Abstract
The prevalent renal transplant population presents an opportunity to observe the adaptive changes in the alloimmune response over time, but such studies have been limited by uncertainties in the conventional biopsy diagnosis of T cell-mediated rejection (TCMR) and antibody-mediated rejection (ABMR). To circumvent these limitations, we used microarrays and conventional methods to investigate rejection in 703 unselected biopsies taken 3 days to 35 years post-transplant from North American and European centers. Using conventional methods, we diagnosed rejection in 205 biopsy specimens (28%): 67 pure TCMR, 110 pure ABMR, and 28 mixed (89 designated borderline). Using microarrays, we diagnosed rejection in 228 biopsy specimens (32%): 76 pure TCMR, 124 pure ABMR, and 28 mixed (no borderline). Molecular assessment confirmed most conventional diagnoses (agreement was 90% for TCMR and 83% for ABMR) but revealed some errors, particularly in mixed rejection, and improved prediction of failure. ABMR was strongly associated with increased graft loss, but TCMR was not. ABMR became common in biopsy specimens obtained >1 year post-transplant and continued to appear in all subsequent intervals. TCMR was common early but progressively disappeared over time. In 108 biopsy specimens obtained 10.2–35 years post-transplant, TCMR defined by molecular and conventional features was never observed. We conclude that the main cause of kidney transplant failure is ABMR, which can present even decades after transplantation. In contrast, TCMR disappears by 10 years post-transplant, implying that a state of partial adaptive tolerance emerges over time in the kidney transplant population.
Keywords: microarrays, antibody, T cells, kidney transplant, graft rejection
Many people are now alive with functioning kidney transplants in the developed world (about 200,000 in the United States alone),1 and all are at risk for rejection and graft loss.2–5 This creates a need to define the changes in rejection risks over time for the two types of rejection—T cell-mediated rejection (TCMR) and antibody-mediated rejection (ABMR). However, conventional biopsy assessment has limitations for such studies.6–10 Histology is on the basis of expert opinion with considerable interobserver disagreement6,7,11; many inflamed biopsies are called borderline to reflect uncertainty, and diagnosis relies on lesions that are not specific. For example, the TCMR lesions arteritis (v lesions) and tubulitis both occur in other conditions: arteritis in ABMR7,12,13 and tubulitis in some primary renal diseases.14,15 ABMR diagnosis relies on relatively subtle microcirculation changes,16 which are also not specific,17,18 as well as staining for complement split product C4d,19 which is not sensitive.4 A more precise assessment of rejection would facilitate efforts to reduce transplant failure.4,10,20,21
The limitations in conventional diagnostic systems can be addressed by introducing a molecular dimension to biopsy interpretation, which has been successful for breast cancer.22–24 The concept is not to replace existing systems but to add new estimates of diagnosis, activity, stage, and prognosis. This is in keeping with newer approaches to prediction that use both conventional and data-driven methods to estimate the truth—not to prove one approach correct but to increase accuracy by incorporating all valid approaches, including expert opinion, molecular measurements, and statistical calculations.25 Moreover, molecular measurements offer potential mechanistic insights. To achieve this objective, we recently developed microarray-based molecular tests for TCMR7 and ABMR8 and validated them in an independent set of biopsies.9,10
This study evaluated how the addition of central molecular assessment to conventional diagnostic assessments would improve our understanding of the rejection phenotypes operating in the prevalent renal transplant population. We developed a sample of unselected biopsies for clinical indications from experienced North American and European centers to reflect the problem of rejection as it presents in the renal transplant universe in the developed world whether days or decades post-transplant. We were particularly interested in the effect of time post-transplant on the risks of TCMR and ABMR in the prevalent renal transplant population as a reflection of host–graft adaptive changes.
Results
Demographics
We prospectively collected 703 biopsies from 564 kidneys between September of 2004 and March of 2012 taken for clinical indications 3 days to 35 years post-transplant (Supplemental Table 1).
The common indications for biopsy were dysfunction and proteinuria. The median follow-up after biopsy in functioning kidneys was 2.7 years. Progression to failure (censored for patient death with function) occurred in 128 kidneys (23%) at a median of 1.1 years after biopsy. Kidneys that progressed to failure after the biopsy more commonly presented with deteriorating function, had donor-specific HLA antibody, and were from patients not on tacrolimus–mycophenolate immunosuppression.
Conventional methods diagnosed rejection in 205 biopsies: 67 pure TCMR, 110 pure ABMR (80 C4d-negative), and 28 mixed with 89 designated borderline (Table 1). For 423 patients in whom the HLA antibody status was known at transplantation, there was no significant difference in ABMR frequency between those patients who were HLA antibody-positive (121) and those patients who were HLA antibody-negative (302), which is in keeping with previous studies showing the importance of de novo HLA antibody formation in ABMR in this population, almost all of which is late (median time from transplant to diagnosis of ABMR was 6.2 years).26
Table 1.
Conventional diagnoses in 703 biopsies
| Conventional Diagnosis | All Biopsies (n=703) | All Kidneysa (n=564) | Kidneys That Did Not Progress to Failure (n=436) | Kidneys That Progressed to Failure (n=128) | P Valueb (Failed Versus Working) |
|---|---|---|---|---|---|
| Rejection-related processes, n (%) | |||||
| Pure TCMR | 67 (10) | 56 (10) | 50 (11)a | 6 (5) | 0.02 |
| Pure ABMR | 110 (16) | 94 (17) | 47 (11) | 47 (37)a | <0.001 |
| C4d-positive ABMR | 30 (4) | 27 (5) | 13 (3) | 14 (11)a | <0.001 |
| C4d-negative ABMR | 80 (11) | 67 (12) | 34 (8) | 33 (26)a | <0.001 |
| Mixed rejection | 28 (4) | 12 (2) | 8 (2) | 4 (3) | 0.48 |
| Borderline | 89 (13) | 63 (11) | 56 (13)a | 7 (5) | 0.02 |
| Transplant glomerulopathy | 27 (4) | 24 (4) | 16 (4) | 8 (6) | 0.20 |
| Other processes, n (%) | |||||
| PVN | 25 (4) | 16 (3) | 11 (3) | 5 (4) | 0.41 |
| GN | 81 (12) | 75 (13) | 55 (13) | 20 (16) | 0.38 |
| Relatively normal | 180 (26) | 147 (26) | 133 (31)a | 14 (11)c | <0.001 |
| Atrophy fibrosis of unknown significance | 72 (10) | 58 (10) | 44 (10) | 14 (11) | 0.78 |
| Other uncommon diagnosed | 24 (3) | 19 (3) | 16 (4) | 3 (2) | 0.59 |
| Total | 703 | 564 | 436 | 128 |
We selected one random biopsy per kidney. Significant differences in progression to failure highlight that the group is overrepresented in the diagnostic category.
Chi-squared and Fisher exact statistical tests.
Of 14 failures after a study biopsy that showed relatively normal histology, 8 failures were attributable to late events apparently unrelated to the study biopsy (ABMR, GN, or nonadherence), 3 failures were missed diagnoses of ABMR, and 3 failures have incomplete follow-up records.
Other diagnoses included C4d deposition without morphologic evidence for active rejection (n=6), missing diagnosis (n=1), and biopsies with no clear diagnosis, including probable TCMR (n=2), probably normal (n=7), probable atrophy fibrosis of unknown significance (n=5), suspicious for ABMR (n=2), and probable borderline (n=1).
Kidneys that progressed to failure after biopsy were diagnosed with pure ABMR more frequently (37% versus 11%) and diagnosed with pure TCMR less frequently (5% versus 11%) than those that did not fail. Of 56 kidneys that showed no rejection by conventional assessment and subsequently failed, 5 kidneys had polyoma virus nephropathy (PVN), 20 kidneys had other glomerular diseases, 14 kidneys had atrophy fibrosis of unknown significance, and 3 kidneys had indeterminate findings; 14 kidneys with relatively normal study biopsies also failed (Table 1).
Adding Molecular to Conventional Assessment
Molecular tests gave continuous scores between zero and one but for this analysis, were designated positive using published cutoffs: TCMR score>0.10 and ABMR score>0.20.7–10 These cutoffs diagnosed rejection in 228 biopsies (32%): 76 pure TCMR, 124 pure ABMR, and 28 mixed (Table 2). Of 89 biopsies called borderline by conventional assessment, the microarray tests diagnosed 17 biopsies as pure TCMR, 8 biopsies as pure ABMR, 5 biopsies as mixed, and 59 biopsies as no rejection. The molecular diagnoses strongly agreed with the conventional diagnosis, with accuracy of 91% for TCMR (Table 3) and 83% for ABMR (Table 4). In biopsies assessed conventionally as relatively normal, the microarray tests found no rejection in 97%.
Table 2.
Agreement between conventional histology and molecular scores in 703 biopsies
| Conventional Assessment/Molecular Assessment | Pure TCMR | Pure ABMR | Mixed | No Rejection | Total |
|---|---|---|---|---|---|
| Pure TCMR | 34a | 1 | 8 | 24 | 67 |
| Pure ABMR | 5 | 59a | 5 | 41 | 110 |
| Mixed | 4 | 16 | 6a | 2 | 28 |
| Borderline | 17 | 8 | 5 | 59 | 89 |
| Transplant glomerulopathy | 0 | 12 | 0 | 15 | 27 |
| No rejectionb | 16 | 28 | 4 | 334a | 382 |
| Total | 76 | 124 | 28 | 475 | 703 |
| Potential reclassification by molecular scores | 42 | 64 | 22 | 126 | 254 |
The principal areas of agreement between conventional and molecular diagnoses.
Diagnoses include PVN, GN, atrophy fibrosis of unknown significance, no major abnormalities, transplant glomerulopathy, others, and relatively normal.
Table 3.
Association of TCMR score with conventional TCMR diagnosis leaving out mixed and borderline biopsies (Fisher exact test; P value<0.001)
| TCMR Score | TCMRa | No TCMRa | Total |
|---|---|---|---|
| TCMR score>0.10 | 42 | 30 | 72; PPV=0.58 |
| TCMR score<0.10 | 25 | 489 | 514; NPV=0.95 |
| Total | 67; sensitivity=0.63 | 519; specificity=0.94 | 586; accuracy=0.91 |
PPV, positive predictive value; NPV, negative predictive value.
For these calculations, biopsies conventionally assessed as mixed and borderline were not included.
Table 4.
Association of ABMR score with conventional ABMR diagnosis leaving out mixed biopsies (Fisher exact test; P value<0.001)
| ABMR Score | ABMRa | No ABMRa | Total |
|---|---|---|---|
| ABMR score>0.2 | 64 | 66 | 130; PPV=0.49 |
| ABMR score<0.2 | 46 | 499 | 545; NPV=0.92 |
| Total | 110; sensitivity=0.58 | 565; specificity=0.87 | 675; accuracy=0.83 |
PPV, positive predictive value; NPV, negative predictive value.
For these calculations, biopsies conventionally assessed as mixed were not included.
The discrepancies between molecular and conventional diagnoses have previously been analyzed (Discussion).7–10
The molecular diagnoses differed from conventional assessments particularly in 28 biopsies diagnosed conventionally as mixed rejection: only 6 biopsies had both positive TCMR and positive ABMR scores, suggesting that these biopsies need closer examination (see below).
In total, 9 of 25 biopsies (36%) with PVN had positive TCMR scores7,9 but were not called TCMR by conventional assessment. As noted previously,7,9 PVN biopsies often have TCMR-like changes on histology and molecular assessment. This probably reflects true TCMR caused by reduction in immunosuppression, but TCMR-like responses to viral antigens cannot be excluded.7,9
Forty-four percent of biopsies with transplant glomerulopathy had positive ABMR scores.
Progression to Failure
In Cox regression analysis of death-censored 3-year survival, using one random biopsy per patient, kidneys with conventional diagnoses of pure TCMR or borderline changes did not have increased risk of failure compared with relatively normal biopsies (Figure 1A), whereas kidneys with pure ABMR had worse survival than those with relatively normal biopsies. Conventional mixed rejection had increased risk, although there were only 12 patients in this group.
Figure 1.
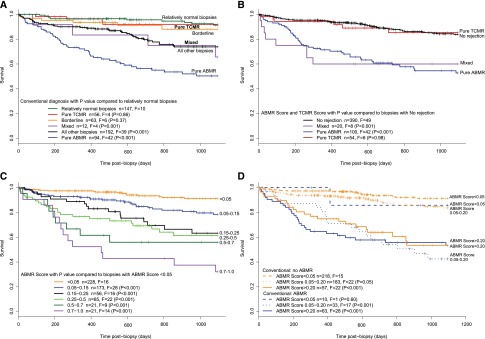
Relationship of biopsy findings to subsequent graft survival. Kaplan–Meier curves for 3-year survival postbiopsy for 564 kidneys with one random biopsy per kidney and censoring death with function at 3 years (n is the total number of kidneys, F is failure, and vertical ticks indicate censoring). The total number of patients at risk at various times postbiopsy was 564 at day of biopsy, 498 at day 200, 435 at day 400, 342 at day 600, 273 at day 800, and 241 at day 1000. (A) Conventional diagnosis showing P values compared with relatively normal biopsies. (B) Molecular diagnosis split by ABMR and TCMR scores in pure ABMR (positive ABMR score and negative TCMR score), pure TCMR (positive TCMR score and negative ABMR score), and mixed (both positive). (C) Survival in groups divided by rising ABMR score. (D) ABMR score combined with the conventional diagnosis of ABMR/mixed versus no ABMR.
Molecular diagnoses independently confirmed these survival patterns (Figure 1B). Kidneys with positive ABMR scores had increased risk of failure, regardless of whether they had pure ABMR (positive ABMR score and negative TCMR score) or mixed (both positive) rejection. Survival of kidneys with pure TCMR (positive TCMR score and negative ABMR score) was similar to kidneys with no rejection.
The ABMR Score Adds Prediction Power to the Conventional ABMR Diagnosis
Each incremental increase in ABMR score from <0.05 to >0.7 was associated with an incremental increase in risk of failure, indicating that the ABMR score provides a quantitative estimate of risk (Figure 1C).
The addition of the ABMR score to the conventional assessment improved the prediction of failure in biopsies assessed conventionally as no ABMR (Figure 1D): biopsies with ABMR scores>0.2 or between 0.05 and 0.20 had increased failure compared with those with the lowest ABMR scores (<0.05). Note that kidneys with a conventional diagnosis of ABMR had a high risk of failure, even when the ABMR score was weakly positive (0.05–0.20).
We used the Net Reclassification Index to determine whether adding the molecular scores to conventional assessment improved prediction of failure. Adding the ABMR scores to the conventional assessment improved the prediction of 3-year graft survival by 23% (95% confidence interval, 10% to 36%; P<0.001). The addition of conventional ABMR assessment to the ABMR score did not improve risk prediction significantly (P=0.12).
Effect of Time Post-Transplant on the Rejection Frequency
For insight into adaptive changes in prevalent kidney transplants, we studied the incidence of TCMR and ABMR activity in biopsies at various times post-transplant (Tables 3 and 4), with boundaries at 7 days, 6 months, and 1, 5, and 10 years. In effect, this groups the biopsies as early (before 1 year), intermediate (from 1 to 5 years), and late (after 5 years).
Conventional and molecular assessments gave similar patterns of TCMR and ABMR activity over time. Pure TCMR diagnoses were frequent early on, uncommon between 1 and 5 years, rare after 5 years, and virtually absent beyond 10 years. Positive TCMR scores were more frequent than conventional TCMR between 6 months and 5 years, reflecting positive TCMR scores in some PVN biopsies. Biopsies presenting with ABMR became common after 1 year post-transplant and continued to present through all subsequent times.
The molecular scores and conventional diagnoses for individual biopsies over time are shown in Figure 2 (comparing the TCMR scores [Figure 2, A and B] and the ABMR scores [Figure 2, C and D]). Conventional diagnoses are indicated by symbol colors for each biopsy, with TCMR in red, pure ABMR in blue, mixed in yellow, borderline in orange, and PVN in green. The log scale (Figure 2, A and C) shows the early detail, and the linear scale (Figure 2, B and D) shows the late detail. The last conventional ABMR activity was at 31 years (day 11,453), and the last positive ABMR score was at 28 years (day 9889).
Figure 2.
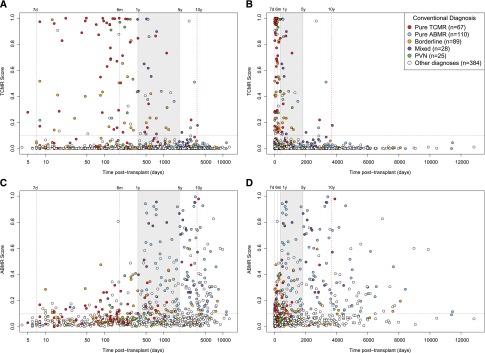
Relationship between time of the biopsy post-transplant and biopsy findings. Distribution of TCMR and ABMR scores and conventional diagnoses in 703 biopsies by the time of the biopsy post-transplant. Each biopsy is represented by a symbol colored by conventional diagnosis. (A) TCMR scores on a log scale. (B) TCMR scores on a linear scale. (C) ABMR scores on a log scale. (D) ABMR scores on a linear scale. The intermediate time period (years 1–5) is shaded to separate early (first 1 year) from late (after 5 years). The time periods are further divided into intervals: early at day 7 and 6 months and late at 10 years.
The striking finding was the disappearance of TCMR from indication biopsies after 10 years post-transplant (Figure 2, Table 5), although some patients in this population were probably nonadherent, which is discussed below. In 110 biopsies after 10 years post-transplant, molecular assessment found 0 pure TCMRs, 34 pure ABMRs, and 1 mixed rejection (a biopsy at 10.2 years [day 3717]). Conventional assessment found 1 pure TCMR, 28 pure ABMRs, and 3 mixed rejections. However, three conventional assessments of TCMR activity after 10 years—one pure TCMR at day 3878 and two mixed rejections at days 4445 and 5985—were on the basis of v lesions and had positive ABMR scores but negative TCMR scores. Thus, in 108 unselected indication biopsies at >10.2 years post-transplant, ABMR activity was frequent—31 conventionally diagnosed and 36 molecularly diagnosed—but no biopsy had confirmed TCMR activity.
Table 5.
Incidence of rejection by time interval
| Classes of Biopsies | Number of Biopsies in Each Interval (Percentage of Total for Interval) | Total | |||||
|---|---|---|---|---|---|---|---|
| Early | Intermediate: 1–5 yr (366–1825 d) | Late | |||||
| 0–7 d | 8 d to 6 mo (8–180 d) | 6–12 mo (181–365 d) | 5–10 yr (1826–3650 d) | >10 yr (>3650 d) | |||
| n | 16 | 213 | 69 | 190 | 105 | 110 | 703 |
| Biopsies of kidneys that progressed to failure | 1 (6) | 18 (8) | 9 (13) | 62 (33) | 33 (31) | 45 (41) | 168 |
| Biopsies with rejection | |||||||
| Conventional pure TCMR | 1 (6) | 41 (19) | 11 (16) | 10 (5) | 3 (3) | 1 (1)a | 67 |
| Molecular pure TCMRb | 1 (6) | 42 (20) | 15 (22) | 18 (9) | 0 (0) | 0 (0) | 76 |
| Conventional pure ABMR | 2 (13) | 5 (2) | 2 (3) | 44 (23) | 29 (28) | 28 (25) | 110 |
| Molecular pure ABMR | 0 (0) | 7 (3)c | 3 (4)c | 41 (22) | 39 (37) | 34 (31) | 124 |
| Conventional mixed | 0 (0) | 2 (1) | 1 (1) | 11 (6) | 11 (10) | 3 (3)d | 28 |
| Molecular mixed | 0 (0) | 5 (2) | 6 (9) | 11 (6) | 5 (5) | 1 (1)d | 28 |
One conventional pure TCMR at day 3878 (with heavy scarring that may have obscured ABMR lesions) and two conventional mixed at days 4445 and 5989 were considered to have TCMR activity caused by isolated v lesions, which may be because of pure ABMR. All had negative TCMR scores and positive ABMR scores.
Positive TCMR score>0.1; positive ABMR score>0.2.
Some positive ABMR scores before 1 year occurred in biopsies from patients who were donor-specific HLA antibody-negative with no conventional evidence of ABMR activity and may reflect ABMR-like molecular changes in other diseases.
One biopsy at 3717 days (10.2 years) with conventional and molecular mixed rejection was the last with a positive TCMR score.
Revisiting Mixed Rejection
Because molecular assessment often disagreed with conventional assessment of mixed rejection, we studied these biopsies in more detail (Table 6). Discrepancies were particularly common in the late biopsies: of 14 biopsies beyond 5 years diagnosed conventionally as mixed, all had positive ABMR scores, but 12 biopsies had negative TCMR scores. In 7 of 12 discrepancies, conventional assessment of TCMR activity was on the basis of v lesions, which can occur in pure ABMR.12,13 The others were diagnosed on the basis of tubulitis, which occurs in many progressive kidney diseases and therefore, may occur in pure ABMR.14,15 Thus, most conventionally diagnosed mixed rejection in the late period may be incorrect because of misclassification of pure ABMR caused by the occurrence of TCMR-like lesions in ABMR.
Table 6.
Molecular findings in 28 biopsies with a conventional diagnosis of mixed rejection
| Discrepancies | 0–7 d | 8 d to 6 mo (8–180 d) | 6–12 mo (181–365 d) | 1–5 yr (366–1825 d) | 5–10 yr (1826–3650 d) | >10 yr (>3650 d) |
|---|---|---|---|---|---|---|
| Number in interval | 0 | 2 | 1 | 11 | 11a | 3a |
| Number of biopsies of kidneys that progressed to failure | 0 | 0 | 1 | 5 | 6 | 0 |
| Classification by molecular scores | ||||||
| Pure TCMR (ABMR score negative TCMR score positive) | 0 | 0 | 1 | 3 | 0 | 0 |
| Pure ABMR (ABMR score positive; TCMR score negative) | 0 | 1 | 0 | 3 | 10 | 2 |
| Mixed (ABMR and TCMR scores positive) | 0 | 0 | 0 | 4 | 1 | 1 |
| No rejection (ABMR and TCMR scores negative) | 0 | 1 | 0 | 1 | 0 | 0 |
As discussed in the text, 5 of 11 mixed biopsies from 5 to 10 years and 2 of 3 mixed biopsies in >10 years with negative TCMR scores were diagnosed conventionally as having TCMR activity only on the basis of isolated v lesions, which can occur in pure ABMR. The others had tubulitis, which can occur in various progressive renal diseases.
Discussion
The emergence of a validated molecular system for biopsy assessment provided a unique opportunity to re-examine the disease states in renal transplants in two dimensions. We studied the incidence, timing, and consequences of rejection in a sample of disturbed kidneys biopsied 3 days to 35 years post-transplant in the current era of immunosuppression since 2004 and in experienced centers.27 The molecular tests strongly agreed with conventional assessment (accuracy of 91% for pure TCMR and 83% for pure ABMR), and the pattern of rejection over time was similar by conventional and molecular assessments. Pure ABMR was rare before 1 year but then became common—about 23%–28% of biopsies—with new cases presenting even 30 years post-transplant. TCMR activity was common in the early and intermediate periods but became rare after 5 years and disappeared after 10 years. Some conventional diagnoses of TCMR activity after 5 years were questionable, because they were on the basis of ambiguous lesions that can occur in pure ABMR. Graft loss was increased after biopsy diagnosis of pure ABMR but not pure TCMR, and mixed rejection behaved like pure ABMR. The ABMR score quantitatively predicted risk of failure, and the addition of the ABMR score to conventional assessment improved the prediction of graft loss. We conclude that combining molecular with conventional assessment improves understanding of troubled kidney transplants, confirming many conventional diagnoses, clarifying borderline biopsies, highlighting potential errors, and improving prediction of failure. The unexpected finding that TCMR activity was virtually unknown after 10.2 years, despite continued emergence of new ABMR cases, indicates that all patients with renal transplant eventually develop an adaptive state that precludes TCMR while permitting ABMR.
The rarity of pure TCMR after 5 years and its virtual disappearance after 10 years in this collection of unselected indication biopsies occurs despite the high probability that some patients were nonadherent. Medical record reviews in a subset of these patients found that many were suspected of nonadherence by their clinicians, including some presenting late; however, TCMR was absent after 10 years.3 Thus, pure ABMR may be a common presentation of nonadherence in late kidney transplants. Nonadherence is difficult to assess, because it is often denied by the patients, but it will be an important target for research, because it could account for a significant amount of late rejection and graft loss.
The absence of confirmed TCMR activity in late kidney transplants is reminiscent of the experience with immunosuppressive drug withdrawal in liver transplantation. Many late recipients of liver transplants can be withdrawn from immunosuppression without TCMR or ABMR, whereas late recipients of kidney transplants cannot be withdrawn safely, with the exception of very few recipients, some of whom have unusual B-cell changes.28–30 The strongest factor determining success of withdrawal of immunosuppression in stable recipients of liver transplants is time post-transplant beyond 10.6 years, similar to the time of the last TCMR activity of 10.2 years in this study. The mechanism of the T-cell adaption in recipients of liver transplants is unknown: changes in natural killer cells31 and iron homeostasis genes32 have been reported, but regulatory B or T cells have not been incriminated,33,34 and the B-cell changes seen in the rare tolerant kidney recipients have not been observed.
Our hypothesis is that all long-term organ transplant recipients on immunosuppression undergo partial adaptive T-cell tolerance that depletes their ability to generate TCMR against the graft, while preserving their capacity to generate anti-HLA antibodies and ABMR. Liver transplants differ from kidney transplants (and probably, other organ transplants) in being relatively resistant to ABMR, perhaps because of their large mass, regenerative ability, and unique microcirculation. However, some investigators contend that liver transplants are not as resistant to ABMR as previously believed, because ABMR phenotypes are being missed.35 Disappearance of late TCMR, despite continuing high risk of ABMR, may reflect exhaustion of the precursors of effector T cells needed for TCMR, despite persistence of the follicular T-helper cells needed for antibody responses. The rare patients with kidney transplants who remain stable without immunosuppression must have the universal TCMR adaptive change plus a rare second defect preventing antidonor antibody responses, which are compatible with the B-cell abnormalities reported in these patients.
Potential limitations of this study include the focus on prevalent rather than de novo transplants, the lack of protocol biopsies of stable patients, and the reliance on local histology assessment without central review. We elected to focus on prevalent transplants, because it is not feasible to acquire biopsies from kidneys two or three decades post-transplant in a study that begins at the time of transplant, just as the study that discovered the importance of the 10.6-year time point for liver tolerance used prevalent patients. We did not include protocol biopsies, because exposing large numbers of stable patients to regular protocol biopsies for decades is difficult to justify and not the standard of care. Our choice of local diagnosis in experienced centers with excellent diagnostic support as the standard for conventional assessment was on the basis of our experience with central histology reading, which is not the standard of care, offers no mechanism for resolving disagreement with the local centers, and is often incomplete, because local centers are reluctant to share some specimens.9,10 In our view, the local interpretation must be the primary comparator, because it determines management, which is why local diagnosis of rejection has always been the end point for trials of immunosuppressive drugs,36–38 despite inclusion of central histology readings in some studies. The possible role of late subclinical rejection (i.e., with no phenotype) cannot be assessed in an indication biopsy study.
Although analyses of large databases have often found that grafts diagnosed with rejection had reduced long-term survival compared with grafts with no recorded rejection,39,40 this biopsy-based analysis indicates that graft loss after TCMR is similar to that after biopsies with relatively normal findings. This does not indicate that TCMR is always benign but only that TCMR is well treated in experienced centers, consistent with recent experience in clinical trials of belatacept, where increased TCMR was not accompanied by increased late graft loss.41,42 The effect of rejection on registry analyses cannot be compared with this study, because registries do not require biopsies. Moreover, rejection recorded in registries can be a marker for conditions interrupting immunosuppression, such as infection or medical illness.
The availability of two independent phenotyping systems clarifies uncertainty, highlights potential errors, reveals unexpected complexity, and can serve to refine both systems by focusing on the discrepancies. Comparison of TCMR scores with histology found that many disagreements were in situations where conventional assessment has known deficiencies (e.g., biopsies with v lesions or heavy scarring).7,9 In ABMR, both systems seem to be necessary to detect the range of phenotypes: with ABMR score-negative biopsies, conventional positive biopsies may reflect a true ABMR state with low molecular activity, whereas with ABMR score-positive biopsies, conventional negative biopsies reflect ABMR currently missed by conventional criteria.8–10 The ABMR score also adds insight in transplant glomerulopathy, which may be either molecularly active ABMR or inactive. The unique strength of molecular assessments for predicting future failure will be particularly welcomed. We envision a new diagnostic classification that reflects the conventional and molecular features, is probabilistic, updatable, and consensus-seeking,25 and is developed through a consensus process among clinicians, pathologists, molecular researchers, and statisticians. The emergence of a new classification combining central molecular testing with local conventional assessment to triangulate on the true disease state (with no a priori assumption that either platform is correct) can potentially serve as a prototype for developing precision diagnostics for other diseases.43
Concise Methods
Biopsies were collected in two consecutive prospective multicenter studies of consented patients: 403 patients from the Genome Canada study and 300 patients from the International Collaborative Microarray (INTERCOM) trial (ClinicalTrials.gov NCT01299168).7–10 Patients were consented under protocols approved by local institutional review boards (participating centers are listed in Supplemental Table 1). To be enrolled, patients had to have biopsies suitable for histology reporting and microarray analysis. Every biopsy was included to represent all injury and disease states commonly affecting kidney transplants.8
Conventional Biopsy Assessment
Details have been published.7–10 Biopsy processing was performed as the standard of care for the center, and all centers agreed to follow the principles of the Banff system. Conventional biopsy interpretation for 403 biopsies in the Genome Canada study led to the discovery of C4d-negative ABMR, resulting in the new Alberta Transplant Applied Genomics Centre (ATAGC) Reference Standard classification (http://atagc.med.ualberta.ca/Documents/) that introduced changes now proposed for the Banff classification.12 INTERCOM biopsies were interpreted locally, and the pathology reports were forwarded to the ATAGC and classified by a study pathologist to reflect the findings by the local center in the context of the new classification.9,10
In reporting biopsies, the pathologists sometimes noted deficiencies that limited scoring. For TCMR lesions, 10 biopsies lacked scores for tubulitis, 10 biopsies lacked scores for infiltrate, and 30 biopsies lacked scores for endothelialitis (v lesions). For ABMR lesions, 10 biopsies lacked scores for glomerulitis, 23 biopsies lacked scores for peritubular capillaritis, and 12 biopsies lacked scores for glomerular double contours.
HLA antibody testing at biopsy was recommended in the study, but the decision to perform it and the method were determined by the local standard of care. Antibody specificities were usually determined by either LABScreen or FlowPRA single-antigen beads (One Lambda, Canoga Park, CA). No testing was reported for 77 patients, and donor-specific HLA antibody was not reported in 11 patients positive for HLA antibody.
Microarray Analyses
One extra biopsy core was collected for microarray analysis, placed immediately in RNALater (Ambion, Inc.), kept at 4°C for 4–24 hours, and stored at −20°C until transfer to the ATAGC; 703 biopsies were adequate for microarray assessment, and 59 biopsies were discarded because of improper sample collection.
RNA extraction, labeling, and processing of HG_U133_Plus_2.0 GeneChip (Affymetrix, Santa Clara, CA) were described previously.7–10,44 Microarray files were analyzed using the R software package, version 3.0.2 (64 bit) with various libraries from Bioconductor 2.13. Microarray expression files are posted on the Gene Expression Omnibus website (accession nos. GSE36059 and GSE48581). Microarrays from 703 indication biopsies and 8 nephrectomies were normalized with Robust Multiarray Averaging. Because 403 biopsy-set RNA were labeled with the 3′ IVT One-Cycle Target Labeling and Control Kit and the INTERCOM biopsy-set RNA were labeled with the 3′ IVT Express Kit, we corrected for batch effects using the Ratio-G method.45 Interquartile range filtering was used to eliminate probe sets with low variation.46
As detailed previously,7–10 the molecular classifiers were built by linear discriminant analysis implemented in the classification function of the CMA library of R using 10 group crossvalidation. The 703 samples were split randomly into 10 approximately equally sized groups, with one group left out as a test set and nine groups used to train the classifier.7 This procedure was applied to all 10 groups. This process was performed 50 times. The median of 50 scores for each biopsy was the final score (between zero and one). The TCMR score classifier was generated by comparing conventional TCMR biopsies with everything else, and the ABMR score classifier was generated by comparing conventional ABMR biopsies (C4d-positive or -negative) with everything else.
Statistical Analyses
Chi-squared, Fisher exact, and nonparametric t tests were used to compare variables as indicated in the tables. For Kaplan–Meier survival analysis, we selected one random biopsy per kidney when more than one biopsy was available. For the Net Reclassification Index, the R package survIDIRI was used.
Disclosures
P.F.H. has shares in Transcriptome Sciences Inc., a University of Alberta spinoff company to develop molecular diagnostics.
Supplementary Material
Acknowledgments
This research was supported by funding and/or resources from the Roche Organ Transplant Research Foundation, Novartis Pharma AG, the Canada Foundation for Innovation, Genome Canada, the University of Alberta Hospital Foundation, Roche Molecular Systems, Hoffmann-La Roche Canada Ltd., the Alberta Ministry of Advanced Education and Technology, and Astellas. P.F.H. held a Canada Research Chair in Transplant Immunology until 2008 and currently holds the Muttart Chair in Clinical Immunology.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Begin at the Beginning to Prevent the End,” on pages 1483–1485.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014060588/-/DCSupplemental.
References
- 1.Collins AJ, Foley RN, Chavers B, Gilbertson D, Herzog C, Ishani A, Johansen K, Kasiske BL, Kutner N, Liu J, St. Peter W, Guo H, Hu Y, Kats A, Li S, Li S, Maloney J, Roberts T, Skeans M, Snyder J, Solid C, Thompson B, Weinhandt E, Xiong H, Yusuf A, Zaun D, Arko C, Chen SC, Daniels F, Ebben J, Frazier E, Johnson R, Sheets D, Wang X, Forrest B, Berrini D, Constantini E, Everson S, Eggers P, Agodoa L: US Renal Data System 2013 Annual Data Report. Am J Kidney Dis 63[1 Suppl]: A7, 2014 [DOI] [PubMed] [Google Scholar]
- 2.El-Zoghby ZM, Stegall MD, Lager DJ, Kremers WK, Amer H, Gloor JM, Cosio FG: Identifying specific causes of kidney allograft loss. Am J Transplant 9: 527–535, 2009 [DOI] [PubMed] [Google Scholar]
- 3.Sellarés J, de Freitas DG, Mengel M, Reeve J, Einecke G, Sis B, Hidalgo LG, Famulski K, Matas A, Halloran PF: Understanding the causes of kidney transplant failure: The dominant role of antibody-mediated rejection and nonadherence. Am J Transplant 12: 388–399, 2012 [DOI] [PubMed] [Google Scholar]
- 4.Einecke G, Sis B, Reeve J, Mengel M, Campbell PM, Hidalgo LG, Kaplan B, Halloran PF: Antibody-mediated microcirculation injury is the major cause of late kidney transplant failure. Am J Transplant 9: 2520–2531, 2009 [DOI] [PubMed] [Google Scholar]
- 5.Gaston RS, Cecka JM, Kasiske BL, Fieberg AM, Leduc R, Cosio FC, Gourishankar S, Grande J, Halloran P, Hunsicker L, Mannon R, Rush D, Matas AJ: Evidence for antibody-mediated injury as a major determinant of late kidney allograft failure. Transplantation 90: 68–74, 2010 [DOI] [PubMed] [Google Scholar]
- 6.Furness PN, Taub N, Assmann KJ, Banfi G, Cosyns JP, Dorman AM, Hill CM, Kapper SK, Waldherr R, Laurinavicius A, Marcussen N, Martins AP, Nogueira M, Regele H, Seron D, Carrera M, Sund S, Taskinen EI, Paavonen T, Tihomirova T, Rosenthal R: International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am J Surg Pathol 27: 805–810, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Reeve J, Sellarés J, Mengel M, Sis B, Skene A, Hidalgo L, de Freitas DG, Famulski KS, Halloran PF: Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. Am J Transplant 13: 645–655, 2013 [DOI] [PubMed] [Google Scholar]
- 8.Sellarés J, Reeve J, Loupy A, Mengel M, Sis B, Skene A, de Freitas DG, Kreepala C, Hidalgo LG, Famulski KS, Halloran PF: Molecular diagnosis of antibody-mediated rejection in human kidney transplants. Am J Transplant 13: 971–983, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Halloran PF, Pereira AB, Chang J, Matas A, Picton M, De Freitas D, Bromberg J, Serón D, Sellarés J, Einecke G, Reeve J: Potential impact of microarray diagnosis of T cell-mediated rejection in kidney transplants: The INTERCOM study. Am J Transplant 13: 2352–2363, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Halloran PF, Pereira AB, Chang J, Matas A, Picton M, De Freitas D, Bromberg J, Serón D, Sellarés J, Einecke G, Reeve J: Microarray diagnosis of antibody-mediated rejection in kidney transplant biopsies: An international prospective study (INTERCOM). Am J Transplant 13: 2865–2874, 2013 [DOI] [PubMed] [Google Scholar]
- 11.Furness PN, Taub N, Convergence of European Renal Transplant Pathology Assessment Procedures (CERTPAP) Project : International variation in the interpretation of renal transplant biopsies: Report of the CERTPAP Project. Kidney Int 60: 1998–2012, 2001 [DOI] [PubMed] [Google Scholar]
- 12.Haas M, Sis B, Racusen LC, Solez K, Glotz D, Colvin RB, Castro MCR, David DSR, David-Neto E, Bagnasco SM, Cendales LC, Cornell LD, Demetris AJ, Drachenberg CB, Farver CF, Farris AB, 3rd, Gibson IW, Kraus E, Liapis H, Loupy A, Nickeleit V, Randhawa P, Rodriguez ER, Rush D, Smith RN, Tan CD, Wallace WD, Mengel M, Banff meeting report writing committee : Banff 2013 meeting report: Inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am J Transplant 14: 272–283, 2014 [DOI] [PubMed] [Google Scholar]
- 13.Lefaucheur C, Loupy A, Vernerey D, Duong-Van-Huyen JP, Suberbielle C, Anglicheau D, Vérine J, Beuscart T, Nochy D, Bruneval P, Charron D, Delahousse M, Empana JP, Hill GS, Glotz D, Legendre C, Jouven X: Antibody-mediated vascular rejection of kidney allografts: A population-based study. Lancet 381: 313–319, 2013 [DOI] [PubMed] [Google Scholar]
- 14.Iványi B, Marcussen N, Olsen S: Tubulitis in primary vascular and glomerular renal disease. Pathol Res Pract 191: 1245–1257, 1995 [DOI] [PubMed] [Google Scholar]
- 15.Berden AE, Jones RB, Erasmus DD, Walsh M, Noël LH, Ferrario F, Waldherr R, Bruijn JA, Jayne DR, Bajema IM, European Vasculitis Society : Tubular lesions predict renal outcome in antineutrophil cytoplasmic antibody-associated glomerulonephritis after rituximab therapy. J Am Soc Nephrol 23: 313–321, 2012 [DOI] [PubMed] [Google Scholar]
- 16.Halloran PF, Wadgymar A, Ritchie S, Falk J, Solez K, Srinivasa NS: The significance of the anti-class I antibody response. I. Clinical and pathologic features of anti-class I-mediated rejection. Transplantation 49: 85–91, 1990 [DOI] [PubMed] [Google Scholar]
- 17.Sis B, Jhangri GS, Riopel J, Chang J, de Freitas DG, Hidalgo L, Mengel M, Matas A, Halloran PF: A new diagnostic algorithm for antibody-mediated microcirculation inflammation in kidney transplants. Am J Transplant 12: 1168–1179, 2012 [DOI] [PubMed] [Google Scholar]
- 18.Baid-Agrawal S, Farris AB, 3rd, Pascual M, Mauiyyedi S, Farrell ML, Tolkoff-Rubin N, Collins AB, Frei U, Colvin RB: Overlapping pathways to transplant glomerulopathy: Chronic humoral rejection, hepatitis C infection, and thrombotic microangiopathy. Kidney Int 80: 879–885, 2011 [DOI] [PubMed] [Google Scholar]
- 19.Feucht HE, Felber E, Gokel MJ, Hillebrand G, Nattermann U, Brockmeyer C, Held E, Riethmüller G, Land W, Albert E: Vascular deposition of complement-split products in kidney allografts with cell-mediated rejection. Clin Exp Immunol 86: 464–470, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solez K, Colvin RB, Racusen LC, Sis B, Halloran PF, Birk PE, Campbell PM, Cascalho M, Collins AB, Demetris AJ, Drachenberg CB, Gibson IW, Grimm PC, Haas M, Lerut E, Liapis H, Mannon RB, Marcus PB, Mengel M, Mihatsch MJ, Nankivell BJ, Nickeleit V, Papadimitriou JC, Platt JL, Randhawa P, Roberts I, Salinas-Madriga L, Salomon DR, Seron D, Sheaff M, Weening JJ: Banff ‘05 Meeting Report: Differential diagnosis of chronic allograft injury and elimination of chronic allograft nephropathy (‘CAN’). Am J Transplant 7: 518–526, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Halloran PF, Langone AJ, Helderman JH, Kaplan B: Assessing long-term nephron loss: Is it time to kick the CAN grading system? Am J Transplant 4: 1729–1730, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Kao KJ, Chang KM, Hsu HC, Huang AT: Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: Implications for treatment optimization. BMC Cancer 11: 143, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solin LJ, Gray R, Baehner FL, Butler SM, Hughes LL, Yoshizawa C, Cherbavaz DB, Shak S, Page DL, Sledge GW, Jr., Davidson NE, Ingle JN, Perez EA, Wood WC, Sparano JA, Badve S: A multigene expression assay to predict local recurrence risk for ductal carcinoma in situ of the breast. J Natl Cancer Inst 105: 701–710, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartmann S, Gerber B, Elling D, Heintze K, Reimer T: The 70-gene signature as prognostic factor for elderly women with hormone receptor-positive, HER2-negative breast cancer. Breast Care (Basel) 7: 19–24, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silver N: The Signal and the Noise: Why So Many Predictions Fail–but Some Don't, 1st Ed., New York, The Penguin Press, 2012 [Google Scholar]
- 26.Hidalgo LG, Campbell PM, Sis B, Einecke G, Mengel M, Chang J, Sellares J, Reeve J, Halloran PF: De novo donor-specific antibody at the time of kidney transplant biopsy associates with microvascular pathology and late graft failure. Am J Transplant 9: 2532–2541, 2009 [DOI] [PubMed] [Google Scholar]
- 27.Halloran PF: Immunosuppressive drugs for kidney transplantation. N Engl J Med 351: 2715–2729, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, Burlingham WJ, Marks WH, Sanz I, Lechler RI, Hernandez-Fuentes MP, Turka LA, Seyfert-Margolis VL, Immune Tolerance Network ST507 Study Group : Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest 120: 1836–1847, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sagoo P, Perucha E, Sawitzki B, Tomiuk S, Stephens DA, Miqueu P, Chapman S, Craciun L, Sergeant R, Brouard S, Rovis F, Jimenez E, Ballow A, Giral M, Rebollo-Mesa I, Le Moine A, Braudeau C, Hilton R, Gerstmayer B, Bourcier K, Sharif A, Krajewska M, Lord GM, Roberts I, Goldman M, Wood KJ, Newell K, Seyfert-Margolis V, Warrens AN, Janssen U, Volk HD, Soulillou JP, Hernandez-Fuentes MP, Lechler RI: Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. J Clin Invest 120: 1848–1861, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Londoño MC, Danger R, Giral M, Soulillou JP, Sánchez-Fueyo A, Brouard S: A need for biomarkers of operational tolerance in liver and kidney transplantation. Am J Transplant 12: 1370–1377, 2012 [DOI] [PubMed] [Google Scholar]
- 31.Martínez-Llordella M, Lozano JJ, Puig-Pey I, Orlando G, Tisone G, Lerut J, Benítez C, Pons JA, Parrilla P, Ramírez P, Bruguera M, Rimola A, Sánchez-Fueyo A: Using transcriptional profiling to develop a diagnostic test of operational tolerance in liver transplant recipients. J Clin Invest 118: 2845–2857, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bohne F, Martínez-Llordella M, Lozano JJ, Miquel R, Benítez C, Londoño MC, Manzia TM, Angelico R, Swinkels DW, Tjalsma H, López M, Abraldes JG, Bonaccorsi-Riani E, Jaeckel E, Taubert R, Pirenne J, Rimola A, Tisone G, Sánchez-Fueyo A: Intra-graft expression of genes involved in iron homeostasis predicts the development of operational tolerance in human liver transplantation. J Clin Invest 122: 368–382, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benítez C, Londoño MC, Miquel R, Manzia TM, Abraldes JG, Lozano JJ, Martínez-Llordella M, López M, Angelico R, Bohne F, Sese P, Daoud F, Larcier P, Roelen DL, Claas F, Whitehouse G, Lerut J, Pirenne J, Rimola A, Tisone G, Sánchez-Fueyo A: Prospective multicenter clinical trial of immunosuppressive drug withdrawal in stable adult liver transplant recipients. Hepatology 58: 1824–1835, 2013 [DOI] [PubMed] [Google Scholar]
- 34.Sanchez-Fueyo A: Tolerance profiles and immunosuppression. Liver Transpl 19[Suppl 2]: S44–S48, 2013 [DOI] [PubMed] [Google Scholar]
- 35.O’Leary JG, Kaneku H, Susskind BM, Jennings LW, Neri MA, Davis GL, Klintmalm GB, Terasaki PI: High mean fluorescence intensity donor-specific anti-HLA antibodies associated with chronic rejection Postliver transplant. Am J Transplant 11: 1868–1876, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ekberg H, Tedesco-Silva H, Demirbas A, Vítko S, Nashan B, Gürkan A, Margreiter R, Hugo C, Grinyó JM, Frei U, Vanrenterghem Y, Daloze P, Halloran PF, ELITE-Symphony Study : Reduced exposure to calcineurin inhibitors in renal transplantation. N Engl J Med 357: 2562–2575, 2007 [DOI] [PubMed] [Google Scholar]
- 37.Vincenti F, Larsen C, Durrbach A, Wekerle T, Nashan B, Blancho G, Lang P, Grinyo J, Halloran PF, Solez K, Hagerty D, Levy E, Zhou W, Natarajan K, Charpentier B, Belatacept Study Group : Costimulation blockade with belatacept in renal transplantation. N Engl J Med 353: 770–781, 2005 [DOI] [PubMed] [Google Scholar]
- 38.Vincenti F, Kirkman R, Light S, Bumgardner G, Pescovitz M, Halloran P, Neylan J, Wilkinson A, Ekberg H, Gaston R, Backman L, Burdick J, Daclizumab Triple Therapy Study Group : Interleukin-2-receptor blockade with daclizumab to prevent acute rejection in renal transplantation. N Engl J Med 338: 161–165, 1998 [DOI] [PubMed] [Google Scholar]
- 39.Matas AJ, Gillingham KJ, Payne WD, Najarian JS: The impact of an acute rejection episode on long-term renal allograft survival (t1/2). Transplantation 57: 857–859, 1994 [DOI] [PubMed] [Google Scholar]
- 40.Meier-Kriesche H-U, Ojo AO, Hanson JA, Cibrik DM, Punch JD, Leichtman AB, Kaplan B: Increased impact of acute rejection on chronic allograft failure in recent era. Transplantation 70: 1098–1100, 2000 [DOI] [PubMed] [Google Scholar]
- 41.Durrbach A, Pestana JM, Pearson T, Vincenti F, Garcia VD, Campistol J, Rial MC, Florman S, Block A, Di Russo G, Xing J, Garg P, Grinyó J: A phase III study of belatacept versus cyclosporine in kidney transplants from extended criteria donors (BENEFIT-EXT study). Am J Transplant 10: 547–557, 2010 [DOI] [PubMed] [Google Scholar]
- 42.Vincenti F, Charpentier B, Vanrenterghem Y, Rostaing L, Bresnahan B, Darji P, Massari P, Mondragon-Ramirez GA, Agarwal M, Di Russo G, Lin CS, Garg P, Larsen CP: A phase III study of belatacept-based immunosuppression regimens versus cyclosporine in renal transplant recipients (BENEFIT study). Am J Transplant 10: 535–546, 2010 [DOI] [PubMed] [Google Scholar]
- 43.Mirnezami R, Nicholson J, Darzi A: Preparing for precision medicine. N Engl J Med 366: 489–491, 2012 [DOI] [PubMed] [Google Scholar]
- 44.Mueller TF, Einecke G, Reeve J, Sis B, Mengel M, Jhangri GS, Bunnag S, Cruz J, Wishart D, Meng C, Broderick G, Kaplan B, Halloran PF: Microarray analysis of rejection in human kidney transplants using pathogenesis-based transcript sets. Am J Transplant 7: 2712–2722, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Luo J, Schumacher M, Scherer A, Sanoudou D, Megherbi D, Davison T, Shi T, Tong W, Shi L, Hong H, Zhao C, Elloumi F, Shi W, Thomas R, Lin S, Tillinghast G, Liu G, Zhou Y, Herman D, Li Y, Deng Y, Fang H, Bushel P, Woods M, Zhang J: A comparison of batch effect removal methods for enhancement of prediction performance using MAQC-II microarray gene expression data. Pharmacogenomics J 10: 278–291, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scholtens D, von Heydebreck A: Statistics for Biology and Health: Bioinformatics and Computational Biology Solutions Using R and Bioconductor., XII, edited by Gentleman R, Carey VJ, Huber W, Irizarray R, Dudoit S, New York, Springer, 2005, pp 229–248 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


