Abstract
Background
The receptor tyrosine kinase ROS1 is a driver gene in the non-small cell lung cancer (NSCLC) with promising target treatment potential. The clinical features of NSCLC patients harboring ROS1 fusion gene were not fully understood due to small-to-modest sample sizes of these association studies.
Methods
We systematically searched PubMed, EMBASE, and the Cochrane Central Register of Controlled Trials (CENTRAL) from their inception to March 31, 2015. We analyzed the association between ROS1 fusion genes and four common clinical variables, i.e., gender, smoking status, pathological type and clinical stage.
Results
Eighteen studies consisting of 9,898 NSCLC patients were included in this meta-analysis. Pooled results showed that significantly higher rate of ROS1 fusion gene was detected in female NSCLC patients (OR =1.54, 95% CI: 1.02-2.34, P=0.042), patients without a smoking history (OR =3.27, 95% CI: 1.44-7.45, P=0.005), patients with adenocarcinomas NSCLC (OR =10.24, 95% CI: 5.13-20.40, P<0.001), and patients with an advanced clinical stages III-IV (OR =2.57, 95% CI: 1.78-3.71, P<0.001). The pooled prevalence of ROS1 fusion gene was 2.4% (95% CI: 1.8-3.1%) in adenocarcinoma and a significantly lower (0.2%) in non-adenocarcinoma tumors.
Conclusions
ROS1 rearrangement was more prevalent in female patients, patients without a smoking history, patients with adenocarcinoma, and patients on more advanced stages (stages III to IV).
Keywords: ROS1, clinicopathologic features, non-small cell lung cancer (NSCLC), meta-analysis
Introduction
Lung cancer remains the leading cause of cancer death worldwide and non-small cell lung cancer (NSCLC) accounts for more than 80% of lung cancer cases. Since the last decade, the identification of key driver genes in NSCLC [epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK), etc.] and the promising results of tyrosine kinase inhibitors (TKIs) targeting these driver genes to treat NSCLC have rapidly facilitated the development of targeted therapy and precision medicine (1-3). In the era of precision medicine, molecular testing became extremely important in both the classification and treatment of lung cancer (4). ROS1 is a receptor tyrosine kinase of the insulin receptor family. ROS1 fusion proteins were firstly demonstrated to be involved in NSCLC in 2007 through global survey of phosphotyrosine signaling of NSCLC cell lines and tumors (5,6). ROS1 fusion kinase is constitutively activated and leads to activation of downstream oncogenic pathways (STAT3, PI3K/AKT/mTOR, RAS-MAPK/ERK pathways) which controls cell proliferation, survival and cell cycling, and eventually results in cell transformation (7,8).
It’s striking that preliminary results from two independent studies showed that crizotinib, an inhibitor of ALK but also effective on ROS1, was highly active at treating patients who had ROS1 rearrangement, showing a response rate of 72-80% (9,10). These promising results highlighted the necessity of thorough investigation of ROS1 fusion gene in NSCLC patients, but the clinical features of ROS1-fusion-gene-harboring patients, which was the base of further research, was not fully understood: the vast majority of studies had small-to-modest sample sizes, which comprised the detection power of each individual study. Therefore in this study, we performed a meta-analysis to determine the clinicopathologic features of NSCLC patients with ROS1 rearrangements.
Materials and methods
Literature search
We adopted guidelines from the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement to perform our meta-analysis (11). We systematically searched PubMed, EMBASE, and the Cochrane Central Register of Controlled Trials (CENTRAL) from their inception to March 31, 2015, using the following search terms and key words: [“carcinoma, non-small-cell lung” (Mesh) OR “lung neoplasm” OR “lung cancer” OR “lung carcinoma” OR “pulmonary neoplasm” OR “pulmonary cancer” OR “pulmonary carcinoma” OR NSCLC] and (ROS1 OR ROS-1 OR “ROS 1”). We also manually checked references of the identified reports and relevant reviews. No language restrictions were applied.
Study selection
Two investigators (Q Zhu and X Zhang) performed study selection independently, with disagreements resolved by consensus. To be included, studies had to meet all the following criteria: (I) included NSCLC patients, regardless of the pathological phenotypes; (II) genotyped whether ROS1 fusion gene is present in NSCLC patients; (III) provided the total number of patients and number of patients harboring ROS1 fusion gene in two categorical groups stratified by one or more of these clinical variables, i.e., gender, smoking status, pathological type and clinical stage; and (IV) were published as full-text articles. We excluded studies without sufficient data to estimate odds ratios (ORs) and their corresponding 95% confidence intervals (CIs).
Outcomes
The pre-specified primary endpoints were to investigate whether there was any association between ROS1 fusion gene and these clinical variables in overall population. The secondary endpoints were to determine whether these associations were different among different ethnic populations.
Data collection and quality assessment
For each study, the following information was independently extracted by two investigators (Q Zhu and X Zhang): first author, year of publication, the performing country of the study, number of patients, ethnicity, age, pathological type of tumors, genotyping methods, and the number of ROS1 fusion genes. For each study, we also recorded the number of ROS1 fusion genes in categorical groups stratified by clinical parameters stated above.
Statistical analysis
In NSCLC patients, we analyzed the association between ROS1 fusion genes and four common clinical variables, i.e., gender, smoking status, pathological type and clinical stage. ORs and their corresponding 95% CIs were pooled across studies using random-effects models in the presence of significant heterogeneity, or fixed-effects model in case no significant heterogeneity was detected (12). We used both the I2 statistic and χ2-based Q test to evaluate the heterogeneity, with I2 higher than 50% or P value of Q statistic’s test below 0.10 indicating significant heterogeneity (13,14). Studies in which ROS1 fusion gene was not detected in any of the two groups were excluded in the analysis. For studies in which ROS1 fusion gene was not detected in only one of the two groups, we calculated OR and its 95% CI by adding 0.5 to each cell of the 2×2 table for that study (15,16). Publication bias was analyzed by Begg’s test and Egger’s test (17), and sensitivity analyses by omitting one study at one time. Subgroup analyses were performed according to the ethnicity of NSCLC patients. All analyses were performed with the STATA version 12.0 (STATA Corporation, College Station, TX, USA) software.
Results
Study selection and characteristics
The flow diagram of the meta-analysis was shown in Figure 1. Our systematic literature search generated 603 studies. Five hundred and seventy studies were excluded as duplicate publications and reviews etc, leaving 33 studies for full-text review. Fifteen studies were further excluded because outcomes of interest were not reported. Thus, 18 studies presented were included in the meta-analysis, encompassing 9,898 NSCLC patients (18-35). Twelve studies were carried out in Asian population (18-29), and six were in Caucasian population (30-35). Baseline characteristics of individual study were shown in Table 1. All studies were published between 2011 and 2015. The number of patients ranged from 108 to 1,478, the mean age from 54 to 65 years. The detection methods included fluorescence in situ hybridization (FISH), reverse transcriptase PCR (RT-PCR) and immunohistochemistry (IHC), and FISH was the most commonly used method.
Figure 1.
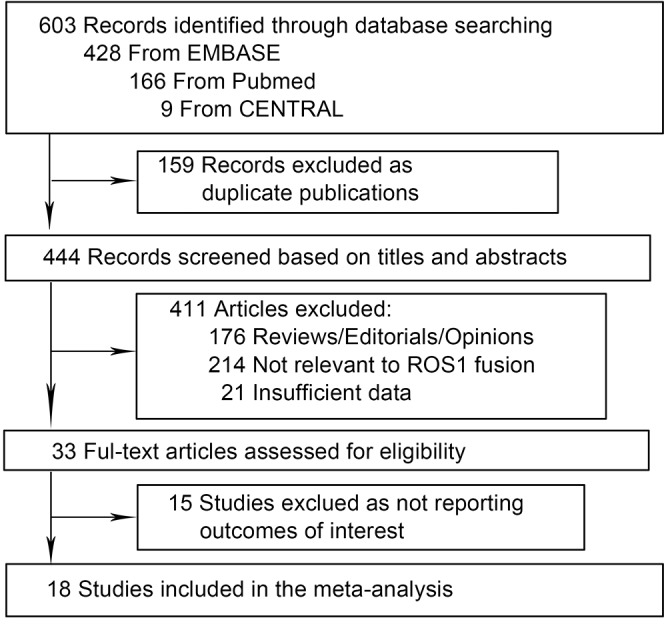
Flow diagram of the study selection in the meta-analysis.
Table 1. Characteristics of included studies.
| Study/first author | Year | Country | Ethnicity | No. of patients | Age [range] | ROS1 fusion | Detection methods | Tumor type |
|---|---|---|---|---|---|---|---|---|
| Pan et al. (27) | 2014 | China | Asian | 1,139 | 59.4±10.8 | 11 | RT-PCR, FISH | Lung adenocarcinoma |
| Kim et al. (26) | 2014 | Korea | Asian | 162 | 60 [42-75] | 5 | FISH | Lung adenocarcinoma |
| Chen et al. (24) | 2014 | China | Asian | 492 | 65 [27-95] | 12 | IHC | Lung adenocarcinoma |
| Go et al. (20) | 2013 | Korea | Asian | 451 | 62 [34-87] | 8 | FISH | NSCLC |
| Zhao et al. (28) | 2014 | China | Asian | 108 | NA | 2 | ARMS PCR | Lung adenocarcinoma |
| Cheng et al. (25) | 2014 | China | Asian | 1,652 | 60 [31-87] | 53 | FISH/RT-PCR | NSCLC |
| Davies et al. (31) | 2012 | America | Caucasian | 447 | 66 [33-86] | 5 | FISH | NSCLC |
| Mescam-Mancini et al. (33) | 2014 | France | Caucasian | 121 | 62 [31-88] | 9 | FISH/IHC | Lung adenocarcinoma |
| Jurmeister et al. (35) | 2015 | Germany | Caucasian | 473 | 54 [29-75] | 4 | FISH | NSCLC |
| Cai et al. (19) | 2013 | China | Asian | 392 | 60 [27–83] | 8 | multiplex RT-PCR | NSCLC |
| Jin et al. (29) | 2015 | Korea | Asian | 375 | 63 [21-84] | 3 | FISH | NSCLC |
| Sholl et al. (32) | 2013 | America | Caucasian | 220 | 64 [29-94] | 9 | IHC/FISH | Lung adenocarcinoma |
| Bergethon et al. (30) | 2012 | America | Mixed | 1,073 | 62 [32-87] | 18 | FISH | NSCLC |
| Yoshida et al. (22) | 2013 | Japan | Asian | 570 | 61 [28-88] | 15 | FISH | NSCLC |
| Cha et al. (23) | 2014 | Korea | Asian | 330 | 61 [28-86] | 13 | FISH | Lung adenocarcinoma |
| Kim et al. (21) | 2013 | Korea | Asian | 208 | 58 [30-78] | 7 | FISH | Lung adenocarcinoma |
| Warth et al. (34) | 2014 | Germany | Caucasian | 1,478 | NA | 9 | FISH | NSCLC |
| Li et al. (18) | 2011 | China | Asian | 202 | 57 | 2 | RT-PCR | Lung adenocarcinoma |
RT-PCR, reverse transcriptase PCR; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; NSCLC, non-small cell lung cancer.
Association between ROS1 fusion gene and gender in NSCLC patients
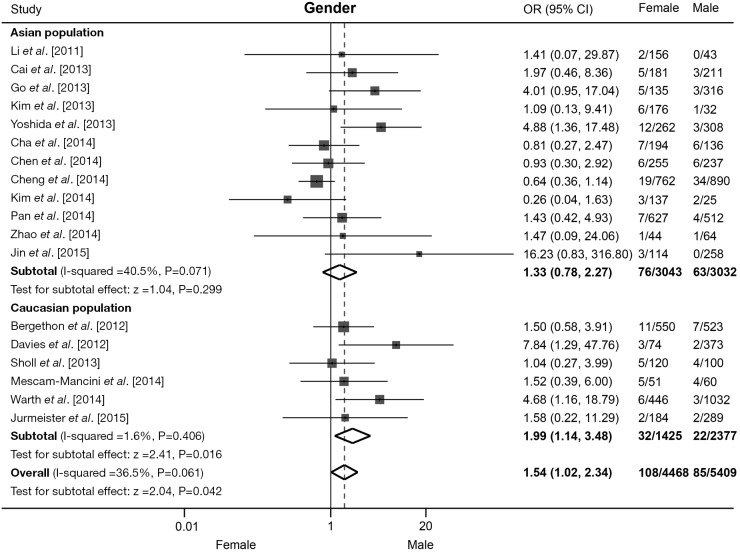
All 18 studies contributed to the analysis of association between ROS1 rearrangement and gender. A total of 5,409 male patients and 4,468 female patients were included, and ROS1 rearrangement was detected in 85 males (1.57%) and 108 females (2.42%) patients. Pooled results showed that female NSCLC patients were associated with an increased rate of ROS1 fusion gene compared with male patients (OR =1.54, 95% CI: 1.02-2.34, P=0.042; Figure 2). There was moderate heterogeneity across these studies (I2=36.5%, P=0.06). We then performed subgroup analyses based on the ethnic origin of participants. Twelve studies including 6,075 patients were involved in the analysis of Asian population, and meta-analysis showed no significant difference in ROS1 rearrangement rate between male and female patients (OR =1.33, 95% CI: 0.78-2.27, P=0.299; Figure 2). Six studies including 3,802 patients were involved in the analysis of Caucasian population, pooled analysis showed a significantly higher rate of ROS1 fusion gene in female patients (OR =1.99, 95% CI: 1.14-3.48, P=0.016; Figure 2). A moderate level of heterogeneity was detected in Asian population (I2=40.5%, P=0.071), but not in Caucasian population (I2=1.6%, P=0.406). No publication bias was observed in the overall analysis, as demonstrated by Begg’s test (P=0.363).
Figure 2.
Association between ROS1 fusion gene and gender in NSCLC patients. Forest plot of odds ratios (ORs) and 95% confidence intervals (CI) from each study, subgroup and overall analysis were shown. Subgroup analyses were stratified by ethnicity. NSCLC, non-small cell lung cancer.
Association between ROS1 fusion gene and smoking status in NSCLC patients
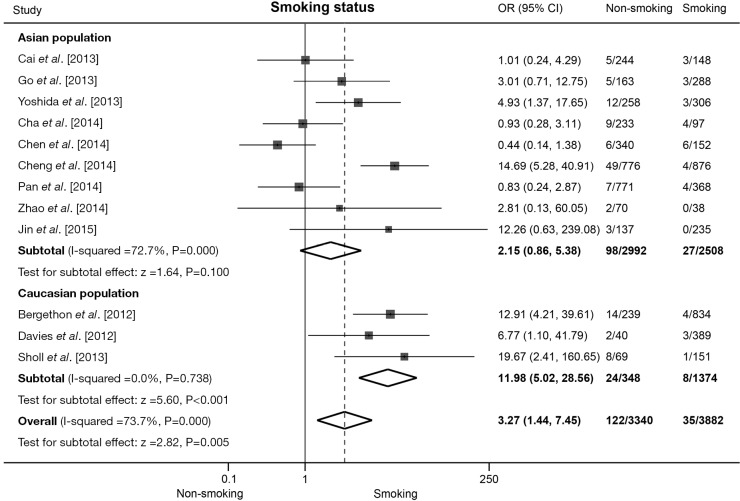
Twelve studies presented clinical data for association analysis between ROS1 fusion gene and smoking status. The pooled frequency of ROS1 fusion gene was 0.90% (35/3,882) and 3.65% (122/3,340) in smoking and non-smoking patients respectively. Significant heterogeneity was found across these studies (I2=73.7%, P<0.001). Pooled results from random effect model showed that non-smoking NSCLC patients were associated with an increased rate of ROS1 fusion gene compared with smoking patients (OR =3.27, 95% CI: 1.44-7.45, P=0.005; Figure 3). Nine studies including 5,500 patients were involved in the analysis of Asian population, and meta-analysis showed no significance but a trend that non-smoking NSCLC patients were associated with increased rate of ROS1 rearrangement (OR =2.15, 95% CI: 0.86-5.38, P=0.100; Figure 3) and the sensitivity analysis showed that the association did become significant (OR =2.70, 95% CI: 1.11-6.67) without the data from Chen et al. (24). Six studies including 1,722 patients were involved in the analysis of Caucasian population, pooled analysis showed a significantly higher rate of ROS1 fusion gene in non-smoking patients (OR =11.98, 95% CI: 5.02-28.56, P<0.001; Figure 3). A significant heterogeneity was detected in Asian population (I2=72.7%, P=0.000), but not in Caucasian population (I2=0.0%, P=0.738). No publication bias was observed in the overall analysis with Begg’s test (P=0.837).
Figure 3.
Association between ROS1 fusion gene and smoking status in NSCLC patients. Forest plot of odds ratios (ORs) and 95% confidence intervals (CI) from each study, subgroup and overall analysis were shown. Subgroup analyses were stratified by ethnicity. NSCLC, non-small cell lung cancer.
Association between ROS1 fusion gene and pathological type in NSCLC patients
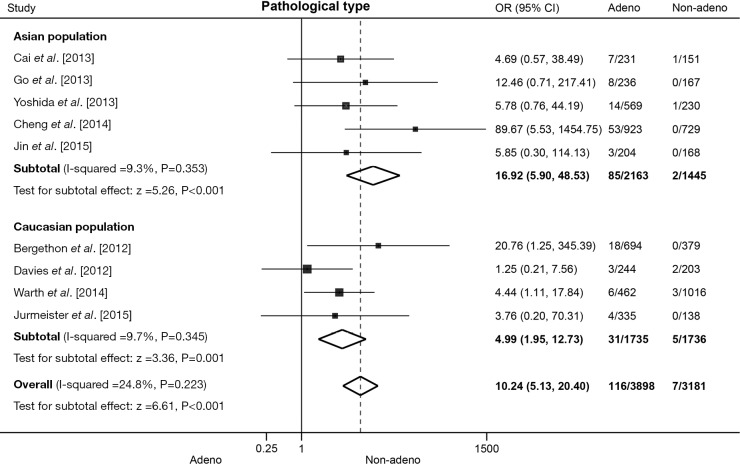
Nine literatures that addressed the frequency of the ROS1 fusion gene in adenocarcinomas and non-adenocarcinomas were included, including 3,898 cases of adenocarcinomas and 3,181 cases of non-adenocarcinomas. The pooled frequency of ROS1 fusion gene was 2.98% (116/3,898) and 0.22% (7/3,181) in adenocarcinomas and non-adenocarcinomas, respectively. No significant heterogeneity was observed (I2=24.8%, P=0.223; Figure 4), thus fixed-effects model was chosen. The pooled results showed that adenocarcinomas was associated with a significantly higher ROS1 fusion gene positivity rate (OR =10.24, 95% CI: 5.13-20.40, P<0.001; Figure 4). Five studies including 3,608 patients were involved in the analysis of Asian population, and four studies including 3,471 patients in Caucasian population. The pooled analysis showed a significantly higher rate of ROS1 rearrangement in adenocarcinomas both in Asian (OR =16.92, 95% CI: 5.90-48.53, P<0.001; Figure 4) and Caucasian (OR =4.99, 95% CI: 1.95-12.73, P=0.001; Figure 4) populations. No significant heterogeneity was detected in Asian population (I2=9.3%, P=0.353), nor in Caucasian population (I2=9.7%, P=0.345). No publication bias was observed in the overall analysis, as demonstrated by Begg’s test (P=0.466).
Figure 4.
Association between ROS1 fusion gene and pathological type in NSCLC patients. Forest plot of odds ratios (ORs) and 95% confidence intervals (CI) from each study, subgroup and overall analysis were shown. Subgroup analyses were stratified by ethnicity. NSCLC, non-small cell lung cancer.
Association between ROS1 fusion gene and clinical stage in NSCLC patients
There are 13 studies detecting the ROS1 fusion gene in NSCLC patients with clear clinical stage. The pooled frequency of ROS1 fusion gene was 1.27% (45/3,541) in patients of clinical stage I-II and 3.56% (105/2,949) in stage III-IV. We found no significant heterogeneity (I2=0.0%, P=0.568; Figure 5). The pooled results showed that an advanced clinical stage (III-IV) was associated with a significantly higher ROS1 rearrangement rate (OR =2.57, 95% CI: 1.78-3.71, P<0.001; Figure 5). Nine studies including 4,324 patients were involved in the analysis of Asian population, and four studies including 2,166 patients in Caucasian population. The pooled analysis showed a significantly higher rate of ROS1 fusion gene in advanced stage patients in both Asian (OR =2.44, 95% CI: 1.61-3.69, P<0.001; Figure 5) and Caucasian (OR =3.06, 95% CI: 1.39-6.74, P=0.005; Figure 5) populations. No publication bias was observed in the overall analysis, as demonstrated by Begg’s test (P=1.000).
Figure 5.
Association between ROS1 fusion gene and clinical stage in NSCLC patients. Forest plot of odds ratios (ORs) and 95% confidence intervals (CI) from each study, subgroup and overall analysis were shown. Subgroup analyses were stratified by ethnicity. NSCLC, non-small cell lung cancer.
Prevalence of ROS1 fusion gene in NSCLC
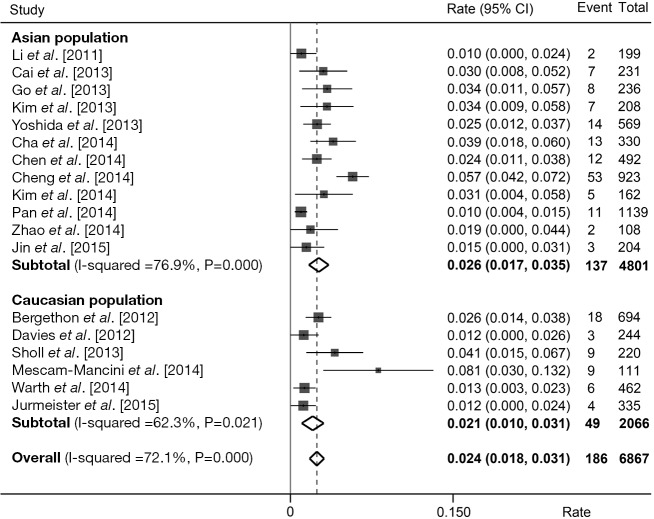
We estimated the prevalence of ROS1 fusion gene based on these 18 studies by taking pathological type into account. A total of 6,868 adenocarcinoma patients were analyzed and 186 were positive for ROS1 fusion gene. Pooled analysis of all 18 studies showed a prevalence of 2.4% (95% CI: 1.8-3.1%; Figure 6) in adenocarcinoma. A relatively higher rate was observed in Asian population (137/4,801, rate =2.6%, 95% CI: 1.7-3.5%; Figure 6) than Caucasian population (49/2,066, rate =2.1%, 95% CI: 1.0-3.1%; Figure 6). Whereas in non-adenocarcinomas patients, the prevalence was substantially lower, with only 7 of 3,181 (0.2%) patients harboring ROS1 fusion gene.
Figure 6.
Prevalence of ROS1 fusion gene in NSCLC patients with adenocarcinoma. Forest plot of rates and 95% confidence intervals (CI) from each study, subgroup and overall analysis were shown. Subgroup analyses were stratified by ethnicity. NSCLC, non-small cell lung cancer.
Discussion
To our knowledge, this was the first meta-analysis performed to investigate the clinic pathologic characteristics of NSCLC patients with a ROS1 fusion gene. Our meta-analysis included 18 studies and a total of 9,898 NSCLC patients. Pooled analyses from these studies revealed that ROS1 rearrangement was more prevalent in female patients, patients without a smoking history, patients with adenocarcinoma, and patients on more advanced stages (stage III to IV). Our meta-analysis also showed a prevalence of 2.4% (95% CI: 1.8-3.1%) of ROS1 fusion gene in adenocarcinoma and a significantly lower rate (0.2%) in non-adenocarcinoma tumors.
Our study provided evidence to guide the prescreening of NSCLC patients to select a more enriched population who are more likely to harbor this specific rearrangement. The prescreening process is critical and important for several reasons. First, ROS1 fusion gene was rare in NSCLC patients. In the first study describing ROS1 fusion gene in NSCLC, Bergethon and colleagues reported 18 of 1,073 (1.7%) NSCLC tumors were ROS1 rearranged, and all 18 ROS1-positive tumors were adenocarcinomas [2.5%, (18/694)] (30), which was similar to our meta-analysis showing a prevalence of 2.4% in adenocarcinoma and extremely rare in non-adenocarcinoma patients. Therefore, systematic testing of ROS1 rearrangement was challenging and unreasonable due to extremely high probability of negative results. Second, the detection method of ROS1 fusion gene remains to be defined. Currently, FISH and next-generation sequencing were the most powerful approach and provided the most solid results, whereas IHC and RT-PCR had a number of limitations (8,23,33). However, FISH and next-generation sequencing were more technically demanding and more expensive. A prescreen process could substantially reduce the number of NSCLC patients enrolling for ROS1 rearrangement test and therefore reduce the whole health cost. Evaluation of the clinic pathologic features of NSCLC patients was the first step of prescreening. Based on our meta-analysis and other studies, young female patients without a smoking history and having tumors with adenocarcinoma histology on advanced clinical stage were more likely to harbor ROS1 fusion gene and should be genetically tested. IHC could also be used as a prescreening method (9).
It’s notable that, according to our meta-analysis and other studies, NSCLC patients with ROS1 fusion gene shared many clinic pathologic features with patients harboring ALK rearrangements (36,37). Similar pathogenesis might exist in these two subtypes of NSCLCs, supported by both structural and functional evidences: the kinase domains of ALK and ROS1 share 77% sequence homology (10,38), and ROS1 signaling and cell viability was substantially inhibited in cell lines expressing ROS1 fusion genes, by crizotinib, an inhibitor of ALK (30,39). Two independent studies conducted on NSCLC patients harboring ROS1 fusion gene, one perspective and one retrospective, showed a promising antitumor activity of crizotinib in these molecularly enriched patients: the response rates were 72% and 80% respectively and the median progression-free survival (PFS) was 9 to 19 months (9,10). Other more effective inhibitors of ROS1 have been developed in the past few years, which were able to overcome crizotinib resistance in vitro and in animal models. A series of clinical trials regarding these inhibitors including crizotinib on ROS1-rearrangment NSCLC were ongoing or being planned (NCT02183870, NCT01970865 and NCT01945021, etc.). Favorable results are expected in near future. Our findings could facilitate the patient selection process for ROS1 inhibitor targeted therapy.
Several limitations in our study should be considered when interpreting these results. First, potential publication bias could exist in our analysis. Although we performed a systemic search of numerous databases and Begg’s test did not shown evidence of significant publication bias, we still could not rule out the possibility that relevant studies might have been missed. Second, although we enrolled 9,898 NSCLC patients in our study, the sample size might still be not large enough provided that the rate of ROS1 fusion gene was small. However, our meta-analysis remained the most powerful study ever by detecting nearly 200 NSCLC patients with ROS1 fusion gene. Third, moderate to significant heterogeneity was observed in several comparisons. We attempted to investigate the heterogeneity by performing subgroup analyses, and found that heterogeneity still existed in Asian population. Particularly when comparing ROS1 fusion gene in patients with different smoking status, sensitivity analysis showed that after omitting study of Chen et al., the direction of pooled results changed in Asian population. Therefore, results should be interpreted with caution removing study of Chen et al.
Conclusions
Our meta-analysis including 9,898 NSCLC patients showed that ROS1 rearrangement was more prevalent in female patients, patients without a smoking history, patients with adenocarcinoma, and patients on more advanced stages (stage III to IV). The prevalence of ROS1 fusion gene was 2.4% (95% CI: 1.8-3.1%) in adenocarcinoma and a significantly lower (0.2%) in non-adenocarcinoma tumors.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- 1.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popper HH, Ryska A, Tímár J, et al. Molecular testing in lung cancer in the era of precision medicine. Transl Lung Cancer Res 2014;3:291-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [DOI] [PubMed] [Google Scholar]
- 6.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [DOI] [PubMed] [Google Scholar]
- 7.Chin LP, Soo RA, Soong R, et al. Targeting ROS1 with anaplastic lymphoma kinase inhibitors: a promising therapeutic strategy for a newly defined molecular subset of non-small-cell lung cancer. J Thorac Oncol 2012;7:1625-30. [DOI] [PubMed] [Google Scholar]
- 8.Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res 2013;19:4040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazières J, Zalcman G, Crinò L, et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: results from the EUROS1 cohort. J Clin Oncol 2015;33:992-9. [DOI] [PubMed] [Google Scholar]
- 10.Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 2009;339:b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177-88. [DOI] [PubMed] [Google Scholar]
- 13.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539-58. [DOI] [PubMed] [Google Scholar]
- 14.Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schömig A, Mehilli J, de Waha A, et al. A meta-analysis of 17 randomized trials of a percutaneous coronary intervention-based strategy in patients with stable coronary artery disease. J Am Coll Cardiol 2008;52:894-904. [DOI] [PubMed] [Google Scholar]
- 16.Sterne JA, Bradburn MJ, Egger M. Meta-analysis in Stata. In: Egger M, Smith GD, Altman D, editors. Systematic Reviews in Health Care. London: Blackwell BMJ Books, 2001:357. [Google Scholar]
- 17.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, Fang R, Sun Y, et al. Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One 2011;6:e28204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai W, Li X, Su C, et al. ROS1 fusions in Chinese patients with non-small-cell lung cancer. Ann Oncol 2013;24:1822-7. [DOI] [PubMed] [Google Scholar]
- 20.Go H, Kim DW, Kim D, et al. Clinicopathologic analysis of ROS1-rearranged non-small-cell lung cancer and proposal of a diagnostic algorithm. J Thorac Oncol 2013;8:1445-50. [DOI] [PubMed] [Google Scholar]
- 21.Kim HR, Lim SM, Kim HJ, et al. The frequency and impact of ROS1 rearrangement on clinical outcomes in never smokers with lung adenocarcinoma. Ann Oncol 2013;24:2364-70. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida A, Kohno T, Tsuta K, et al. ROS1-rearranged lung cancer: a clinicopathologic and molecular study of 15 surgical cases. Am J Surg Pathol 2013;37:554-62. [DOI] [PubMed] [Google Scholar]
- 23.Cha YJ, Lee JS, Kim HR, et al. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PLoS One 2014;9:e103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YF, Hsieh MS, Wu SG, et al. Clinical and the prognostic characteristics of lung adenocarcinoma patients with ROS1 fusion in comparison with other driver mutations in East Asian populations. J Thorac Oncol 2014;9:1171-9. [DOI] [PubMed] [Google Scholar]
- 25.Cheng H, Ye L, Xue L. Detection of ROS1 gene rearrangement by FISH and analysis of its clinical features in non-small cell lung cancer patients. Zhonghua Zhong Liu Za Zhi 2014;36:751-4. [PubMed] [Google Scholar]
- 26.Kim MH, Shim HS, Kang DR, et al. Clinical and prognostic implications of ALK and ROS1 rearrangements in never-smokers with surgically resected lung adenocarcinoma. Lung Cancer 2014;83:389-95. [DOI] [PubMed] [Google Scholar]
- 27.Pan Y, Zhang Y, Li Y, et al. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: a comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer 2014;84:121-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhao C, Li X, Li J, et al. Detecting ALK, ROS1 and RET Fusion Genes in Cell Block Samples. Transl Oncol 2014;7:363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin Y, Sun PL, Kim H, et al. ROS1 gene rearrangement and copy number gain in non-small cell lung cancer. Virchows Arch 2015;466:45-52. [DOI] [PubMed] [Google Scholar]
- 30.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res 2012;18:4570-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sholl LM, Sun H, Butaney M, et al. ROS1 immunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am J Surg Pathol 2013;37:1441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mescam-Mancini L, Lantuéjoul S, Moro-Sibilot D, et al. On the relevance of a testing algorithm for the detection of ROS1-rearranged lung adenocarcinomas. Lung Cancer 2014;83:168-73. [DOI] [PubMed] [Google Scholar]
- 34.Warth A, Muley T, Dienemann H, et al. ROS1 expression and translocations in non-small-cell lung cancer: clinicopathological analysis of 1478 cases. Histopathology 2014;65:187-94. [DOI] [PubMed] [Google Scholar]
- 35.Jurmeister P, Lenze D, Berg E, et al. Parallel screening for ALK, MET and ROS1 alterations in non-small cell lung cancer with implications for daily routine testing. Lung Cancer 2015;87:122-9. [DOI] [PubMed] [Google Scholar]
- 36.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 2009;27:4247-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solomon B. Validating ROS1 rearrangements as a therapeutic target in non-small-cell lung cancer. J Clin Oncol 2015;33:972-4. [DOI] [PubMed] [Google Scholar]
- 38.Huber KV, Salah E, Radic B, et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature 2014;508:222-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDermott U, Iafrate AJ, Gray NS, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res 2008;68:3389-95. [DOI] [PubMed] [Google Scholar]