution.
Discussion
Aberrant fusion of normal cells offers an appealing mechanism for catastrophic change in the genome and hence for oncogenesis. By causing tetraploidy, and possibly allowing endoreplication or failure of cytokinesis, cell fusion might initiate chromosomal instability and oncogenesis.51,61,62 However, cell fusion, by itself, has never been found to originate cancer.11,20 We report that normal diploid cells, deliberately fused, can undergo cellular and chromosomal changes typical of cancer and can exhibit transformation and the capacity to form tumors. We trace these changes to diverse clonal populations established in proximity to the fusion event.
The fusion of normal cells during development eventuates in marked changes in morphology, biochemistry, and biology of the progeny.12,63 Fusion of cells in injured, inflamed, and/or infected tissues occurs often and is postulated to promote repair and host defense.19,64,65 However, in development and disease, tight cellular and molecular regulation and censoring of progeny with aneuploidy, DNA damage, or replicative stress is thought to avoid untoward consequences.43,44,63,66
The paucity of direct experimental evidence that cell fusion per se can originate malignancy reflects in part the regulation of fusion and censoring of defective progeny. However, even the most tightly regulated systems fail, if only rarely, and hence the question of whether cell fusion by itself can initiate tumors remains. We addressed that question using an experimental system that avoided some limitations mentioned above. We used IEC-6 intestinal epithelial cells, which repeatedly have proven to be subject to the normal regulation of genome integrity and cell replication, to maintain a stable karyotype and to fail to form tumors over many generations. We fused these cells using PEG, which is nonmutagenic67 and inhibits transformation and oncogenesis of epithelial cells.32,68 Although we cannot exclude the possibility that IEC-6 cells harbor some defect that renders them uniquely susceptible to transformation and oncogenesis caused by PEG and/or cell fusion distinct from other normal cells, such a defect has not been described previously. Therefore, our finding that the fusion of these cells can induce transformation and malignancy and that these changes occur abruptly on fusion provides a step toward identifying controls of cell replication and genetic integrity vulnerable in this common event.
Once formed, tumors are thought to evolve via one of the three pathways. In one pathway, ongoing chromosomal and genomic instability generates phenotypic plasticity and the most adaptable and/or prolific of which expand, thus modifying the repertoire of tumor cells over time.4,58–60 Another pathway of tumor evolution involves repeated episodes of manifold genetic change generating bursts of heterogeneity from which the best-adapted cells are selected.69 A third pathway involves ongoing selection of malignant cells from a diverse population generated by a catastrophic event, such as cell fusion.7,69 The stability or reversion toward diploidy of clonal karyotypes in the system we explored and the findings that rate of growth and histopathology were established within the first passages after cell fusion are most consistent with the third mechanism: ongoing selection from a diverse population established by a catastrophic event, such as cell fusion. These findings are consistent with genomic analyses of human colorectal cancers, suggesting a genomic crisis followed by a relative stability of tumor genome.69
The temporal sequence of events linking cell fusion with the karyotypic changes and oncogenesis is still uncertain. The double-strand DNA damage we detected might suggest chromothripsis, the abrupt fragmentation or shattering and rejoining in one or more chromosomes, as a driving mechanism.7,69,70 However, some fused cell clones exhibited normal karyotypes, and approximately half of the cells in clones with abnormal karyotypes had no detectable DNA damage. Tetraploid binuclear cells may become tetraploid mononuclear cells through nuclear fusion or synchronous mitosis. We observed only synchronous mitosis followed by cytokinesis and did not observe nuclear fusion (or exchange of chromosomal DNA between nuclei). Because the mononuclear hybrid cells are usually indistinguishable microscopically from normal diploid cells, the frequency of cell fusion in cancer and other conditions may be underestimated.
Acknowledgments
We thank Beth Moore for discussions and Lucas Stein, Ines Silva, and Shoichiro Tsuji for technical assistance.
X.Z. designed and conducted and/or supervised experiments, interpreted results, and wrote the manuscript; K.M. and W.L. performed experiments and interpreted results; J.P.G. evaluated pathology; M.C. critically analyzed and interpreted results; J.L.P. conceived, initiated, and supervised the research, interpreted results, and edited the manuscript. J.L.P. takes responsibility for the accuracy and integrity of the work.
Footnotes
See related Commentary on page 1846
Supported by NIH grants HL52297 (J.P.) and T32HL007749 (X.Z.) and the University of Michigan Comprehensive Cancer Center.
Disclosures: None declared.
References
- 1.Land H., Parada L.F., Weinberg R.A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stratton M.R., Campbell P.J., Futreal P.A. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davoli T., de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 6.Li R., Sonik A., Stindl R., Rasnick D., Duesberg P. Aneuploidy vs. gene mutation hypothesis of cancer: recent study claims mutation but is found to support aneuploidy. Proc Natl Acad Sci U S A. 2000;97:3236–3241. doi: 10.1073/pnas.040529797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., McLaren S., Lin M.L., McBride D.J., Varela I., Nik-Zainal S., Leroy C., Jia M., Menzies A., Butler A.P., Teague J.W., Quail M.A., Burton J., Swerdlow H., Carter N.P., Morsberger L.A., Iacobuzio-Donahue C., Follows G.A., Green A.R., Flanagan A.M., Stratton M.R., Futreal P.A., Campbell P.J. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty A., Lazova R., Davies S., Backvall H., Ponten F., Brash D., Pawelek J. Donor DNA in a renal cell carcinoma metastasis from a bone marrow transplant recipient. Bone Marrow Transplant. 2004;34:183–186. doi: 10.1038/sj.bmt.1704547. [DOI] [PubMed] [Google Scholar]
- 9.Yilmaz Y., Lazova R., Qumsiyeh M., Cooper D., Pawelek J. Donor Y chromosome in renal carcinoma cells of a female BMT recipient: visualization of putative BMT-tumor hybrids by FISH. Bone Marrow Transplant. 2005;35:1021–1024. doi: 10.1038/sj.bmt.1704939. [DOI] [PubMed] [Google Scholar]
- 10.Lazova R., Laberge G.S., Duvall E., Spoelstra N., Klump V., Sznol M., Cooper D., Spritz R.A., Chang J.T., Pawelek J.M. A Melanoma Brain Metastasis with a Donor-Patient Hybrid Genome following Bone Marrow Transplantation: first Evidence for Fusion in Human Cancer. PLoS One. 2013;8:e66731. doi: 10.1371/journal.pone.0066731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duelli D., Lazebnik Y. Cell-to-cell fusion as a link between viruses and cancer. Nat Rev Cancer. 2007;7:968–976. doi: 10.1038/nrc2272. [DOI] [PubMed] [Google Scholar]
- 12.Ogle B.M., Cascalho M., Platt J.L. Biological implications of cell fusion. Nat Rev Mol Cell Biol. 2005;6:567–575. doi: 10.1038/nrm1678. [DOI] [PubMed] [Google Scholar]
- 13.Bjerkvig R., Tysnes B.B., Aboody K.S., Najbauer J., Terzis A.J. Opinion: the origin of the cancer stem cell: current controversies and new insights. Nat Rev Cancer. 2005;5:899–904. doi: 10.1038/nrc1740. [DOI] [PubMed] [Google Scholar]
- 14.Rizvi A.Z., Swain J.R., Davies P.S., Bailey A.S., Decker A.D., Willenbring H., Grompe M., Fleming W.H., Wong M.H. Bone marrow-derived cells fuse with normal and transformed intestinal stem cells. Proc Natl Acad Sci U S A. 2006;103:6321–6325. doi: 10.1073/pnas.0508593103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogle B.M., Butters K.B., Plummer T.B., Ring K.R., Knudsen B., Litzow M.R., Cascalho M., Platt J.L. Spontaneous fusion of cells between species yields transdifferentiation and retroviral in vivo. FASEB J. 2004;18:548–550. doi: 10.1096/fj.03-0962fje. [DOI] [PubMed] [Google Scholar]
- 16.Harkness T., Weaver B.A., Alexander C.M., Ogle B.M. Cell fusion in tumor development: accelerated genetic evolution. Crit Rev Oncog. 2013;18:19–42. doi: 10.1615/critrevoncog.v18.i1-2.30. [DOI] [PubMed] [Google Scholar]
- 17.Powell A.E., Anderson E.C., Davies P.S., Silk A.D., Pelz C., Impey S., Wong M.H. Fusion between Intestinal epithelial cells and macrophages in a cancer context results in nuclear reprogramming. Cancer Res. 2011;71:1497–1505. doi: 10.1158/0008-5472.CAN-10-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duelli D., Lazebnik Y. Cell fusion: a hidden enemy? Cancer Cell. 2003;3:445–448. doi: 10.1016/s1535-6108(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 19.Berndt B., Zanker K.S., Dittmar T. Cell fusion is a potent inducer of aneuploidy and drug resistance in tumor cell/normal cell hybrids. Crit Rev Oncog. 2013;18:97–113. doi: 10.1615/critrevoncog.v18.i1-2.60. [DOI] [PubMed] [Google Scholar]
- 20.Lu X., Kang Y. Cell fusion as a hidden force in tumor progression. Cancer Res. 2009;69:8536–8539. doi: 10.1158/0008-5472.CAN-09-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogle B.M., Knudsen B.E., Nishitai R., Ogata K., Platt J.L. Toward the development of human T cells in swine for potential use in adoptive T cell immunotherapy. Tissue Eng. 2009;15:1031–1040. doi: 10.1089/ten.tea.2008.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies P.S., Powell A.E., Swain J.R., Wong M.H. Inflammation and proliferation act together to mediate intestinal cell fusion. PLoS One. 2009;4:e6530. doi: 10.1371/journal.pone.0006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan A.W., Hickey R.D., Paulk N.K., Culberson A.J., Olson S.B., Finegold M.J., Grompe M. Ploidy reductions in murine fusion-derived hepatocytes. PLoS Genet. 2009;5:e1000385. doi: 10.1371/journal.pgen.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nygren J.M., Liuba K., Breitbach M., Stott S., Thoren L., Roell W., Geisen C., Sasse P., Kirik D., Bjorklund A., Nerlov C., Fleischmann B.K., Jovinge S., Jacobsen S.E. Myeloid and lymphoid contribution to non-haematopoietic lineages through irradiation-induced heterotypic cell fusion. Nat Cell Biol. 2008;10:584–592. doi: 10.1038/ncb1721. [DOI] [PubMed] [Google Scholar]
- 25.Wang X., Willenbring H., Akkari Y., Torimaru Y., Foster M., Al-Dhalimy M., Lagasse E., Finegold M., Olson S., Grompe M. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature. 2003;422:897–900. doi: 10.1038/nature01531. [DOI] [PubMed] [Google Scholar]
- 26.Quaroni A., Wands J., Trelstad R.L., Isselbacher K.J. Epithelioid cell cultures from rat small intestine: characterization by morphologic and immunologic criteria. J Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boucher M.J., Jean D., Vezina A., Rivard N. Dual role of MEK/ERK signaling in senescence and transformation of intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2004;286:G736–G746. doi: 10.1152/ajpgi.00453.2003. [DOI] [PubMed] [Google Scholar]
- 28.Duhamel S., Hebert J., Gaboury L., Bouchard A., Simon R., Sauter G., Basik M., Meloche S. Sef downregulation by Ras causes MEK1/2 to become aberrantly nuclear localized leading to polyploidy and neoplastic transformation. Cancer Res. 2012;72:626–635. doi: 10.1158/0008-5472.CAN-11-2126. [DOI] [PubMed] [Google Scholar]
- 29.Soubeyran P., Andre F., Lissitzky J.C., Mallo G.V., Moucadel V., Roccabianca M., Rechreche H., Marvaldi J., Dikic I., Dagorn J.C., Iovanna J.L. Cdx1 promotes differentiation in a rat intestinal epithelial cell line. Gastroenterology. 1999;117:1326–1338. doi: 10.1016/s0016-5085(99)70283-0. [DOI] [PubMed] [Google Scholar]
- 30.Moucadel V., Totaro M.S., Dell C.D., Soubeyran P., Dagorn J.C., Freund J.N., Iovanna J.L. The homeobox gene Cdx1 belongs to the p53-p21(WAF)-Bcl-2 network in intestinal epithelial cells. Biochem Biophys Res Commun. 2002;297:607–615. doi: 10.1016/s0006-291x(02)02250-7. [DOI] [PubMed] [Google Scholar]
- 31.Patterson A.P., Chen Z., Rubin D.C., Moucadel V., Iovanna J.L., Brewer H.B., Jr., Eggerman T.L. Developmental regulation of apolipoprotein B mRNA editing is an autonomous function of small intestine involving homeobox gene Cdx1. J Biol Chem. 2003;278:7600–7606. doi: 10.1074/jbc.M201601200. [DOI] [PubMed] [Google Scholar]
- 32.Bailon P., Won C.Y. PEG-modified biopharmaceuticals. Expert Opin Drug Deliv. 2009;6:1–16. doi: 10.1517/17425240802650568. [DOI] [PubMed] [Google Scholar]
- 33.Scherer W.F., Syverton J.T., Gey G.O. Studies on the propagation in vitro of poliomyelitis viruses, IV: viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med. 1953;97:695–710. doi: 10.1084/jem.97.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaroszeski M.J., Gilbert R., Heller R. Detection and quantitation of cell-cell electrofusion products by flow cytometry. Anal Biochem. 1994;216:271–275. doi: 10.1006/abio.1994.1041. [DOI] [PubMed] [Google Scholar]
- 35.Manders E.M., Kimura H., Cook P.R. Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J Cell Biol. 1999;144:813–821. doi: 10.1083/jcb.144.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimonjic D.B., Rezanka L., DiPaolo J.A., Popescu N.C. Refined localization of the erbB-3 proto-oncogene by direct visualization of FISH signals on LUT-inverted and contrast-enhanced digital images of DAPI-banded chromosomes. Cancer Genet Cytogenet. 1995;80:100–102. doi: 10.1016/0165-4608(94)00161-4. [DOI] [PubMed] [Google Scholar]
- 37.Ito M., Hiramatsu H., Kobayashi K., Suzue K., Kawahata M., Hioki K., Ueyama Y., Koyanagi Y., Sugamura K., Tsuji K., Heike T., Nakahata T. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002;100:3175–3182. doi: 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- 38.Tomayko M.M., Reynolds C.P. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 39.Uetake Y., Sluder G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint.”. J Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi Q., Zhao X., Huang Y., Ma T., Zhang Y., Hou H., Cooke H.J., Yang D.Q., Wu M., Shi Q. p53 dependent centrosome clustering prevents multipolar mitosis in tetraploid cells. PLoS One. 2011;6:e27304. doi: 10.1371/journal.pone.0027304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujiwara T., Bandi M., Nitta M., Ivanova E.V., Bronson R.T., Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 42.Ganem N.J., Godinho S.A., Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreassen P.R., Lohez O.D., Lacroix F.B., Margolis R.L. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson S.L., Compton D.A. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L., Li J., Rao J.N., Li M., Bass B.L., Wang J.Y. Inhibition of polyamine synthesis induces p53 gene expression but not apoptosis. Am J Physiol. 1999;276:C946–C954. doi: 10.1152/ajpcell.1999.276.4.C946. [DOI] [PubMed] [Google Scholar]
- 46.Matsukawa Y., Marui N., Sakai T., Satomi Y., Yoshida M., Matsumoto K., Nishino H., Aoike A. Genistein arrests cell cycle progression at G2-M. Cancer Res. 1993;53:1328–1331. [PubMed] [Google Scholar]
- 47.Vitale I., Senovilla L., Jemaa M., Michaud M., Galluzzi L., Kepp O., Nanty L., Criollo A., Rello-Varona S., Manic G., Metivier D., Vivet S., Tajeddine N., Joza N., Valent A., Castedo M., Kroemer G. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J. 2010;29:1272–1284. doi: 10.1038/emboj.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burrell R.A., McClelland S.E., Endesfelder D., Groth P., Weller M.C., Shaikh N., Domingo E., Kanu N., Dewhurst S.M., Gronroos E., Chew S.K., Rowan A.J., Schenk A., Sheffer M., Howell M., Kschischo M., Behrens A., Helleday T., Bartek J., Tomlinson I.P., Swanton C. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris M. Polyploid series of mammalian cells. Exp Cell Res. 1971;66:329–336. doi: 10.1016/0014-4827(71)90685-9. [DOI] [PubMed] [Google Scholar]
- 50.Gordon D.J., Resio B., Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 51.Janssen A., van der Burg M., Szuhai K., Kops G.J., Medema R.H. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 52.Bonner W.M., Redon C.E., Dickey J.S., Nakamura A.J., Sedelnikova O.A., Solier S., Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins A.R. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 54.Schvartzman J.M., Sotillo R., Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holland A.J., Cleveland D.W. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petitjean A., Mathe E., Kato S., Ishioka C., Tavtigian S.V., Hainaut P., Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 57.Cahill D.P., Kinzler K.W., Vogelstein B., Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–60. [PubMed] [Google Scholar]
- 58.Loeb L.A. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lengauer C., Kinzler K.W., Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 60.Shah S.P., Roth A., Goya R., Oloumi A., Ha G., Zhao Y. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crasta K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weaver B.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 63.Oren-Suissa M., Podbilewicz B. Cell fusion during development. Trends Cell Biol. 2007;17:537–546. doi: 10.1016/j.tcb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 64.Duelli D.M., Padilla-Nash H.M., Berman D., Murphy K.M., Ried T., Lazebnik Y. A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr Biol. 2007;17:431–437. doi: 10.1016/j.cub.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 65.Clawson G.A. Cancer: fusion for moving. Science. 2013;342:699–700. doi: 10.1126/science.1244270. [DOI] [PubMed] [Google Scholar]
- 66.Zhou X., Platt J.L. Molecular and cellular mechanisms of mammalian cell fusion. Adv Exp Med Biol. 2011;713:33–64. doi: 10.1007/978-94-007-0763-4_4. [DOI] [PubMed] [Google Scholar]
- 67.Wangenheim J., Bolcsfoldi G. Mouse lymphoma L5178Y thymidine kinase locus assay of 50 compounds. Mutagenesis. 1988;3:193–205. doi: 10.1093/mutage/3.3.193. [DOI] [PubMed] [Google Scholar]
- 68.Corpet D.E., Parnaud G., Delverdier M., Peiffer G., Tache S. Consistent and fast inhibition of colon carcinogenesis by polyethylene glycol in mice and rats given various carcinogens. Cancer Res. 2000;60:3160–3164. [PubMed] [Google Scholar]
- 69.Kloosterman W.P., Hoogstraat M., Paling O., Tavakoli-Yaraki M., Renkens I., Vermaat J.S., van Roosmalen M.J., van Lieshout S., Nijman I.J., Roessingh W., van 't Slot R., van de Belt J., Guryev V., Koudijs M., Voest E., Cuppen E. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol. 2011;12:R103. doi: 10.1186/gb-2011-12-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Forment J.V., Kaidi A., Jackson S.P. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]